Abstract
We propose herein a new method of DNA distribution, whereby DNA clones or PCR products are printed directly onto the pages of books and delivered to users along with relevant scientific information. DNA sheets, comprising water-soluble paper onto which DNA is spotted, can be bound into books. Readers can easily extract the DNA fragments from DNA sheets and amplify them using PCR. We show that DNA sheets can withstand various conditions that may be experienced during bookbinding and delivery, such as high temperatures and humidity. Almost all genes (95%–100% of randomly selected RIKEN mouse cDNA clones) were recovered successfully by use of PCR. Readers can start their experiments after a 2-h PCR amplification without waiting for the delivery of DNA clones. The DNA Book thus provides a novel method for delivering DNA in a timely and cost-effective manner. A sample DNA sheet (carrying RIKEN mouse cDNA clones encoding genes of enzymes for the TCA cycle) is included in this issue for field-testing. We would greatly appreciate it if readers could attempt to extract DNA and report the results and whether the DNA sheet was shipped to readers in good condition.
Developing genome technologies allows the continued elucidation of the genomes of many species, including human and mouse (International Human Genome Sequencing Consortium 2001; Venter et al. 2001; Mouse Genome Sequencing Consortium 2002). However, genome sequences are not sufficient to allow a true understanding of the final mRNA, protein, or even regulatory elements encoded in the genomic sequence. For example, information on mRNA structures, alternative splicings, anti-sense RNA, noncoding RNA, and promoters are essential for the analysis of gene function. To reveal these elusive targets, collection and analysis of cDNA must be undertaken. Furthermore, functional genomics for post-genome sequence research requires exact physical clones of cDNA for protein synthesis and cDNA microarrays.
RIKEN has been constructing the Mouse Genome Encyclopedia to establish an infrastructure for functional genomics. The Mouse Genome Encyclopedia is a comprehensive collection of the mouse transcriptome comprising (1) a full set of mouse full-length cDNA clones, (2) nucleotide sequences of clones, and (3) functional annotation of clones. To achieve this goal, RIKEN has developed a series of full-length cDNA technologies (Carninci and Hayashizaki 1999; Carninci et al. 2000, 2001), high-throughput sequencing pipeline (Shibata et al. 2000), and created a large database with a highly functional annotation by organizing two international annotation meetings, FANTOM (RIKEN Genome Exploration Research Group Phase II Team and FANTOM Consortium 2001; The FANTOM Consortium and The RIKEN Genome Exploration Research Group Phase I and II Team 2002). Through these activities, the RIKEN Mouse Genome Encyclopedia is growing rapidly day by day, and we are now facing the next problem, resource distribution.
Currently, the Internet allows wide and simple transfer and distribution of scientific information, including nucleotide sequences and gene annotation, as well as scientific papers. The RIKEN FANTOM database of sequences and annotation has been released and is accessible through the Internet (http://fantom2.gsc.riken.go.jp/). Leading to this, a different situation exists regarding clone distribution. In addition to scientific information, scientific journals have always either requested or strongly recommended authors to make their DNA clone resources available to the scientific community for promotion of further research. At present, authors generally voluntarily provide genes in response to such requests by depositing them into public gene banks, or entrusting commercial companies for the distribution. DNA clones are typically shipped as transformed Escherichia coli or DNA in tubes or titer plates. However, these methods are barely capable of handling the huge amount of clone resources that are now being produced. Actually, difficulties arise in meeting the numerous clone requests from around the world and distributing resources. The handling, preparation, packaging, and shipment of countless clones require substantial amounts of time, effort, and money. Thus, researchers will still have to wait for the delivery of the clones they have requested before starting experiments.
In addition, organizations that constitutively distribute DNA clone resources from genome and transcriptome science must demonstrate the following characteristics: (1) establishment of an economical basis for autonomous activity; (2) ability to handle legal issues such as intellectual property rights; and (3) technologies for resource distribution. However, as mentioned above, traditional protocols for clone distribution do not fulfill the recent requirements for DNA clone distribution produced by transcriptome analysis.
To improve this situation, we propose a novel concept of DNA distribution—the DNA Book. If the sheets on which DNA clones or PCR products are printed are attached to the pages of books and journals, DNA could be delivered to users along with the relevant scientific articles and information. Researchers could easily extract and use this DNA for their own experiments. The DNA Book offers the following advantages: (1) simultaneous delivery of scientific information and associated DNA clones or PCR products themselves; (2) easy extraction of DNA using PCR; (3) a cheap method of DNA distribution; and (4) stable preservation in the laboratory environment.
The present work describes the feasibility of DNA sheets and the DNA Book according to the results of various tests. In addition, we have bound one DNA sheet into this issue as a sample. The final aim of this study is to conduct a field test of the DNA Book. We are eager to learn whether the DNA attached to the page can be amplified successfully when the DNA Book is received by readers worldwide. We would very much appreciate if readers could try to extract the DNA and report the results to our Web page, http://genome.gsc.riken.go.jp/DNA-Book/.
RESULTS
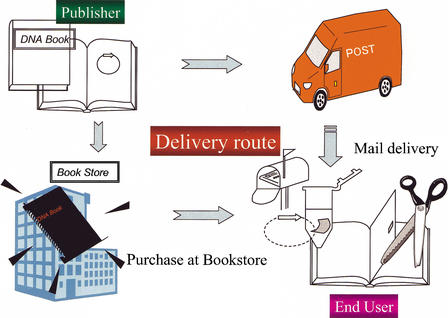
Figure 1 shows the concept behind the DNA Book. Papers are printed as usual, with letters and figures describing genetic research in the conventional way. DNA solutions are then spotted at defined positions on the same sheet of paper. The DNA sheets are bound into the book or journal and delivered to readers through bookstores or by courier as printed matter. Researchers, students, and readers who are interested in the enclosed genes can extract, amplify, and immediately apply DNA from the sheets to their research. To produce the DNA Book, several issues must be considered. (1) DNA should be extractable by readers easily, with a high success rate; (2) DNA on the sheets should be able to be preserved in a stable form during bookbinding and shipping; and (3) costs should not be expensive. Either the cDNA clones or PCR fragments of cDNA could be used for DNA printing onto sheets. In the Results section, we show the data based on the use of full-length cDNAs for DNA printing. In practical use, PCR fragments are also recommended as suitable materials to be printed.
Figure 1.
Concept of the “DNA Book.” DNA of genes are attached to pages of the DNA Book, and delivered to readers through conventional distribution channels, such as couriers and bookstores. Readers can extract and amplify DNA from the pages.
Producing DNA Sheets and DNA Extraction
To achieve the goals of the DNA Book technology, we decided to use water-soluble paper to form the DNA sheet. We selected 60MDP paper (Mishima Paper Co., Ltd.), which dissolves in water at room temperature within 35 sec.
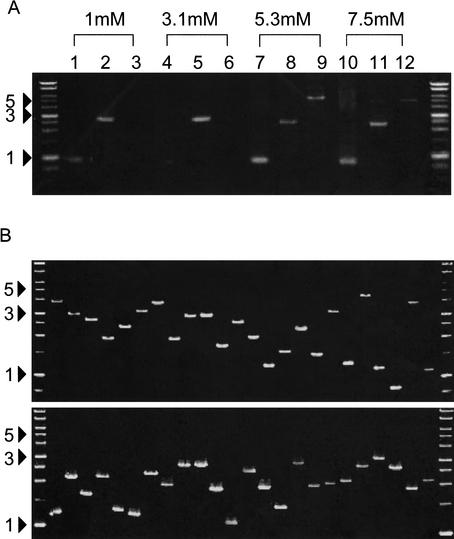
Plasmid DNA solutions were transferred onto 60MDP paper using a 96-pin tool (Multi 96-multiblot replicator), which was originally designed for the creation of high-density filters, and allowed us to spot defined amounts of DNA solution onto specific positions on the paper. Spotted positions can be identified easily by use of a red dye mixed in the DNA solution. We used 0.05 microG of DNA solution for each spot. After drying the paper in air for more than 30min, we extracted DNA from the sheet using the following method. The piece of 60MDP paper (4 mm × 4 mm) containing the DNA spot was placed into a 0.2-mL PCR tube. We added 50 microL of PCR solution containing all of the necessary reagents (primers, polymerase, dNTPs, etc.), and initiated the PCR reaction as described in the Methods. We tested three RIKEN cDNA clones with various insert sizes (722, 2418, and 5438 bp). The cDNA inserts were amplified successfully at higher Mg2+ concentrations of around 5.3 mM (Fig. 2A). This result is consistent with the fact that 60MDP binds cations (Y. Ishino and Y. Kimura, unpubl.). The amount of DNA spotted on the paper should be at least 5 ng. Conditions for spotting and extracting DNA were thus established. In addition, we tested several other PCR conditions. A relatively small volume of PCR solution (25 microL) demonstrated reduced DNA amplification. Increased amounts of polymerase had no effect. Addition of BSA and DMSO did not improve the PCR reaction. Several other kinds of papers also can be used for the DNA sheets with optimized conditions (data not shown).
Figure 2.
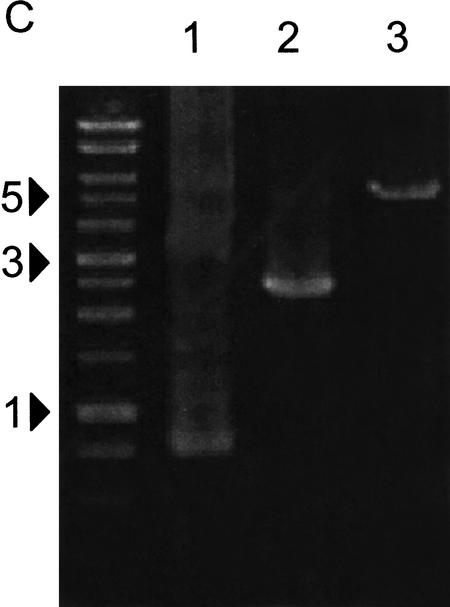
Extraction of DNA from DNA sheets by PCR. (A) PCR conditions for DNA extraction. Fifty nanograms of three RIKEN mouse cDNAs (722 bp, lanes 1,4,7,10; 2418 bp, lanes 2,5,8,11; 5438 bp, lanes 3,6,9,12) in 0.5 μL plasmid solution were spotted on water-soluble 60MDP paper (Mishima Co., Ltd.). After dryingin air for 30 min, insert DNA was amplified from paper pieces (4 mm × 4 mm) using PCR with vector-specific primer in the presence of various concentrations of Mg2+. DNA marker sizes are shown in kilobases. (B) Various DNA inserts can be recovered from DNA sheets. Random selections of 93 plasmids of RIKEN mouse cDNA clones, ranging in size from 732–4896 bp, were extracted from DNA sheets. In two independent experiments, almost all cDNA inserts (95% and 100% of 93 clones) were amplified successfully. Representative results for 24 samples are shown. Marker sizes are shown in kilobases. (C) DNA sheets can tolerate rubbingat 37°C in 70% humidity overnight. DNA sheets spotted with ∼50 ngof cDNA plasmids were kept in a humidified incubator or were inserted into an issue of Genome Research, and shaken overnight at 180 rpm under conditions of 37°C and 70% humidity. DNA inserts of cDNA were extracted from DNA sheets using PCR, as described in the Methods. Lanes 1, 2, and 3 represent 722, 2418, and 5438 bp cDNA inserts, respectively.
We also tested a higher density of DNA spots per unit area, as with a 384-well plate. Successful recovery of DNA by PCR was achieved without any contamination from neighboring DNAs.
Furthermore, as an alternative, an aliquot of solution containing the dissolved DNA sheet can undergo either PCR in a separate tube or E. coli transformation. Readers can keep any remaining solution as backup or for other experiments. Please see the Methods section for a protocol of E. coli transformation. Users do not need to undertake any purification of DNA after dissolution of the DNA sheet, and can apply aliquots directly to transformation. Transformation efficiency from the DNA sheet is equivalent to the control plasmid (data not shown). When PCR errors must be avoided for downstream experiments such as protein expression, DNA recovery by E. coli transformation is recommended.
Success Rate of DNA Extraction
Figure 2B shows the success rates for extracting various DNA from the DNA sheet. We randomly selected 93 RIKEN mouse cDNA clones of various insert sizes (732–4896 bp), spotted them on 60MDP sheets, and performed DNA extraction using PCR. Two independent experiments revealed that almost all cDNA inserts (95% and 100% of 93 clones) were successfully amplified. Representative results from 24 of the 93 clones are shown in Figure 2B. These results suggest that almost all cDNA clones can be applied to DNA sheets.
Preservation of DNA Sheets
DNA sheets and the DNA Book must be able to endure various conditions present throughout the bookbinding process at the publisher, shipment to readers, and storage by readers in ordinary rooms. Temperature, pressure, humidity, light, and physical rubbing are the major problems with the potential to qualitatively affect the DNA Book or DNA sheets. No problems were envisaged with the effects of usual temperature. DNA sheets treated at 140°C for several seconds, 100°C for 10 min, or -40°C for 14 h still allowed successful recovery of cDNA inserts using PCR. In addition, high-pressure conditions from 8.8 to 17 MPa had no effect on the recovery of DNA (data not shown). These results suggest that DNA sheets remain undamaged by the high temperatures and pressures that may be experienced during bookbinding, and by the low temperatures that may be experienced during air transport
Next, we observed successful recovery of DNA from DNA sheets kept overnight under conditions of 70%–100% humidity at 37°C. We tested whether rubbing stress on DNA sheets could cause problems, such as inhibition of PCR amplification, or cross-contamination with neighboring DNA. DNA sheets spotted with three kinds of cDNA plasmids were stored in a humidified incubator, or inserted into an issue of Genome Research and shaken strongly using a rotating shaker (180 rpm) at 37°C with 70% humidity for 14 h. DNA inserts were then extracted by PCR as described above. All DNA inserts (722, 2418, and 5438 bp) were recovered (Fig. 2C). No contamination of DNA spots with neighboring spots was observed. The 60MDP paper used here for the DNA sheet comprises two kinds of fibers, wood pulp and carboxyl-methyl cellulose (CMC). Water-soluble CMC allows 60MDP to dissolve in water and DNA might be incorporated and trapped by the CMC gel when spotted. In addition, CMC might be able to keep DNA bound tightly within the DNA sheet after drying, and thus prevent the release or transfer of spotted DNA.
Figure 3.
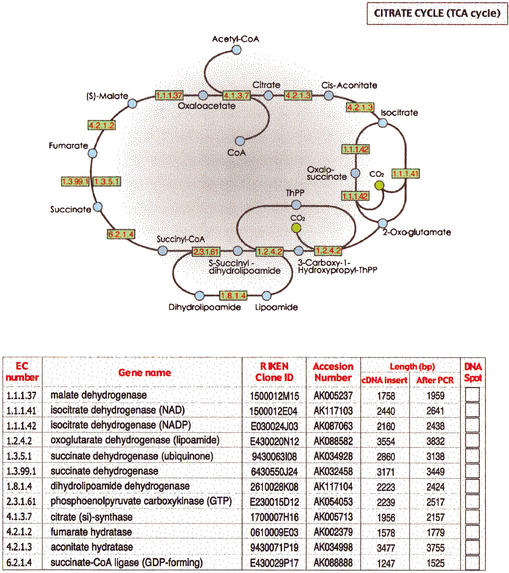
A DNA sheet is attached as a sample for field testing. Twelve cDNA plasmids of the RIKEN Mouse Genome Encyclopedia annotated as genes involved in TCA cycles (Bono et al. 2003) are attached in the right column of the annotation table. EC numbers and gene names are used according to the KEGG database (Kanehisa et al. 2002). The cDNAs were cloned into the pFLC vector (Carninci et al. 2001), which is identical to the pBluescript except for the modified cloningsites. A total of 100 ngof plasmids were spotted on water-soluble MDP60 paper (Mishima Co., Ltd.), and dried. Positions of attached plasmids are shown in red. TCA cycle and EC number of genes are shown at top. To amplify cDNA inserts, cut out the 2 mm × 2 mm area spotted with DNA, place it into a PCR tube, add 50 μL of PCR solution, centrifuge the resulting solution, and then initiate PCR cycle. PCR solution contains 1.5 U of KOD Plus DNA Polymerase (TOYOBO Co., Ltd.), and 1× KOD Plus PCR buffer together with 0.2 μM of PCR primers (-21M13; 5′-TGTAAAACGACGGCCAGT-3′, 1233-Rv; 5′-AGCGGATAACAATTTCACACAGGA-3′), 0.2 mM each of dATP, dGTP, dCTP and dTTP, and 5.3 mM MgCl2. After centrifuging the resulting solution, the PCR cycle is initiated. PCR cycles comprise 2 min at 94°C; 29 cycles of denaturing(94°C, 1 min), annealing(60°C, 1 min) and extension (68°C, 75 sec), and 15 min at 74°C. Alternatively, an aliquot of soluble-paper solution can be subjected to E. coli transformation. This sample DNA sheet is attached for large-scale field testing of the DNA Book, to test how well the DNA sheet is delivered to readers. We would gratefully appreciate readers attempts to extract genes printed on this sheet. Feedback can be input on our Web page, http://genome.gsc.riken.go.jp/DNA-Book/. Results from readers will be reported as a separate paper in the near future. Patent for DNA Book pending.
J. Kawai and Y. Hayashizaki
DNA Sheet Sample and the Field Test
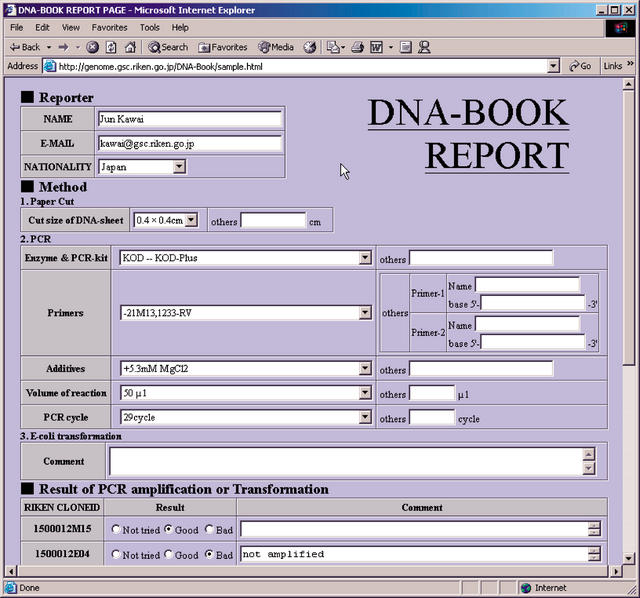
In practical use, DNA Books are likely to be piled up in bookstore warehouses, and are sometimes thrown around during the delivery process. In these processes, mechanical sliding stress at high pressure is quite likely to be exerted on the surface of the DNA sheet. Although we have tested the tolerance of the DNA sheet to temperature, high pressure, humidity, and rubbing stress, it is still not equivalent to the full range of situations and conditions DNA sheets will encounter in practical use. Because publication of this work offers the best opportunity to test such parameters, we ask for readers' contributions to examine the practical use of DNA sheets by testing DNA amplification and sending data to us on our Web site. An actual test sample for the DNA sheet is included in Figure 3. Twelve DNA samples of RIKEN mouse cDNA clones, annotated as genes of the TCA cycle, (The FANTOM Consortium and The RIKEN Genome Exploration Research Group Phase I and II 2002; Bono et al. 2003), have been attached in the right column of the annotation table of Figure 3 together with a description. We would greatly appreciate the efforts of readers in recovering DNA from the page and reporting the results on our Web page, http://www.gsc.riken.go.jp/DNA-Book/. The reporting page is shown in Figure 4. Conditions of readers' PCR experiments including polymerase, buffer compositions, volumes, primer sequences, additives, results, and any comments would be helpful in evaluating the DNA Book and suggesting future improvements. In the case of transformation experiments, similar information from readers' experiments would prove invaluable. See Methods and the legend to Figure 3 for the protocol to recover DNA. After summarizing readers' results, the data will be reported in the future.
Figure 4.
Reporting page at http://genome.gsc.riken.go.jp/DNA-Book/ for field-testing of the DNA Book. The authors appreciate readers' efforts to supply feedback of results for DNA recovery.
DISCUSSION
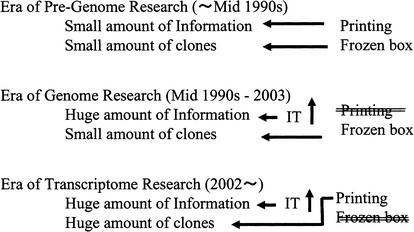
Figure 5 shows historical transitions in the role of printing media. In the pre-genome era of research for conventional molecular biology, printed materials, books, and journals, were the vehicles to convey information regarding genes and scientific research. Thus, printed matter can also carry sequence information (database). However, DNA clones were distributed as frozen transformants of E. coli and/or as DNAs. The amount of sequence information and number of DNA clones were small enough to be handled using such methods in the pre-genome research era. From the middle of the 1990s, development of high-throughput genome technology such as automated fluorescence sequencers allowed production of huge amounts of genome sequence information for various species. Genome sequence information started to be distributed worldwide via the Internet, allowing instantaneous dissemination of information. Transfer of genome sequence information in an electronic form through the Internet is extremely convenient for downstream research, as investigators had to search for and analyze large amounts of genomic information with the aid of the exponentially growing power of computers, following Moore's law. Furthermore, scientific publications have also started to be distributed over the Internet, utilizing the substantial advantages of rapid, worldwide dissemination and cost-effectiveness. These changes have rendered printed material less significant as a vehicle of information. Conversely, tubes and plates have remained in use as vehicles of distribution for DNA clones or PCR products, even after the advent of the genome era. Although the need for genomic DNA clones is growing, the number of users requesting such clones has so far stayed small enough for shipping by use of traditional methods to remain feasible. The reason is that genome sequences are much more frequently used than genomic clones, due to the design of current research in molecular biology.
Figure 5.
As we enter the era of transcriptome research, printing and books will play a novel role in the distribution of biomaterials.
However, the situation has started to undergo massive changes with the advent of the era of transcriptome research, wherein mRNA transcribed from genomes and cDNA synthesized in vitro represent the major targets of analysis examining gene function. Information obtained from cDNA analysis is essential to understanding the structures of transcriptomes and proteomes. The structure of mRNA, with starting points of transcription, splicing, and termination, and anti-sense RNA and protein noncoding RNA cannot yet be predicted exactly from genomic sequences using only bioinformatic tools such as exon prediction software. Furthermore, functional studies of genes such as comprehensive expression profiling with cDNA microarrays and determination of three-dimensional protein structures require physical cDNA. Systematic analysis of transcriptomes on the basis of full-length cDNA technology is producing a vast number of cDNA clones every day. Our group has actually developed full-length cDNAs and sequences for 60,770 mouse cDNA clones (FANTOM Consortium and The RIKEN Genome Exploration Research Group Phase I and II 2002, Arabidopsis thaliana cDNA clones from RIKEN (Seki et al. 2002), and rice cDNA clones from the National Institute of Agrobiological Sciences (The Rice Full-length cDNA Consortium, in prep.), and part of ∼15,000 human and mouse cDNA clones for the Mammalian Gene Collection (MGC) Program Team 2002. These are only the numbers of fully sequenced cDNA clones, with many more clones (100-fold more) isolated for the initial screening of clustering. These cDNA clone resources are being made public. The requirements for distributing so many cDNA clones in a timely and cost-effective manner have led to the need for a solution beyond the capabilities of traditional distribution methods using tubes and plates.
The DNA Book we describe herein represents a potential solution to this problem. The DNA Book is a paper-based vehicle for the transfer of genetic material, whereby DNA is attached to book pages and shipped together with the relevant scientific information. Clearly, the DNA Book has a strong potential to replace tubes or plates as a vehicle of DNA clones, and offers several advantages:
. It allows DNA distribution with fewer tasks and at lower cost, as simultaneous printing of numerous cDNA clones or PCR fragments in small amounts on the pages allows DNA distribution in a less laborious and more cost-effective manner. Every user can then obtain all of the necessary information and material in a bookstore.
. DNA is very stable in printed form and this characteristic allows bookbinding and shipment by courier.
. Researchers can easily extract DNA clones from the DNA Book within a short time.
. Both information and physical DNA clones or PCR fragments are conveyed simultaneously. Researchers can start an experiment immediately after planning what they wish to investigate, without requesting clones or waiting for the arrival of materials from providers.
The DNA Book could be applied to a variety of situations. (1) For example, physical cDNA fragments can simply be printed onto standard textbooks of molecular biology. A comprehensive set of yeast genes ∼6000 can be printed on a volume of DNA Book with low cost. Similar concepts can also be applied to the higher organism. In the near future, the RIKEN Mouse Genome Encyclopedia can be released as a DNA Book for wide distribution. The 60,770 mouse full-length cDNA clones of FANTOM2 (FANTOM Consortium and The RIKEN Genome Exploration Research Group Phase I and II Team 2002) could be printed on 80DNA sheets with 768 clones per page. (2) Alternatively, cDNA sets focusing on specific biological areas of interest could be printed as specialized DNA books, such as “Human Apoptosis Genes” and “Mouse Signal Transduction Genes”. Researchers with an interest in a specific field could choose one of a DNA Book series without additional payment for genes that are of no interest to them. Furthermore, a series of such separate volumes would reduce the burden on providers, as collection and rearraying of DNA clones is very laborious and error prone. (3) Furthermore, publishers will be able to publish scientific journals with relevant DNA fragments accompanying the articles. The concept of the DNA Book thus provides printed materials with a new role – distribution of genetic material (Fig. 5).
To establish the DNA Book, one problem remains to be addressed, namely the evaluation of stable preservation of the DNA Book. Although we have demonstrated that DNA sheets can tolerate conditions likely to be encountered during bookbinding and shipping (Fig. 2C), further testing is warranted. The final aim of this work is, therefore, to enlist the help of readers in a large-scale field-test of the DNA Book. We have attached a sample DNA sheet accompanying this article (Fig. 3). We would greatly appreciate readers' attempts to extract the genes printed on Figure 3, and feedback can be input on our Web page, http://genome.gsc.riken.go.jp/DNA-Book/. Results received from readers will be reported in a separate paper in the near future.
METHODS
DNA Preparation for Printing and Spotting
Plasmid DNA of RIKEN mouse cDNA was purified using a Qiagen Spin Miniprep Kit (QIAGEN) or the ultra-centrifugation method (Sambrook and Russell 2001). DNA was dissolved in TE (10mM Tris-HCl at pH 8.0, 1 mM EDTA) and DNA concentration was adjusted to 0.1 μg/μL. A small amount of DNA solution (∼0.1 μL each) was taken by the VP409 96-multiblot replicator (Bio Medical Science), and placed on the target DNA sheet. DNA solution was spotted five times for each spot, for a total of about 0.5 μL (0.05 μg) of DNA. Letters and figures had been printed on the sheet prior to spotting the DNA. The 60MDP paper used for DNA sheets was kindly provided by Mishima Paper Co., Ltd.
Alternatively, PCR fragments of cDNA can be used for printing. Thus, cDNA inserts were amplified from 1 ng of cDNA plasmids with KOD Plus DNA Polymerase (TOYOBO Co., Ltd.). PCR conditions were 1× KOD Plus PCR buffer, together with 0.2 μM of PCR primers (-21M13; 5′-TGTAAAACGACGGCCAGT-3′, 1233-Rv; 5′-AGCGGATAACAATTTCACACAGGA-3′) and 0.2 mM each of dATP, dGTP, dCTP, and dTTP. PCR cycles comprised 2 min at 94°C; 29 cycles of denaturing (94°C, 1 min), annealing (60°C, 1 min) and extension (68°C, 75 sec), and 15 min at 74°C. After purification by PEG precipitation, 50ng of PCR fragments can be spotted onto the DNA sheet as described above.
Sample DNA Sheet
A sample DNA sheet, to which 12 RIKEN cDNA plasmids of TCA cycle genes have been attached, is bound to this article (Fig. 3). The cDNA was cloned in pFLC vectors (Carninci et al. 2001). Vector structures are identical to pBluescript (Stratagene), except for the modified cloning sites. Clone IDs (DDBJ accession numbers) are 1500012M15 (AK005237), 1500012E04 (AK117103), E030024J03 (AK087063), E430020N12 (AK088582), 9430063I08 (AK034928), 6430550J24 (AK032458), 2610028K08 (AK117104), E230015D12 (AK054053), 1700007H16 (AK005713), 0610009E03 (AK002379), 9430071P19 (AK034998), and E430029P17 (AK088888). Spots of red dye on the right column of the annotation table represent 0.1 μg of plasmids.
Extraction and Recovery of DNAs
A piece of DNA sheet (∼4 mm × 4 mm) was cut out and placed into a PCR tube, followed by addition of 50μL of PCR solution. PCR solution contained 1.5 U of KOD Plus DNA Polymerase (TOYOBO Co., Ltd.), and 1× KOD Plus PCR buffer together with 0.2 μM of PCR primers (-21M13 and 1233-Rv, described above), 0.2 mM each of dATP, dGTP, dCTP and dTTP, and 5.3 mM MgCl2. After centrifuging the resulting solution, the PCR cycle was initiated. PCR cycles were comprised of 2 min at 94°C, 29 cycles of denaturing (94°C, 1 min), annealing (60°C, 1 min) and extension (68°C, 75 s), and 15 min at 74°C. Aliquots of PCR solutions were analyzed using 1% agarose gel electrophoresis.
Alternatively, aliquots of dissolved-paper solution can be subjected to E. coli transformation. As described above, a piece of DNA sheet (∼4 mm × 4 mm) was cut out and placed in a tube, followed by addition of 100 μL of TE. An aliquot (1–5 μL) was utilized to transform E. coli such as DH10B as described (Sambrook and Russell 2001). The solution in which the DNA sheet is dissolved does not have to be further purified. Transformed E. coli should be selected using ampicillin (50μg/mL).
Acknowledgments
We thank Mitsue Nakamura, Noriko Yokota, Shinya Sawata, Koichi Imotani, Hisamichi Ogura, Akiko Tomotsugu, Tsugumi Kawashima, Shigeyasu Yoshida, and Ryo Ishimoto for their technical assistance, Kazuhiro Shibata, Masayoshi Itoh, and Hiromi Torigoe for their kind support, and Hidemasa Bono and Katsunaga Sakai for selection of TCA cycle genes. We also thank the Mishima Paper Co., Ltd. for kindly providing the water-soluble paper. This study was supported by a Research Grant for the RIKEN Genome Exploration Research Project from the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government to Y.H.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.914203.
Footnotes
A sample DNA sheet, to which 12 RIKEN cDNA plasmids of TCA cycle genes have been attached, is bound to this article (Fig.3). Clone IDs (DDBJ accession numbers) are 1500012M15 (AK005237), 1500012E04 (AK117103), E030024J03 (AK087063), E430020N12 (AK088582), 9430063I08 (AK034928), 6430550J24 (AK032458), 2610028K08 (AK117104), E230015D12 (AK054053), 1700007H16 (AK005713), 0610009E03 (AK002379), 9430071P19 (AK034998), and E430029P17 (AK088888)
References
- Bono, H., Nikaido, I., Kasukawa, T., Hayashizaki, Y., RIKEN GER Group and GSL Members, and Okazaki, Y. 2003. Comprehensive analysis of the mouse metabolome based on the transcriptome. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Carninci, P. and Hayashizaki, Y. 1999. High-efficiency full-length cDNA cloning. Methods Enzymol. 303: 19-44. [DOI] [PubMed] [Google Scholar]
- Carninci, P., Shibata, Y., Hayatsu, N., Sugahara, Y., Shibata, K., Itoh, M., Konno, H., Okazaki, Y., Muramatsu, M., and Hayashizaki, Y. 2000. Normalization and subtraction of cap-trapper-selected cDNAs to prepare full-length cDNA libraries for rapid discovery of new genes. Genome Res. 10: 1617-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci, P., Shibata, Y., Hayatsu, N., Itoh, M., Shiraki, T., Hirozane, T., Watahiki, A., Shibata, K., Konno, H., Muramatsu, M., et al. 2001. Balanced-size and long-size cloning of full-length, cap-trapped cDNAs into vectors of the novel λ-FLC family allows enhanced gene discovery rate and functional analysis. Genomics 77: 79-90. [DOI] [PubMed] [Google Scholar]
- The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I and II Team. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563-573. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Kanehisa, M., Goto, S., Kawashima, S., and Nakaya, A. 2002. The KEGG databases at GenomeNet. Nucleic Acids Res. 30: 42-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammalian Gene Collection (MGC) Program Team. 2002. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. 99: 16899-16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- RIKEN Genome Exploration Research Group Phase II Team and FANTOM Consortium. 2001. Functional annotation of a full-length mouse cDNA collection. Nature 409: 685-690. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D.W. 2001. Molecular cloning: A laboratory manual, 3rd edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Seki, M., Narusaka, M., Kamiya, A., Ishida, J., Satou, M., Sakurai, T., Nakajima, M., Enju, A., Akiyama, K., Oono, Y., et al. 2002. Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141-145. [DOI] [PubMed] [Google Scholar]
- Shibata, K., Itoh, M., Aizawa, K., Nagaoka, S., Sasaki, N., Carninci, P., Konno, H., Akiyama, J., Nishi, K., Kitsunai, T., et al. 2000. RIKEN integrated sequence analysis (RISA) system—384-format sequencing pipeline with 384 multicapillary sequencer. Genome Res. 10: 1757-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://fantom2.gsc.riken.go.jp/; FANTOM home page.
- http://genome.gsc.riken.go.jp/DNA-Book/; DNA Book home page.