Abstract
The MRG gene family (also known as SNSR) belongs to the G-protein-coupled receptor (GPCR) superfamily, is expressed specifically in nociceptive neurons, and is implicated in the modulation of nociception. Here, we show that Ka/Ks (the ratio between nonsynonymous and synonymous substitution rates) displays distinct profiles along the coding regions of MRG, with peaks (Ka/Ks > 1) corresponding to extracellular domains, and valleys (Ka/Ks < 1) corresponding to transmembrane and cytoplasmic domains. The extracellular domains are also characterized by a significant excess of radical amino acid changes. Statistical analysis shows that positive selection is by far the most suitable model to account for the nucleotide substitution patterns in MRG. Together, these results demonstrate that the extracellular domains of the MRG receptor family, which presumably partake in ligand binding, have experienced strong positive selection. Such selection is likely directed at altering the sensitivity and/or selectivity of nociceptive neurons to aversive stimuli. Thus, our finding suggests pain perception as an aspect of the nervous system that may have experienced a surprising level of adaptive evolution.
The MRG gene family has been identified independently by two groups (Dong et al. 2001; Lembo et al. 2002). The first group isolated mouse and human members, and named them MRG for Mas-related genes (Dong et al. 2001). The second group identified rat and human members, and named them SNSR for sensory-neuron-specific G-protein-coupled receptors (Lembo et al. 2002). To avoid confusion in the two naming systems, we will adhere to the MRG naming convention. Intriguingly, this gene family is expressed exclusively in a highly specialized neuronal population—nociceptive sensory neurons of the dorsal root and trigeminal ganglia (Dong et al. 2001; Lembo et al. 2002). Additionally, different MRG genes are expressed in distinct subsets of nociceptive cells (Dong et al. 2001), which would establish a rich array of nociceptive neurons distinguishable by their MRG expression status (a situation analogous to the expression of individual olfactory receptors in distinct subsets of olfactory neurons). Members of the MRG receptor family can be potently activated by peptide ligands such as RF-amides, and the opioid peptides BAM22 and γ2-MSH (Dong et al. 2001; Han et al. 2002; Lembo et al. 2002). Based on the above observations, these receptors were assumed to be involved in modulating nociceptive sensitivity and/or selectivity via interactions with peptide ligands such as opioids (Dong et al. 2001; Han et al. 2002; Lembo et al. 2002).
The mouse and human MRG gene family have been classified into four major subfamilies, including MRGX in human, and MrgA, MrgB, and MrgC in mouse (please refer to the original reports for the phylogenetic relationship of all known MRG genes). Human MRGX and murine MrgA may have an orthologous relationship, whereas murine MrgB and MrgC appear to be specific to the mouse without clear human orthologs (Dong et al. 2001; Lembo et al. 2002). Among the four subfamilies, the murine MrgC consists mostly of pseudogenes with perhaps one exception (Han et al. 2002), whereas the other subfamilies contain multiple functional genes as well as pseudogenes.
In this study, we performed evolutionary analysis on the MRG gene family of both human and mouse. We show that this gene family displays clear signatures of adaptive evolution in the putative ligand-binding domains. Implications of such adaptive evolution on organismal biology, particularly that relating to pain perception, is discussed.
RESULTS
Evidence of Positive Selection
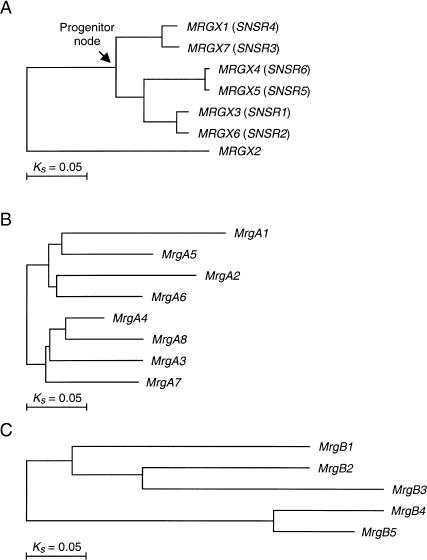
The phylogenies of the functional genes in MRGX, MrgA, and MrgB are shown in Figure 1. Pairwise comparisons of evolutionary distances were carried out on these genes. They showed that human MRGX genes are closely related to one another (average pairwise Ks is 0.15; Fig. 1A). The Ks between even the most distantly related MRGX genes is much lower than the average Ks between human and mouse orthologs estimated at 0.47 (Makolowski and Boguski 1998). Hence, the human MRGX subfamily likely arose from recent amplifications that postdated human-mouse divergence. In particular, MRGX4 and 5 show very little synonymous divergence (Ks = 0.006), indicating that they are likely the result of a duplication postdating human-chimp divergence (average Ks between human and chimp orthologs is around 0.015). High levels of relatedness were also observed among murine MrgA genes (average Ks = 0.20; Fig. 1B). In contrast, murine MrgB genes are more distantly related with one another (average Ks = 0.46; Fig. 1C), suggesting that they arose from much earlier amplifications, perhaps around the time of human-mouse divergence. We did not detect any evidence of gene conversion, such as conversion tract, between MRG paralogs (data not shown).
Figure 1.
Phylogenies of human MRGX (A), murine MrgA (B), and murine MrgB (C). Horizontal branch length is scaled to Ks.
We next calculated pairwise Ka/Ks for all of the human MRGX genes, and separately, murine MrgA and MrgB. Many pairwise comparisons yielded Ka/Ks values greater than 1 (data not shown). The average pairwise Ka/Ks is 1.10 for human MRGX, 0.92 for murine MrgA, and 0.70 for murine MrgB. These high Ka/Ks values, especially those of human MRGX, indicate that these genes have evolved very rapidly at the protein level, perhaps as a consequence of positive selection (Ka/Ks greater than unity is an indicator of positive selection; Li 1997). However, the pairwise Ka/Ks values are not significantly greater than 1. We therefore cannot rule out the possibility that these genes are pseudogenes (Ka/Ks should be around unity if genes are evolving free of constraint, as in the case of pseudogenes). To distinguish between these two possibilities, we reasoned that if these genes are indeed pseudogenes, nonsynonymous substitutions should be distributed more or less randomly across their coding regions. On the other hand, if they are functional and their high Ka/Ks values are due to positive selection, nonsynonymous substitutions might be significantly more concentrated within specific regions that are under positive selection, but relatively scarce in regions that are under constraint. These predictions were investigated by the sliding-window analysis of Ka/Ks between each pair of human MRGX, and separately, murine MrgA and MrgB. As shown in Supplementary Figure S1, this analysis revealed distinct peaks (Ka/Ks » 1) and valleys (Ka/Ks « 1), consistent with positive selection having operated on specific regions of these receptors (i.e., the peaks), and functional constraint being dominant in the other regions (the valleys). Thus, the sliding-window analysis revealed clear signs of positive selection, which were masked by purifying selection when gene-average Ka and Ks were considered.
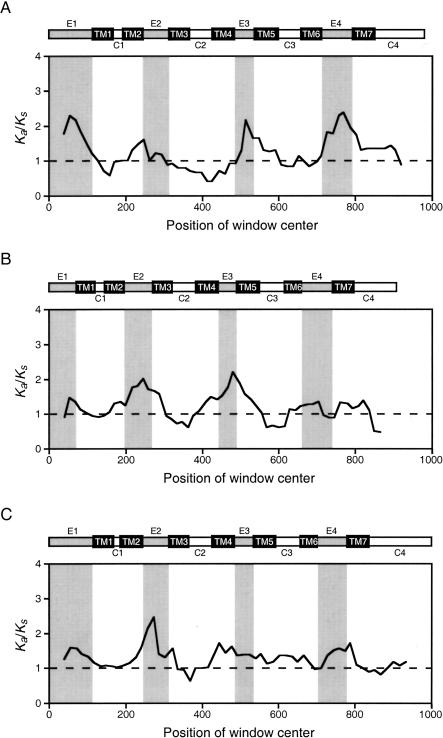
Importantly, as demonstrated by representatively pairwise comparisons in Figure 2A-C, most of the peaks correspond to extracellular domains known to partake in ligand binding, whereas the valleys lie within transmembrane and cytoplasmic domains that typically contribute to receptor anchoring and signal propagation (Bockaert and Pin 1999). An exception is the cytoplasmic domain at the C-terminus, which also has high Ka/Ks values in some comparisons. The exceedingly high Ka/Ks values found consistently around the extracellular domains of MRG provide strong evidence that the ligand-binding portions of these peptide receptors have experienced positive selection. As a control, we also conducted sliding-window analysis between MrgC pseudogenes. In contrast to functional genes, pseudogene comparisons produced Ka/Ks profiles that appear quite stochastic and bear very weak correlation to the domain structure (Fig. 2D). The weak correlation may be due to random noise, or the fact that these pseudogenes had once been functional genes under a similar selective regime. Comparable observations were made with other MRG pseudogenes (data not shown).
Figure 2.
Sliding-window analysis of Ka/Ks performed on representative functional members of human MRGX (A), murine MrgA (B), murine MrgB (C), and pseudogene members of murine MrgC (D). Protein domains are drawn schematically above each graph. Gray blocks in graphs correspond to extracellular domains. E1-E4: extracellular domains 1-4. TM1-TM7: transmembrane domains 1-7. C1-C4: cytoplasmic domains 1-4. Definitions of domain boundaries are as published (Dong et al. 2001; Lembo et al. 2002). Not all pairwise comparisons are shown due to the large number of possible pairs; data for all possible pairs are available in Supplemental Fig. S1.
A potential problem in the pairwise comparison is the fact that different comparisons are not always independent because they may share internal segments of the phylogenetic tree. This calls into question whether the Ka/Ks peaks and valleys found in the pairwise comparisons are representative of the entire gene family. To address this problem, we calculated Ka and Ks values for the entire phylogenetic tree of a subfamily instead of only between pairs of subfamily members (see Methods). We found that the resulting Ka/Ks profiles in the sliding window analysis are not qualitatively different from that in the pairwise comparisons (Fig. 3), indicating that the characteristic Ka/Ks peaks and valleys are a consistent feature in the evolution of these genes.
Figure 3.
Sliding-window analysis of Ka/Ks performed on MRGX (A), murine MrgA (B), and murine MrgB (C), where Ka and Ks were the sum divergence values across all segments of the corresponding phylogenetic tree.
We next performed more detailed analyses of human MRGX by first reconstructing the sequence at the progenitor node of this subfamily (indicated by arrow in Fig. 1A), and then comparing each gene to this deduced progenitor. This allowed us to examine whether positive selection has operated in similar manners on all of the MRGX genes since they parted from the progenitor. Sliding-window analysis between each of the MRGX genes and the progenitor yielded four relatively distinct profiles (Fig. 4). These four profiles are similar to one another in that their Ka/Ks peaks are almost always located in extracellular domains. However, they differ in the number and height of the peaks, suggesting that different MRGX genes may have experienced somewhat different selective regimes since they evolved away from the progenitor. It is also noticeable that the Ka/Ks between the progenitor and MRGX3 (or MRGX6) hovers around 1, and lacks domain-correlated peaks and valleys (Fig. 4). This raises the possibility that MRGX3 and 6 may be pseudogenes despite having intact open reading frames. It is also possible that these two genes are functional, but have experienced selective regimes quite different from the other members of MRGX.
Figure 4.
Sliding-window analysis of Ka/Ks performed between each human MRGX gene and the deduced sequence of the progenitor node (indicated in Fig. 1A). The four panels represent four distinct profiles in the seven comparisons. Vertical arrows at the top indicate positions of codons under positive selection as revealed by the Maximum Likelihood analysis (Yang 1998; these positions with respect to MRGX1 are 11, 12, 18, 32, 43, 78, 79, 82, 86, 91, 99, 108, 136, 153, 171, 208, 236, 239, 240, 244, 246, 247, 262, 298, 303, 305, 307, and 319).
Excess Amino Acid Replacements Over Neutral Expectation
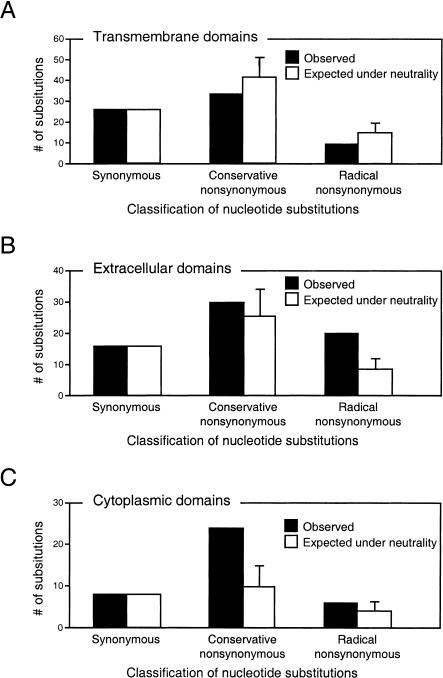
We wished to examine the statistical significance by which the number of amino acid replacements in individual protein domains deviates from neutrality. To this end, we performed simulations to obtain the number of amino acid replacements that would have occurred under complete neutrality; we then compared the simulated results with the observed amino acid changes. To better appreciate the biochemical nature of the amino acid replacements, we divided all of the replacements into two classes: those that are biochemically conservative and those that are radical (see Methods). When this analysis was performed on the MRGX genes, we found that the transmembrane domains have fewer observed amino acid replacements relative to neutral expectation, which is consistent with purifying selection (Fig. 5A). In contrast, the extracellular domains exhibit more replacements than neutral expectation (Fig. 5B). This excess is especially significant for radical replacements (P < 0.0001). The cytoplasmic domains also show more replacements than neutral expectation, with conservative changes showing the most significant excess (Fig. 5C). Interestingly, the extra amino acid replacements in cytoplasmic domains are contributed primarily by the C-terminal domain, where there is high Ka/Ks (16 of the 24 conservative changes and six of six radical changes observed in the cytoplasmic domains fall in the C-terminus). The above observations argue strongly that the extracellular domains and the last of the cytoplasmic domains of MRGX have evolved under positive selection. Particularly noteworthy is the fact that the extracellular domains—the sites where receptor-ligand interactions presumably take place—show an excess of radical amino acid replacements over neutrality. It is generally assumed that radical replacements are much more likely to alter protein function than conservative changes (Grantham 1974; Hughes et al. 2000; Dagan et al. 2002). Hence, positive selection operating on the extracellular domains was most likely directed at creating novel biochemical properties within these domains. The outcome is likely significant alterations in the affinity or specificity of the MRGX receptors for their ligands. A significant excess of radical amino acid replacements was also observed for murine MrgA genes (data not shown).
Figure 5.
Comparison of the observed number of amino acid replacements with that expected under neutrality (i.e., no selection) in human MRGX. The transmembrane, extracellular, and cytoplasmic domains are considered in A, B, and C, respectively. To obtain the number of amino acid replacements under neutral expectation, simulation was performed using sequence of the deduced MRGX progenitor (indicated in Fig. 1A) as the starting sequence (see Methods). Error bar represents one standard deviation of the simulated results.
Distribution of Putative Selected Sites
Another statistical test for positive selection is the Likelihood Ratio test, which has been applied to the detection of positive selection in a number of genes (Nielsen and Yang 1998; Yang 1998, 2000, 2001). In essence, it calculates the likelihood that a particular evolutionary model will produce the observed pattern of nucleotide substitutions. It then assesses the probability that two models should differ in log likelihood as much as that observed. The significance level of this P-value allows the identification of the model that shows the best fit for the data (see Methods for more detail). Here, we considered three basic models: (1) the one-ratio model, which assumes the same strength of selection at all codon sites (Ka/Ks = constant); (2) the neutral model, which assumes two types of codons, those conserved by functional constraint (Ka/Ks = 0), and those not subject to constraint (Ka/Ks = 1); and (3) the selection model, which assumes three types of codons, including the two in the neutral model, and a third type that is under positive selection (Ka/Ks > 1). We first calculated the likelihood of observing the substitution patterns in the MRGX genes under each of these three models (Table 1). We then obtained the P-value that any two models should differ in log likelihood as much as that observed, given the degree of freedom (Table 2). This analysis showed that the selection model fits the observed substitution patterns significantly better than either of the other two models that do not involve positive selection (P « 0.0001; see Table 1). We also analyzed the likelihood of additional models where some sites were allowed to evolve under varying degrees of partial constraint (1 > Ka/Ks > 0), with or without positively selected sites. They all led to the same conclusion, that is, models that incorporate positive selection are consistently and significantly better than models that exclude positive selection (data not shown).
Table 1.
Likelihood Estimates of Different Evolutionary Models
| Evolutionary model | # of parameters | Ω estimate (frequency of sites) a | Log likelihood |
|---|---|---|---|
| One-ratio model (MO) | 13 | ω = 0.93 (100%) | −2910.02 |
| Neutral model (M1) | 13 | ω1 = 0.00 (31%) | −2891.93 |
| ω2 = 1.00 (69%) | |||
| Selection model (M2) | 15 | ω1 = 0.00 (27%) | −2876.49 |
| ω2 = 1.00 (60%) | |||
| ω3 = 4.91 (13%) b |
ω stands for Ka/Ks.
See Fig. 4 for positions of positively selected sites.
Table 2.
Likelihood Ratio Between Different Evolutionary Models
| Evolutionary model | Degree of freedom | X2 | P |
|---|---|---|---|
| M0 vs. M2 | 2 | 67.06 | ≪0.0001 |
| M1 vs. M2 | 2 | 30.88 | ≪0.0001 |
Degree of freedom is the difference in the number of parameters between models.
χ2 is twice the difference of log likelihood between models.
P is the probability that two models should differ in log likelihood, as much as that observed, given the degree of freedom.
The Likelihood Ratio analysis also identified a set of specific codon sites in human MRGX that may have experienced positive selection (indicated by arrows in Fig. 4). Not surprisingly, these sites are concentrated in and around extracellular domains and the C-terminal cytoplasmic domain, where Ka/Ks often greatly exceeds 1. Of the 28 sites revealed by the analysis to have likely experienced positive selection, 16 fall within extracellular domains, even though extracellular domains represent only 27% of the total protein. This enrichment of selected sites in the extracellular domains relative to the rest of the proteins is statistically highly significant (P = 0.0007 by the two-tailed Fisher's exact test). The same analyses were performed on the murine Mrg genes, which similarly revealed the existence of positive selection in extracellular domains (data not shown). It is worth noting, however, that the Likelihood Ratio analysis can yield false positive results in the ascertainment of selected sites (Suzuki and Nei 2002). Therefore, the significance of this analysis lies less in the identification of specific selected sites, and more in the detection of general trends such as a strong presence of positive selection and the concentration of selected sites in the extracellular domains.
Comparison to Other Nociception-Related Genes
Members of the MRG receptors, including MRGX1, MRGX7, and MrgA1, can be potently activated by pain-modulating opioid peptides such as BAM22 (derived from proenkephalin) and γ2-MSH (derived from proopiomelanocortin; Han et al. 2002; Lembo et al. 2002). Based on this information, we speculated that strong positive selection might have also operated on other opioid receptors, and by extension, their peptide ligands. To examine this, we analyzed a number of well known opioid receptors such as δ, κ, μ, and the orphanin receptors (all of which belong to the GPCR superfamily), and the classical opioid peptide precursors including proenkephalin A, proopiomelanocortin, and prodynorphin (Dores et al. 1990; Mansour et al. 1995; Henderson and McKnight 1997). We also analyzed Tachykinin (precursor of substance P, a pain-modulating peptide) and its receptors. We did not uncover any evidence of positive selection in these opioid receptors or their ligands (data not shown). Thus, the strong signature of positive selection seen in MRG is not broadly shared by other opioid receptors.
DISCUSSION
The recently discovered MRG gene family encodes a large group of G-protein-coupled receptors in mammals. These receptors are implicated in the modulation of nociception by virtue of their specific expression in nociceptive sensory neurons and their responsiveness to opioid peptides (Dong et al. 2001; Lembo et al. 2002). In this study, we demonstrate that human and mouse members of the MRG family have experienced positive selection. In particular, we show that the most intensely selected sites of these receptors reside in or near the extracellular domains. It has been shown that the binding between G-protein-coupled receptors and their peptide ligands occur within the receptors' extracellular domains and their immediate vicinities in the superior parts of the transmembrane domains (Bockaert and Pin 1999). It is therefore reasonable to suppose that positive selection on MRG is directed at creating novel receptor-ligand interactions. It is possible that MRG has evolved rapidly in both copy number and protein sequence in response to the rapid diversification of their ligands. Alternatively, different MRG genes might be selected for varying affinities to the same set of ligands. Regardless of which scenario is correct, the rapid evolution of MRG in both copy number and protein sequence has likely resulted in altered nociceptive properties of the host organisms. Positive selection is also evident in the C-terminal cytoplasmic domain of some MRG members. The corresponding domain in other G-protein-coupled receptors has been implicated in signal propagation via binding to the PDZ-motif of downstream effector proteins (Hall et al. 1998). It is therefore likely that rapid evolution in this domain may alter the dynamics of signal propagation emanating from the MRG receptors.
Our study places the MRG family in a growing list of mammalian genes or gene families linked to positive selection (Hill and Hastie 1987; Stewart et al. 1987; Hughes and Nei 1988; Tanaka and Nei 1989; Yokoyama and Yokoyama 1989; Hughes and Hughes 1993; Shyue et al. 1995; Messier and Stewart 1997; Yang 1998; Gilad et al. 2000, 2002; Sharon et al. 2000; Johnson et al. 2001; Liberles et al. 2001; Tishkoff et al. 2001; Ding et al. 2002; Enard et al. 2002; Sabeti et al. 2002; Zhang et al. 2002; for review, see Wolfe and Li 2003). Among this list, MRG carries the distinction of being the first example of nociception-related genes. The perception of hazardous stimuli as being painful—and the subsequent avoidance of such stimuli—are critical to the survival of animals. When a species encounters evolutionary shifts in ecological conditions (such as habitat, climate, diet, predator-prey relationship, and social interactions), or its own internal physiology, previously innocuous stimuli may now impair fitness, and conversely, formerly aversive stimuli may become harmless or even beneficial. In the face of such evolutionary changes, a species would be under selective pressure to tune its nociceptive sensitivity and selectivity, so as to continue to correctly interpret those stimuli that endanger survival as being painful, while remaining undisturbed by innocuous stimuli (Kavaliers 1988). We hypothesize that such selective pressure has operated on MRG to drive its rampant amplification and fast protein evolution. Indeed, nociceptive properties do vary remarkably between species, among individuals of the same species, and between genders (Kavaliers 1988). Humans, for example, exhibit highly variable sensitivity to pain, including drastically different responses to identical injuries or pathologies (Libman 1934; Chen et al. 1989). Exceptionally heightened or reduced nociceptive sensitivity can have severe or even fatal consequences (Indo et al. 1996; Ophoff et al. 1996; Friedberg and Jason 2001). Similarly, closely related laboratory mice can differ by orders of magnitude in their pain threshold to noxious stimuli (Mogil et al. 1999). Such diversity in nociceptive response attests to the dynamic quality in the evolution of nociception (Kavaliers 1988). It will be of interest to see what role MRG plays in between- or within-species differences of nociception.
The highly restricted expression of MRG may also facilitate the rapid evolution of this gene family. As suggested previously (Hastings 1996; Duret and Mouchiroud 2000), genes with tissue-specific expression are likely to experience less evolutionary constraint relative to broadly expressed genes. The expression of MRG is restricted to nociceptive sensory neurons, which is a highly specialized cell population (indeed, each MRG gene appears to be expressed in only a subset of nociceptive neurons; Dong et al. 2001; Han et al. 2002; Lembo et al. 2002). In contrast, many other genes involved in the modulation of nociception, such as classical opioid receptors and their peptide ligands, are expressed broadly within and beyond the nervous system (Hollt et al. 1982; Pittius et al. 1984; Zhu and Pintar 1998; Zhu et al. 1998). This might explain why the MRG family, unlike many other genes implicated in pain modulation, has evolved exceedingly quickly under selection.
According to classical theories of gene family evolution, duplicated genes typically assume one of two evolutionary fates: the extra copy can decay due to redundancy, or the duplicated paralogs can diversify in functionality (Ohno 1970). Functional diversification may be accomplished by either the partitioning of the ancestral function among paralogs (subfunctionalization) or the acquisition of novel function (neofunctionalization; Hughes 1994; Sidow 1996; Force et al. 1999; Prince and Pickett 2002). Subfunctionalization is characterized by relaxation of selective constraint, whereas neofunctionalization requires positive selection. In either case, the outcome could be rapid protein evolution (i.e., high Ka/Ks) following gene duplication. It has indeed been noted that recently duplicated genes in various species tend to have high Ka/Ks values (Lynch and Conery 2000). However, there is considerable debate as to whether rapid protein evolution seen in recently duplicated genes is caused by relaxation of constraint or positive selection (Lynch and Conery 2000; Lynch and Force 2000; Kondrashov et al. 2002; Prince and Pickett 2002). The evolution of the MRG genes offers a salient example of gene family growth under the influence of positive selection. Along with other examples (Hill and Hastie 1987; Hughes et al. 2000; Bielawski and Yang 2001; Yang et al. 2002), it argues that neofunctionalization by positive Darwinian selection can factor prominently in the preservation and functional diversification of duplicated genes.
METHODS
Sequence Collection and Calculation of Divergence
Sequences were obtained from public databases with the following accession numbers: MRGX1 (SNSR4), AF474990; MRGX2, AY042214; MRGX3 (SNSR1), AF474987; MRGX4 (SNSR6), AF474992; MRGX5 (SNSR5), AF474991; MRGX6 (SNSR2), AF474988; MRGX7(SNSR3), AF474989; MrgA1, AY042191; MrgA2, AY042192; MrgA3, AY042193; MrgA4, AY042194; MrgA5, AY042195; MrgA6, AY042196; MrgA7, AY042197; MrgA8, AY042198; MrgB1, AY042199; MrgB2, AY042200; MrgB3, AY042201; MrgB4, AY042202; MrgB5, AY042203; Prodynorphin, NM_024411 (human), AF026537 (mouse), NM_019374 (rat); Proenkephalin A, P01210 (human), P22005 (mouse), P04904 (rat); Proopiomelanocortin, NM_000939 (human), AH005319 (mouse), AF510391 (rat); Tachykinin, P20366 (human), P41539 (mouse), P06767 (rat); TACR1, NM_001058 (human), NM_009313 (mouse); TACR2, NM_001057 (human), NM_009314 (mouse); TACR3 NM_001059 (human); δ-receptor, P41143 (human), P32300 (mouse), P33533 (rat); κ-receptor, P41145 (human), P33534 (mouse), P34975 (rat); μ-receptor, P35372 (human), AF286024 (Macaque), P42866 (mouse); Orphanin receptor, P41146 (human), P35377 (mouse), P35370 (rat). MRG pseudogene sequences were kindly provided by Zylka and colleagues (Dong et al. 2001). The Ka and Ks values were calculated by the Li method (Li 1993) using the Diverge program of the GCG package (Genetics Computing Group, Madison, WI), as well as the Maximum Likelihood method of Goldman and Yang (1994) using the PAML package (Yang 1997). These two methods produced essentially the same results. Data from GCG were presented. To obtain Ka and Ks values of an entire gene subfamily (rather than just in pairwise comparisons), Ka and Ks for individual segments of the subfamily tree were calculated separately and summed to yield values for the entire tree.
Construction of Phylogenetic Tree
Tree topology was obtained by the neighbor-joining method using the MEGA2 program (Mukhopadhyay et al. 1999), available at http://megasoftware.net. The sequence of each node was deduced by the Maximum Parsimony method using the Pamp program of the PAML package (Yang 1997).
Sliding-WindowAnalysis of Ka/Ks
Sliding-window analysis was performed with a window size of 90 bp and a sliding increment of 15 bp. The Ka/Ks of each window was calculated as the ratio between window-specific Ka and gene-average Ks. Although using window-specific Ks produced results that are qualitatively the same, noise in window-specific Ks can sometimes severely hamper the analysis (e.g., the window-specific Ks between closely related paralogs can occasionally drop to 0, which precludes the calculation of Ka/Ks). Because Ks is distributed quite randomly (data not shown), the use of gene-average instead of window-specific Ks should not introduce any systematic bias. For example, the average pairwise Ks among MRGX1, 3, 4, 5, 6, and 7 is 0.060 for the extracellular domains, which is similar to (and in fact slightly lower than) that for the transmembrane and cytoplasmic domains (0.078 and 0.068, respectively). In this case, the use of gene-average Ks in the sliding-window analysis is actually more conservative than window-specific Ks in detecting Ka/Ks peaks within the extracellular domains.
Simulation
The simulation of nucleotide substitutions was done as described (Wyckoff et al. 2000). Substitutions were randomly placed in the sequence using the observed synonymous transition-totransversion ratio. The exact nature of the substitution was recorded, and the simulation was reiterated until the number of synonymous changes reached the observed value. Data from a total of 10,000 replicas were then tallied, with nonsynonymous substitutions classified according to Grantham's amino acid replacement distance matrix (Grantham 1974). Following previous convention (Li et al. 1985; Wyckoff et al. 2000), a Grantham's distance of 100 or less was considered conservative, and otherwise radical.
Statistical Analysis
The fit of different evolutionary models to the observed patterns of nucleotide substitutions was assessed by the Likelihood Ratio analysis (Yang 1998) using the Codeml program in the PAML package (Yang 1997). For technical details of this test, please refer to Yang's original description (Yang 1998). Briefly, the program produced the number of parameters used in the analysis, Ka/Ks values (denoted as ω in PAML), and the log likelihood of each model. With these parameters, the probability that two models should differ in log likelihood as much as that observed, given the degree of freedom, was calculated using the Akaike Information Criterion as described (Posada and Crandall 1998; Yang 1998). If the model allowed positive selection, the program also indicated all of the sites that have likely experienced positive selection (Yang 1998).
Acknowledgments
We thank Steve Dorus, Christine Malcom, Eric Vallender, Gil-Joong Yoon, and Gerald Wyckoff for valuable technical input; Steve Dorus, Sandra Gilbert, Donghyun Park, and Eric Vallender for critical reading of the manuscript; Xinzhong Dong and David Anderson for MRG sequence data; and the Searle Scholarship and the Burroughs Wellcome Fund for partial financial support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1431603.
References
- Bielawski, J.P. and Yang, Z. 2001. Positive and negative selection in the DAZ gene family. Mol. Biol. Evol. 18: 523-529. [DOI] [PubMed] [Google Scholar]
- Bockaert, J. and Pin, J.P. 1999. Molecular tinkering of G protein-coupled receptors: An evolutionary success. EMBO J. 18: 1723-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, A.C., Dworkin, S.F., Haug, J., and Gehrig, J. 1989. Human pain responsivity in a tonic pain model: Psychological determinants. Pain 37: 143-160. [DOI] [PubMed] [Google Scholar]
- Dagan, T., Talmor, Y., and Graur, D. 2002. Ratios of radical to conservative amino acid replacement are affected by mutational and compositional factors and may not be indicative of positive Darwinian selection. Mol. Biol. Evol. 19: 1022-1025. [DOI] [PubMed] [Google Scholar]
- Ding, Y.C., Chi, H.C., Grady, D.L., Morishima, A., Kidd, J.R., Kidd, K.K., Flodman, P., Spence, M.A., Schuck, S., Swanson, J.M., et al. 2002. Evidence of positive selection acting at the human dopamine receptor D4 gene locus. Proc. Natl. Acad. Sci. 99: 309-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, X., Han, S., Zylka, M.J., Simon, M.I., and Anderson, D.J. 2001. A diverse family of GPCRs expressed in specific subsets of nociceptive sensory neurons. Cell 106: 619-632. [DOI] [PubMed] [Google Scholar]
- Dores, R.M., McDonald, L.K., Steveson, T.C., and Sei, C.A. 1990. The molecular evolution of neuropeptides: Prospects for the '90s. Brain Behav. Evol. 36: 80-99. [DOI] [PubMed] [Google Scholar]
- Duret, L. and Mouchiroud, D. 2000. Determinants of substitution rates in mammalian genes: Expression pattern affects selection intensity but not mutation rate. Mol. Biol. Evol. 17: 68-74. [DOI] [PubMed] [Google Scholar]
- Enard, W., Przeworski, M., Fisher, S.E., Lai, C.S., Wiebe, V., Kitano, T., Monaco, A.P., and Paabo, S. 2002. Molecular evolution of FOXP2, a gene involved in speech and language. Nature 418: 869-872. [DOI] [PubMed] [Google Scholar]
- Force, A., Lynch, M., Pickett, F.B., Amores, A., Yan, Y.L., and Postlethwait, J. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg, F. and Jason, L.A. 2001. Chronic fatigue syndrome and fibromyalgia: Clinical assessment and treatment. J. Clin. Psychol. 57: 433-455. [DOI] [PubMed] [Google Scholar]
- Gilad, Y., Segre, D., Skorecki, K., Nachman, M.W., Lancet, D., and Sharon, D. 2000. Dichotomy of single-nucleotide polymorphism haplotypes in olfactory receptor genes and pseudogenes. Nat. Genet. 26: 221-224. [DOI] [PubMed] [Google Scholar]
- Gilad, Y., Rosenberg, S., Przeworski, M., Lancet, D., and Skorecki, K. 2002. Evidence for positive selection and population structure at the human MAO-A gene. Proc. Natl. Acad. Sci. 99: 862-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman, N. and Yang, Z. 1994. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11: 725-736. [DOI] [PubMed] [Google Scholar]
- Grantham, R. 1974. Amino acid difference formula to help explain molecular evolution. Science 185: 862-864. [DOI] [PubMed] [Google Scholar]
- Hall, R.A., Premont, R.T., Chow, C.W., Blitzer, J.T., Pitcher, J.A., Claing, A., Stoffel, R.H., Barak, L.S., Shenolikar, S., Weinman, E.J., et al. 1998. The β2-adrenergic receptor interacts with the Na+/H+-exchanger regulatory factor to control Na+/H+ exchange. Nature 392: 626-630. [DOI] [PubMed] [Google Scholar]
- Han, S.K., Dong, X., Hwang, J.I., Zylka, M.J., Anderson, D.J., and Simon, M.I. 2002. Orphan G protein-coupled receptors MrgA1 and MrgC11 are distinctively activated by RF-amide-related peptides through the Gα q/11 pathway. Proc. Natl. Acad. Sci. 99: 14740-14745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings, K.E. 1996. Strong evolutionary conservation of broadly expressed protein isoforms in the troponin I gene family and other vertebrate gene families. J. Mol. Evol. 42: 631-640. [DOI] [PubMed] [Google Scholar]
- Henderson, G. and McKnight, A.T. 1997. The orphan opioid receptor and its endogenous ligand-nociceptin/orphanin FQ. Trends Pharmacol. Sci. 18: 293-300. [PubMed] [Google Scholar]
- Hill, R.E. and Hastie, N.D. 1987. Accelerated evolution in the reactive centre regions of serine protease inhibitors. Nature 326: 96-99. [DOI] [PubMed] [Google Scholar]
- Hollt, V., Haarmann, I., Grimm, C., Herz, A., Tulunay, F.C., and Loh, H.H. 1982. Proenkephalin intermediates in bovine brain and adrenal medulla: Characterization of immunoreactive peptides related to BAM-22P and peptide F. Life Sci. 31: 1883-1886. [DOI] [PubMed] [Google Scholar]
- Hughes, A.L. 1994. The evolution of functionally novel proteins after gene duplication. Proc. R Soc. Lond. B Biol. Sci. 256: 119-124. [DOI] [PubMed] [Google Scholar]
- Hughes, A.L. and Hughes, M.K. 1993. Adaptive evolution in the rat olfactory receptor gene family. J. Mol. Evol. 36: 249-254. [DOI] [PubMed] [Google Scholar]
- Hughes, A.L. and Nei, M. 1988. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335: 167-170. [DOI] [PubMed] [Google Scholar]
- Hughes, A.L., Green, J.A., Garbayo, J.M., and Roberts, R.M. 2000. Adaptive diversification within a large family of recently duplicated, placentally expressed genes. Proc. Natl. Acad. Sci. 97: 3319-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indo, Y., Tsuruta, M., Hayashida, Y., Karim, M.A., Ohta, K., Kawano, T., Mitsubuchi, H., Tonoki, H., Awaya, Y., and Matsuda, I. 1996. Mutations in the TRKA/NGF receptor gene in patients with congenital insensitivity to pain with anhidrosis. Nat. Genet. 13: 485-488. [DOI] [PubMed] [Google Scholar]
- Johnson, M.E., Viggiano, L., Bailey, J.A., Abdul-Rauf, M., Goodwin, G., Rocchi, M., and Eichler, E.E. 2001. Positive selection of a gene family during the emergence of humans and African apes. Nature 413: 514-519. [DOI] [PubMed] [Google Scholar]
- Kavaliers, M. 1988. Evolutionary and comparative aspects of nociception. Brain Res. Bull. 21: 923-931. [DOI] [PubMed] [Google Scholar]
- Kondrashov, F.A., Rogozin, I.B., Wolf, Y.I., and Koonin, E.V. 2002. Selection in the evolution of gene duplications. Genome Biol. 3: research0008.1-0008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lembo, P.M., Grazzini, E., Groblewski, T., O'Donnell, D., Roy, M.O., Zhang, J., Hoffert, C., Cao, J., Schmidt, R., Pelletier, M., et al. 2002. Proenkephalin A gene products activate a new family of sensory neuron-specific GPCRs. Nat. Neurosci. 5: 201-209. [DOI] [PubMed] [Google Scholar]
- Li, W.H. 1993. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J. Mol. Evol. 36: 96-99. [DOI] [PubMed] [Google Scholar]
- Li, W.H. 1997. Molecular evolution. Sinauer Associates, Sunderland, MA.
- Li, W.H., Wu, C.-I., and Luo, C.C. 1985. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol. Bio. Evo. 2: 150-174. [DOI] [PubMed] [Google Scholar]
- Liberles, D.A., Schreiber, D.R., Govindarajan, S., Chamberlin, S.G., and Benner, S.A. 2001. The adaptive evolution database (TAED). Genome Biol. 2: research0028.1-0028.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libman, E. 1934. Observations on individual sensitiveness to pain. J. Am. Med. Assoc. 102: 335-341. [Google Scholar]
- Lynch, M. and Conery, J.S. 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151-1155. [DOI] [PubMed] [Google Scholar]
- Lynch, M. and Force, A. 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154: 459-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makolowski, W. and Boguski, M.S. 1998. Evolutionary parameters of the transcribed mammalian genome: An analysis of 2,820 orthologous rodent and human sequences. Proc. Natl. Acad. Sci. 95: 9407-9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour, A., Hoversten, M.T., Taylor, L.P., Watson, S.J., and Akil, H. 1995. The cloned μ, δ, and κ receptors and their endogenous ligands: Evidence for two opioid peptide recognition cores. Brain Res. 700: 89-98. [DOI] [PubMed] [Google Scholar]
- Messier, W. and Stewart, C.B. 1997. Episodic adaptive evolution of primate lysozymes. Nature 385: 151-154. [DOI] [PubMed] [Google Scholar]
- Mogil, J.S., Wilson, S.G., Bon, K., Lee, S.E., Chung, K., Raber, P., Pieper, J.O., Hain, H.S., Belknap, J.K., Hubert, L., et al. 1999. Heritability of nociception I: Responses of 11 inbred mouse strains on 12 measures of nociception. Pain 80: 67-82. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay, N., Almasy, L., Schroeder, M., Mulvihill, W.P., and Weeks, D.E. 1999. Mega2, a data-handling program for facilitating genetic linkage and association analyses. Am. J. Hum. Genet. 65: A436. [DOI] [PubMed] [Google Scholar]
- Nielsen, R. and Yang, Z. 1998. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148: 929-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, S. 1970. Evolution by gene duplication. Springer-Verlag, Berlin.
- Ophoff, R.A., Terwindt, G.M., Vergouwe, M.N., van Eijk, R., Oefner, P.J., Hoffman, S.M., Lamerdin, J.E., Mohrenweiser, H.W., Bulman, D.E., Ferrari, M., et al. 1996. Familial hemiplegic migraine and episodic ataxia type-2 are caused by mutations in the Ca2+ channel gene CACNL1A4. Cell 87: 543-552. [DOI] [PubMed] [Google Scholar]
- Pittius, C.W., Seizinger, B.R., Pasi, A., Mehraein, P., and Herz, A. 1984. Distribution and characterization of opioid peptides derived from proenkephalin A in human and rat central nervous system. Brain Res. 304: 127-136. [DOI] [PubMed] [Google Scholar]
- Posada, D. and Crandall, K.A. 1998. MODELTEST: Testing the model of DNA substitution. Bioinformatics 14: 817-818. [DOI] [PubMed] [Google Scholar]
- Prince, V.E. and Pickett, F.B. 2002. Splitting pairs: The diverging fates of duplicated genes. Nat. Rev. Genet. 3: 827-837. [DOI] [PubMed] [Google Scholar]
- Sabeti, P.C., Reich, D.E., Higgins, J.M., Levine, H.Z., Richter, D.J., Schaffner, S.F., Gabriel, S.B., Platko, J.V., Patterson, N.J., McDonald, G.J., et al. 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature 419: 832-837. [DOI] [PubMed] [Google Scholar]
- Sharon, D., Gilad, Y., Glusman, G., Khen, M., Lancet, D., and Kalush, F. 2000. Identification and characterization of coding single-nucleotide polymorphisms within a human olfactory receptor gene cluster. Gene 260: 87-94. [DOI] [PubMed] [Google Scholar]
- Shyue, S.K., Hewett-Emmett, D., Sperling, H.G., Hunt, D.M., Bowmaker, J.K., Mollon, J.D., and Li, W.H. 1995. Adaptive evolution of color vision genes in higher primates. Science 269: 1265-1267. [DOI] [PubMed] [Google Scholar]
- Sidow, A. 1996. Gen(om)e duplications in the evolution of early vertebrates. Curr. Opin. Genet. Dev. 6: 715-722. [DOI] [PubMed] [Google Scholar]
- Stewart, C.B., Schilling, J.W., and Wilson, A.C. 1987. Adaptive evolution in the stomach lysozymes of foregut fermenters. Nature 330: 401-404. [DOI] [PubMed] [Google Scholar]
- Suzuki, Y. and Nei, M. 2002. Simulation study of the reliability and robustness of the statistical methods for detecting positive selection at single amino acid sites. Mol. Biol. Evol. 19: 1865-1869. [DOI] [PubMed] [Google Scholar]
- Tanaka, T. and Nei, M. 1989. Positive Darwinian selection observed at the variable-region genes of immunoglobulins. Mol. Biol. Evol. 6: 447-459. [DOI] [PubMed] [Google Scholar]
- Tishkoff, S.A., Varkonyi, R., Cahinhinan, N., Abbes, S., Argyropoulos, G., Destro-Bisol, G., Drousiotou, A., Dangerfield, B., Lefranc, G., Loiselet, J., et al. 2001. Haplotype diversity and linkage disequilibrium at human G6PD: Recent origin of alleles that confer malarial resistance. Science 293: 455-462. [DOI] [PubMed] [Google Scholar]
- Wolfe, K.H. and Li, W.H. 2003. Molecular evolution meets the genomics revolution. Nat. Genet. 33 Suppl: 255-265. [DOI] [PubMed] [Google Scholar]
- Wyckoff, G.J., Wang, W., and Wu, C.-I. 2000. Rapid evolution of male reproductive genes in the descent of man. Nature 403: 304-309. [DOI] [PubMed] [Google Scholar]
- Yang, J., Huang, J., Gu, H., Zhong, Y., and Yang, Z. 2002. Duplication and adaptive evolution of the chalcone synthase genes of Dendranthema (Asteraceae). Mol. Biol. Evol. 19: 1752-1759. [DOI] [PubMed] [Google Scholar]
- Yang, Z. 1997. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555-556. [DOI] [PubMed] [Google Scholar]
- Yang, Z. 1998. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 15: 568-573. [DOI] [PubMed] [Google Scholar]
- Yang, Z. 2000. Maximum likelihood estimation on large phylogenies and analysis of adaptive evolution in human influenza virus A. J. Mol. Evol. 51: 423-432. [DOI] [PubMed] [Google Scholar]
- Yang, Z. 2001. Maximum likelihood analysis of adaptive evolution in HIV-1 gp120 envgene. Pac. Symp. Biocomput. 226-237. [PubMed]
- Yokoyama, S. and Yokoyama, R. 1989. Molecular evolution of human visual pigment genes. Mol. Biol. Evol. 6: 186-197. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Zhang, Y.P., and Rosenberg, H.F. 2002. Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat. Genet. 30: 411-415. [DOI] [PubMed] [Google Scholar]
- Zhu, Y. and Pintar, J.E. 1998. Expression of opioid receptors and ligands in pregnant mouse uterus and placenta. Biol. Reprod. 59: 925-932. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., Hsu, M.S., and Pintar, J.E. 1998. Developmental expression of the μ, κ, and δ opioid receptor mRNAs in mouse. J. Neurosci. 18: 2538-2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://megasoftware.net; The Web site for the MEGA (Molecular Evolutionary Genetic Analysis) program and related information.