Abstract
The Mouse Genome Sequencing Consortium and the RIKEN Genome Exploration Research grouphave generated large sets of sequence data representing the mouse genome and transcriptome, respectively. These data provide a valuable foundation for genomic research. The challenges for the informatics community are how to integrate these data with the ever-expanding knowledge about the roles of genes and gene products in biological processes, and how to provide useful views to the scientific community. Public resources, such as the National Center for Biotechnology Information (NCBI; http://www.ncbi.nih.gov), and model organism databases, such as the Mouse Genome Informatics database (MGI; http://www.informatics.jax.org), maintain the primary data and provide connections between sequence and biology. In this paper, we describe how the partnership of MGI and NCBI LocusLink contributes to the integration of sequence and biology, especially in the context of the large-scale genome and transcriptome data now available for the laboratory mouse. In particular, we describe the methods and results of integration of 60,770 FANTOM2 mouse cDNAs with gene records in the databases of MGI and LocusLink.
Large-scale sequencing and annotation efforts, such as the human and mouse genome sequencing initiatives (Lander et al 2001; Waterston et al. 2002), the RIKEN full-length enriched cDNA sequencing project (Kawai et al. 2001; Okazaki et al. 2002), and the Mammalian Gene Collection (MGC; Strausberg et al. 1999), have made publicly available a wealth of genomic and transcript information to support diverse research efforts related to understanding mammalian biology and disease. Now more than ever, users need easy access to integrated views of, and analysis tools for, high-quality information about mammalian genes and genomes. The challenge is to develop strategies for integrating these data with continually emerging knowledge about the function, variation, and regulation of genes and other genomic features. The collaboration between the Mouse Genome Informatics (MGI) group8 and the National Center for Biotechnology Information's (NCBI) LocusLink and RefSeq groups (http://www.ncbi.nih.gov/) exemplifies how coordinated efforts facilitate connectivity between sequence and biology in the mouse. The MGI resource provides highly integrated and curated views of genetic, genomic, and biological data for the laboratory mouse. LocusLink, through the Reference Sequence (RefSeq) project, connects biological information to the sequences of reference chromosomes, RNAs, and proteins. The MGI/LocusLink collaboration has its greatest impact through the mutual determination of the sequences which best define mouse genes. Once gene-to-sequence(s) connections are established, and associations to available information about the genes are made, the foundation is set for additional computation, curation, and Internet connectivity for the scientific community.
Both MGI and LocusLink seek to provide access to comprehensive and accurate information about genes defined by sequence. Thus, the collaboration between LocusLink and MGI aims to identify and curate a comprehensive catalog of mouse genes and the nucleotide and protein sequences that define them. Nucleotide and protein sequences are associated with gene records in MGI and LocusLink when, through conservative computational assessment or expert curation, the sequences are found to be derived from a region of the genome that defines a gene. Associating sequences with gene records places these sequences into a gene-centric context, in which they become integrated with other biological information associated with those genes.
If a high-quality mRNA sequence is identified for a protein-coding gene, and it contains a complete coding sequence (CDS), then that sequence is used as the source for RefSeq mRNA and protein accessions. Confusion can arise when multiple sequences for the same gene are submitted to sequence databases with different names (Bult et al. 2000). This problem is lessened, however, if the query results contain sequences that are integrated with corresponding gene records in MGI and LocusLink, which provide standard nomenclature, synonyms, and cross references to other data sources. Thus, the correct association of nucleotide and protein sequences with gene objects, represented in genome databases and instantiated as reference sequences, is key to the integration of sequence and biology.
As new sequence data are generated and gene models defined, it is important to determine which come from previously described genes and which are truly novel. Additional sequences from the same gene may define physiological significant variants. Sequences from novel genes are the foundations for defining those genes and initiating gene records. Therefore, a primary objective for database integration of new sequence data is to determine which sequences correspond to mouse genes in the public databases of MGI and LocusLink. Informatics tools designed to detect sequence similarity on a large scale are essential for this analysis, and serve as a means of robust first-pass sequence annotation. The MGI and LocusLink databases also rely on literature curation and community-wide expert confirmation of sequence and data associations. Human curation is critical for reviewing sequence-to-gene associations for highly related sequences, because biological context may be required in the decision-making process. In addition, manual evaluation of annotation processes that are primarily computational may lead to improved algorithms for more automated analyses.
Here we present the strategies developed collaboratively by MGI and LocusLink to compose a comprehensive catalog of mouse genes with accurate associations to genomic, transcript, and protein sequences. We focus primarily on the RIKEN full-length enriched cDNA clone set, describing the computational and manual curation methods used to distill information from the sequences for 60,770 FANTOM2 cDNA clones to information for 19,980 mouse genes. We discuss how this serves as an entry point to the process by which the MGI and LocusLink/RefSeq groups join sequence information from this comprehensive mouse cDNA project and the mouse genome draft assembly with existing information about mouse biology. The integration of these data sets and other important sequence resources, most notably the MGC cDNAs and finished BAC sequences, demonstrates the synergistic effect that cooperation among public resources has on the quality and usefulness of community data.
RESULTS
Associating FANTOM2 cDNA Clones With Mouse Genes
Cluster Triage
The FANTOM2 data set includes the previously released 21,076 cDNAs (FANTOM1; Kawai et al. 2001) and 39,694 additional cDNAs (FANTOM2-new), totaling 60,770 sequences (Okazaki et al. 2002). Sequence clustering is the first step toward integration with known mouse genes (Fig. 1). Table 1 shows the number of clusters containing FANTOM2 sequences from the three different cluster builds and their cluster unions, and includes for comparison, the resolved numbers of MGI genes and transcriptional units (TUs) from the FANTOM2 Representative Transcript Protein Set (RTPS, version 6.3; Fig. 2; see “Sequence Clustering, Cluster Triage, and Genes Versus TUs”). The RIKEN, NCBI UniGene, and TIGR clustering algorithms approach cluster building differently. The UniGene and RIKEN methods cluster without performing assemblies, which tends to group transcript variants together and produce fewer clusters. The TIGR method performs sequence assemblies and calculates consensus sequences, which tends to separate transcript variants and produce more clusters. In addition, the starting data sets varied. RIKEN clusters contained FANTOM2 sequences only, whereas UniGene and TIGR clusters contained FANTOM2 sequences plus all mouse cDNAs and ESTs. These factors explain the variation in cluster number observed between the three cluster builds (Table 1). The cluster unions calculated across the three builds incorporated this clustering variation and reduced the total number of clusters to consider.
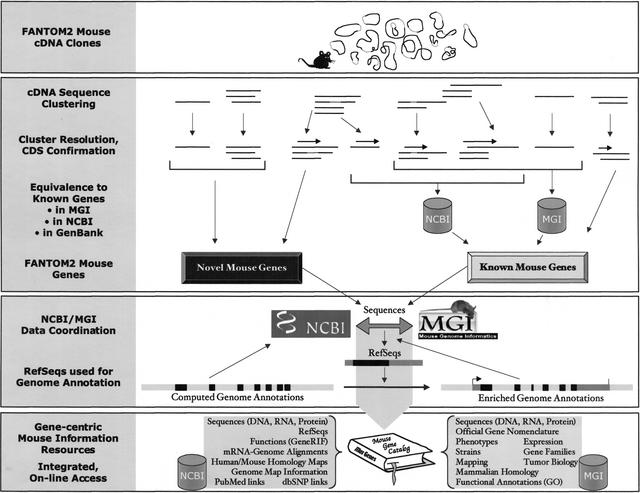
Figure 1.
The flow from FANTOM2 mouse cDNA clones to genes, to their integration with NCBI LocusLink and MGI. Mouse cDNA clones isolated (closed circles in the first panel) and sequenced (horizontal lines in the second panel) by the RIKEN group are clustered computationally (top clusters in the second panel). Computed clusters are then resolved into gene-specific groups by human inspection (bottom clusters in the second panel). Dotted lines represent transcript variation. Computed clusters can group sequences from different genes, such as paralogs and read-through transcripts (third and fourth computed clusters from left, respectively), and other distinct gene sequences that share some region of overlap requiring manual resolution. CDS regions for protein coding genes are indicated (horizontal arrows over clusters). Equivalence of FANTOM2 sequences with known mouse genes in NCBI LocusLink and MGI is detected by incorporation of known sequences in the FANTOM2 clusters or by BLAST (data not shown). LocusLink and MGI contain overlapping but distinct sequence data sets. Some characterized mouse sequences not present in LocusLink or MGI can have sequence identity to FANTOM2 sequences (far right cluster). Remaining FANTOM2 genes are considered novel. The curation of sequences for novel and known mouse genes is coordinated between LocusLink and MGI, and LocusLink establishes RefSeqs (third panel). Genome centers feed predicted gene models to NCBI, but rely on transcript-based evidence in the form of RefSeqs to improve genome annotations. Gene models with enriched annotations link back to gene records in LocusLink and MGI on the basis of integrated sequence accessions. Through data coordination, LocusLink and MGI establish a catalog of mouse genes with accurate sequence associations and integrated biological information.
Table 1.
Clustering of FANTOM2 Sequences
| Sequence groupa | Total groups |
|---|---|
| RIKEN Clusters | 35,957 |
| NCBI UniGene Clusters | 37,782 |
| TIGR Clusters | 45,538 |
| Cluster Unions | 34,526 |
| MGI-associated Cluster Unions | 16,475 |
| MGI Genes | 19,980b |
| RTPS6.3 TUs | 33,409 |
Sequence Groups include FANTOM2 sequences that were not grouped with other FANTOM2 sequences (i.e., singletons)
The MGI gene number represents genes associated with FANTOM2 sequences after the FANTOM2 load into the MGI database. Many (18,794) FANTOM2 sequences were not associated with MGI genes at load time
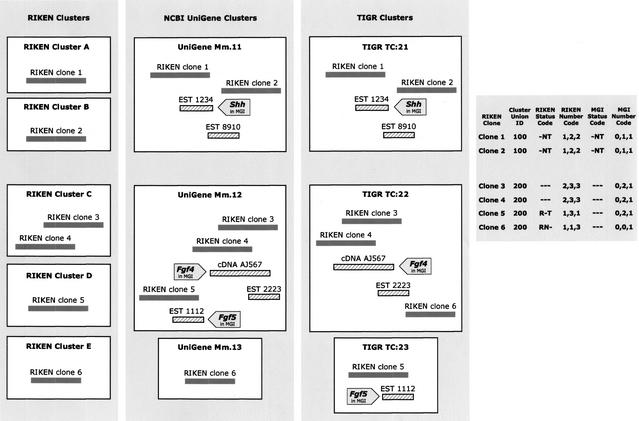
Figure 2.
Schematic showing status and number code assignments for clusters and their use in cluster curation. Hypothetical outputs of the three cluster builds for six FANTOM2 clones are shown. Status and number codes for each clone, as well as cluster union IDs, appear in the table to the right. FANTOM2 clones and non-RIKEN public sequences are shown as solid and open boxes, respectively. Sequences associated with MGI genes are distinguished by block arrows. In the top set of clusters, RIKEN clones 1 and 2 were grouped with the same EST sequences in NCBI UniGene and TIGR clusters, and were assigned the same cluster union ID (100). The RIKEN status code (-NT) for these clones indicates that NCBI Unigene and TIGR clusters are the same for these clones, but that RIKEN clusters A and B are different. The RIKEN number code (1,2,2) indicates that one, two, and two total RIKEN clones were clustered with those clones (including themselves) in the three respective cluster builds, irrespective of clone identities. The MGI status code (-NT) indicates that only the UniGene and TIGR clusters grouped sequences associated with the same number and identity of MGI genes (only MGI gene Shh is represented via EST 1234). The MGI number code (0,1,1) indicates that a single MGI gene is represented in the UniGene and TIGR clusters (irrespective of gene identities) and none in the RIKEN clusters containing these clones. The bottom set shows the clusters containing four additional RIKEN clones (3 through 6). Clones 3, 4, and 5 are grouped in UniGene cluster Mm.12; yet, clones 3, 4, and 6 are grouped in the TIGR cluster TC:22, thus all four clones are assigned to the same cluster union (200). The variability in RIKEN status codes indicates that each clone was grouped differently from the others by the three builds. The MGI number code (0,2,1) for three of the clones (3, 4, and 5) indicates that the UniGene cluster containing them (Mm.12) has grouped sequences associated with two MGI genes (Fgf4 and Fgf5), whereas only one MGI gene is represented in each of the TIGR clusters that contain them (TC:22, and TC:23). The MGI status code (---) for each clone indicates that no two clusters containing them represent exactly the same set of MGI genes. For this example, curators would determine if the sequence associations in UniGene cluster Mm.12 are biologically appropriate; if so, then the MGI gene records involved may need to be merged into a single record.
To determine the number of FANTOM2 clones that are potentially derived from genes represented in MGI, we found all FANTOM2 clusters (cluster unions) containing at least one sequence that was associated with a gene in MGI at the time of the analysis (16,475 clusters; Table 1). A total of 15,658 FANTOM2-new sequences were incorporated into clusters that contained at least one sequence associated with an MGI gene. The total number of RIKEN sequences (FANTOM1 and FANTOM2-new) in these MGI-associated cluster unions was 36,734. A cluster integrity evaluation system was developed to facilitate manual curation of this large data set (Table 2; see Methods). Nearly 64% of the 36,734 MGI-associated FANTOM2 sequences were assigned the highest consistency code (RNT) for either RIKEN clone distribution or MGI gene representation (categories 1 through 5 in Table 2).
Table 2.
Cluster Consistency and Number Codes for MGI-Associated FANTOM2 Clones
| Category | Description | RIKEN status code | RIKEN number code | MGI status code | MGI number code | Total clones |
|---|---|---|---|---|---|---|
| 1 | Singletons: clones unclustered with other clones | RNT | 1,1,1 | RNT | 1,1,1 | 7743 |
| 2 | The same set of clones in each cluster view, and the same single MGI gene represented in each cluster view | RNT | not 1,1,1 | RNT | 1,1,1 | 6769 |
| 3 | Different clones among the three cluster views, but the same single MGI gene represented in the three views | not RNT | * | RNT | 1,1,1 | 6875 |
| 4 | Different clones among the three cluster views, but the same set of MGI genes represented in the three views | not RNT | * | RNT | not 1,1,1 | 903 |
| 5 | The same set of clones in each cluster view, but different MGI genes represented among the three cluster views | RNT | * | not RNT | * | 1187 |
| 6 | Different clones among the three cluster views, but at least two cluster views do not represent an MGI gene | not RNT | * | not RNT | *,0,0 or 0,*,0 or 0,0,* | 3874 |
| 7 | Different clones among the three cluster views, but at least two cluster views represent a single MGI gene | not RNT | * | not RNT | *,1,1 or 1,*,1 or 1,1,* | 6603 |
| 8 | Different clones among the three cluster views, and multiple different MGI genes are represented | not RNT | * | not RNT | all remaining | 2780 |
Any value (wildcard)
Integration of FANTOM2 Data
Integration of RIKEN cDNA data into MGI was conducted in two phases (FANTOM1 and FANTOM2). For FANTOM1 integration, MGI curators established associations between FANTOM1 sequences and MGI genes after evaluating BLAST alignments during the FANTOM1 annotation period. New MGI gene records were created for novel FANTOM1 sequences. Clone source information was incorporated by creating molecular segment records in MGI for all FANTOM1 cDNAs (Kawai et al. 2001). The FANTOM1 phase included curation that took place at MGI and LocusLink between the release dates of FANTOM1 and FANTOM2 data (February 2001 and December 2002, respectively). In MGI, at least 1300 gene record changes were processed that involved FANTOM1 sequences and genes during this period, including updates to nomenclature and other biological information. LocusLink/RefSeq staff processed >1000 additional merges, often involving MGI genes defined by ESTs.
FANTOM2 integration involves both FANTOM1 and FANTOM2-new sequences and is described in the present work. For FANTOM2 phase integration, MGI curators focused on clone-to-gene resolution from a cluster perspective, which complemented the clone-oriented functional annotation by annotators of the Mouse Annotation Teleconference for RIKEN cDNA sequences (MATRICS; Okazaki et al. 2002). Two tools were used to curate clusters efficiently. A cluster visualization tool developed at RIKEN provided curators convenient graphical views of cluster alignments. Additionally, an MGI-integrated FANTOM2 data table developed by the MGI group allowed curators to query by cluster complexity, annotate whole clusters, and view updated associations of sequences to MGI genes (see Methods). Because less complex clusters were targeted and full cluster alignments could be visualized graphically, curators were able to annotate over half of the 36,734 sequences contained in MGI-associated clusters to 6817 MGI genes (Table 3). Most of these annotations were to existing MGI genes, although a few clusters resolved to multiple genes, some of which were novel to MGI. Figure 3 shows how cluster information from these resources was combined during integration with MGI to ensure accurate data representation.
Table 3.
Gene Numbers for Curated Versus Noncurated Clusters and Singletons
| Clone-to-gene association in MGI | MGI genes | Singletons | Total clones |
|---|---|---|---|
| MGI-curateda | 6,817 | 1,221b | 19,575 |
| Not MGI-curatedc | 13,163 | 7,050b | 22,401 |
| None establishedd | — | 16,129e | 18,794 |
| Totals | 19,980 | 24,400 | 60,770 |
Clones associated with existing or novel MGI genes after detailed cluster consideration by MGI curators
For FANTOM2 clones associated with MGI genes, a singleton is defined as the only FANTOM2 clone associated with that particular gene. Some of these singletons were clustered with other FANTOM2 sequences but were separated by curators. Many of these sequences overlap with non-RIKEN transcript sequences
Clones associated with existing or novel MGI genes without detailed cluster analysis by curators. Associations to existing MGI genes were established only in the absence of conflicting relationships to MGI genes for all cluster members. Novel genes were created only for multi-clone clusters in which all cluster members mapped to the same mouse chromosome
Clones loaded without MGI gene associations. All are FANTOM2-new clones, and most are singletons (single clone in a cluster). Non-singleton clones from this set are either part of ambiguous MGI-associated clusters (multiple MGI genes represented) or are novel gene candidates but have chromosome mapping discrepancies for some cluster members
For FANTOM2 clones not associated with MGI genes, a singleton is defined as a cluster containing only one clone
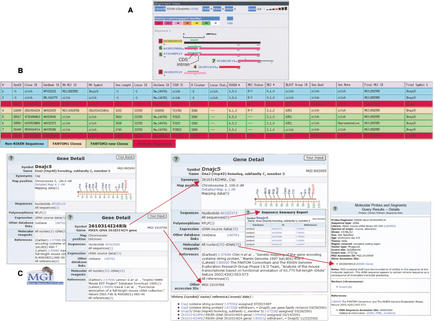
Figure 3.
Combining cluster visualization tools with the MGI FANTOM2 data table for accurate integration in MGI. (A) Alignment view display, a cluster visualization tool available at the FANTOM2 Web interface. FANTOM2 sequences grouped in RIKEN cluster (locus ID) 22339 are shown as colored bars. RIKEN clone IDs are shown to the left of each sequence, as are the corresponding row numbers for the sequences in the MGI FANTOM2 table in B. Sequence alignments are with respect to the top sequence (black), as are various features, including sequence similarity (color-coded as shown) and gaps. The green arrows above the sequences represent predicted CDS regions (shown). The gaps in sequences 5 and 6 (intron) reveal the presence of an unspliced intron in sequences 3 and 8. Note truncation of the CDS at this position in sequences 3 and 8. Sequences 5 and 6 are properly spliced. Sequences 3, 4, and 7 are partial transcripts. Non-RIKEN sequences are not shown in this view. (B) MGI FANTOM2 data table display of the FANTOM2 sequences in A and two non-RIKEN sequences (blue) included in this cluster union (R Cluster 3268). Rows and columns correspond to sequences and sequence features, respectively. Rows are color-coded to reflect sequence origin or other status (as shown). Sequences 3 and 8 are marked as problem sequences because they contain an unprocessed intron (Seq Qual: Problem-in). Sequence 6 was selected as the representative clone (Seq Note: Representative). Sequences 1 and 3 were associated with MGI gene Dnajc5 before the FANTOM2 load, sequence 4 with MGI gene 2610314I24Rik (RA symbol). All sequences are associated with MGI gene Dnajc5 after the FANTOM2 load (Final symbol 1). (C) Integration in MGI. The FANTOM1 clone 2610314I24 (sequence 4 in A, B) does not overlap the coding region of Dnajc5 and was represented as a unique MGI gene during the FANTOM1 load (Symbol: 2610314I24Rik), whereas FANTOM1 clone 1810057D19 (sequence 3 in A, B), which does overlap the CDS, was associated with the Dnajc5 gene. FANTOM2-new sequences reveal that sequence 4 is actually derived from the 3′-UTR region of Dnajc5 and that sequence 3 contains an intron that truncates the CDS. This information triggered a merge in MGI, in which the 2610314I24Rik gene was withdrawn to equal Dnajc5. The MGI accession ID for the previous gene (MGI:1919766) becomes a secondary accession ID for the Dnajc5 gene (shown), and all information previously associated with 2610314I24Rik was migrated to Dnajc5. The nomenclature history for the Dnajc5 gene details this event. The molecular segment record for clone D030049H18 (sequence 8 in A, B), an intron-containing transcript (problem sequence) is shown. A note is attached to molecular segment records of problem sequences to inform users that the sequence has been judged by curators to have some type of problem. Key to MGI FANTOM2 table columns (see Methods for descriptions): SeqID indicates RIKEN Seqid; clone ID, RIKEN cloneid; GenBank ID, DDBJ/EMBL/GenBank seqid; RA MGI ID, MGI ID to which the sequence was associated before the FANTOM2 load; RA symbol, gene symbol corresponding to the RA MGI ID; Seq length, sequence length (bp); locus ID, RIKEN cluster ID; UniGene ID, NCBI UniGene cluster ID; TIGR TC, TIGR cluster ID; R cluster, cluster union ID; locus stat, RIKEN status code; RIKEN #, RIKEN number code; MGI status, MGI status code; MGI #, MGI number code; BLAST group ID; Seq qual, sequence quality; Seq note, sequence note (to designate Representative clone); final MGI ID, MGI ID to which the sequence is associated after the FANTOM2 load; and final symbol 1, gene symbol corresponding to the Final MGI ID.
For sequences in MGI-associated clusters that remained uncurated at the time of the load, associations between FANTOM2 clones and MGI genes were established from the sequence clusters generated by RIKEN computational clustering (or from limited cluster examination by other FANTOM2 annotators). Uncurated associations to existing MGI genes were established only when the sequence clustering resulted in unambiguous relationships to MGI genes (Table 3). There remain a number of complex FANTOM2 clusters in which sequence-to-gene resolution is difficult, even with genomic sequence context. For such clusters, preexisting associations between FANTOM1 clones and MGI genes were maintained without processing gene record rearrangements in MGI, and FANTOM2-new members of these clusters were loaded into MGI without gene associations. Curation of these complex clusters is ongoing. For the 24,036 sequences not included in MGI-associated clusters, novel MGI genes were created only for transcripts supported by multiple clones (i.e., multi-clone clusters), and when all cluster members mapped to the same chromosome in the draft mouse genome sequence (MGSC V3; Okazaki et al. 2002).
Because of the time interval between FANTOM2 sequence clustering and release of the data, and some underclustering (particularly for intron-containing singleton clones), we prioritized minimization of redundant MGI gene records over comprehensive association of FANTOM2 clones to MGI genes; thus, no novel MGI genes were created based on uncurated FANTOM2 singleton sequences for this load. The majority (86%) of FANTOM2 sequences loaded into MGI without MGI gene associations are singletons (Table 3). A concerted effort is underway to integrate these remaining FANTOM2 sequences using the MGSC assembly.
The FANTOM2-new data had a significant impact on gene annotation in MGI, bringing valuable perspective to many FANTOM1 sequences for which MGI gene records were created previously, as illustrated for the Dnajc5 gene in Figure 3. At the time of the load into MGI, FANTOM2 data influenced >1200 MGI gene merges, and ∼200 gene nomenclature updates from mammalian orthologs. Over 9000 unmapped MGI genes were mapped to chromosomes with FANTOM2 data, and >18,000 FANTOM2 gene ontology (GO) annotations (Ashburner et al. 2000) were distributed among 6787 MGI genes. Of the FANTOM2 clones inspected by MGI curators during FANTOM1 and FANTOM2 phases, 3774 were annotated as problem sequences at the time of the FANTOM2 load. The majority of these are intron-containing transcripts (see examples in Fig. 3). All problem sequences in MGI are flagged with a note (Fig. 3), and are available at the public MGI FTP site (ftp://ftp.informatics.jax.org/pub/reports/MGI_ProblemSequence.rpt). As with FANTOM1 data, molecular segment records were created for all FANTOM2-new clones (see Methods). A summary of the integration procedures carried out in MGI for the FANTOM2 data is provided in Table 4.
Table 4.
Summary of FANTOM2 Data Integration in MGI
| Procedure | FANTOM2 clones representing novel MGI genes | FANTOM2 clones representing existing MGI genes |
|---|---|---|
| Gene object created | Yes, for curated sequences and for uncurated multi-clone clusters | No |
| Gene nomenclature | Standardized RIKEN or derived from ortholog or paralog nomenclature | Existing or derived from ortholog or paralog nomenclature |
| DDBJ seqID-to-gene association | Yes, if not a problem sequence | Same as for novel MGI genes |
| Chromosome obtained from genome assembly | Yes, if no mapping conflicts between cluster members | Yes, if no mapping conflicts between cluster members or with existing Chr assignment in MGI |
| Mammalian ortholog record created | Yes, for curated orthologs | Same as for Novel MGI genes |
| Gene reference | FANTOM1 or FANTOM2 reference | Existing gene reference |
| Molecular segment object created | Yes, for all clones | Same as for Novel MGI genes |
| Molecular segment name | “RIKEN clone” RIKEN clonelD | Same as for novel MGI genes |
| Molecular segment attributes | Clone library name, mouse strain, tissue, age, sex | Same as for novel MGI genes |
| Molecular segment-to-gene association | Yes, for all clones associated to genes | Same as for novel MGI genes |
| DDBJ seqID-to-molecular segment association | Yes, for all clones with DDBJ seqIDs | Same as for novel MGI genes |
| Molecular segment reference | FANTOM1 or FANTOM2 reference | Same as for novel MGI genes |
| Molecular segment, problem sequence note | Yes, for problem sequences | Same as for novel MGI genes |
| Gene ontology (GO) annotation | Yes, if annotation source were: InterPro, SWISS-PROT/Trembl, paralogous MGI gene or expert confirmed | Same as for novel MGI genes |
The procedures carried out to integrate FANTOM2 data into MGI are compared for sequences representing novel and existing genes in MGI
The NCBI LocusLink group performed additional steps to integrate novel FANTOM2 gene records before loading these from MGI into LocusLink. Between the time of FANTOM2 data curation and the public release of those data, other genedefining sequences were integrated into the LocusLink and RefSeq databases. These sequences fell into two major categories: (1) cDNA sequences submitted to DDBJ/EMBL/GenBank, particularly those from the MGC, and (2) gene models from NCBI's annotation pipeline (∼37,000). Thus, before loading novel FANTOM2 genes from MGI, additional redundancy checks were performed, including alignment of FANTOM2 sequences to mouse cDNAs released after the FANTOM2 data freeze and to gene models on the MGSC assembly produced from NCBI's annotation pipeline (especially those that are EST-based), as well as mapping the existing model mRNAs from the annotation pipeline (RefSeq accessions of the format XM_123456) to MGI genes. Data with conflicts, or genes with marginal support, were not loaded automatically and are receiving additional curation.
Genes Represented by FANTOM2 Clones
A total of 19,980 MGI genes were represented by FANTOM2 data (Tables 3, 5). Of these, 11,351 (57%) were novel mouse genes to MGI and LocusLink databases. A total of 3016 novel MGI genes were created from FANTOM2-new sequences. The number of MGI genes associated with FANTOM2 data at the time of the load under-represents the total number of genes included in the data set, because nearly half (18,794, 47%) of the 39,694 FANTOM2-new sequences were not associated with MGI genes during the FANTOM2 load (Table 3). An estimate of the total number of genes represented by the FANTOM2 data is provided from the number of TUs these sequences represent in the FANTOM2 RTPS6.3 (33,409, see Table 5 and Discussion). Focused redundancy checks oriented by the genome assembly sequence at MGI and LocusLink will resolve the number of genes represented by the FANTOM2 data over time. Nonetheless, the data from the FANTOM2 Consortium provide a significant increase in the representation of the transcribed mouse genome shared between MGI and LocusLink.
Table 5.
MGI Genes and RTPS6.3 TUs Represented by the FANTOM2 Data
| FANTOM2 clones | MGI genes | RTPS6.3 TUsa | |
|---|---|---|---|
| FANTOM2 clones with MGI gene associations | 41,976 | 19,980 | 17,123 |
| FANTOM2 clones with no MGI gene associations | 18,794 | — | 16,286 |
| Totals | 60,770 | 19,980 | 33,409 |
The numbers of RTPS6.3 TUs that incorporate the corresponding set of FANTOM2 clones are shown
Curated Gene Records in MGI and LocusLink
Curated sequence-to-gene associations drive the synchronization of gene records in the MGI and LocusLink databases. Knowledge of genes can evolve independently until integrated sequence data nucleates the union of biological information and initiates cooperative data curation between databases. This is demonstrated by the history of flexed tail (sideroflexin 1, Sfxn1) gene annotation in MGI (Fig. 4A) and by the current state of annotation for this gene in MGI and LocusLink (Fig. 4B). By contrast to the Sfxn1 gene, for which biology and sequence information are linked, Figure 5 shows MGI representation of a novel FANTOM2 gene, for which our knowledge of the gene is limited to sequence information.
Figure 4.
Gene Representation in MGI and LocusLink. (A) Emerging representation of the flexed tail gene (sideroflexin 1, Sfxn1). A gene record for the flexed tail (f) mouse mutation, described by Hunt et al. (1933), is created in MGI. Over time, MGI captures published information about the flexed tail locus; however, no sequence information is available. Clone 2810002O05, a novel mouse cDNA sequence is released with the FANTOM1 data, and a gene record is created in MGI and LocusLink for the sequence. Sequence-based annotations (GO terms, protein, domains, UniGene) are associated with gene 2810002O05Rik, and the MGI/LocusLink coordinated data exchange begins. LocusLink creates a RefSeq for the gene. After release of FANTOM1 data, Fleming et al. (2001) report the cloning of flexed tail and its sequence. Sequence analysis reveals that the flexed tail sequence is identical to the FANTOM1 cDNA. Gene 2810002O05Rik is merged with the flexed tail gene, and based on Fleming et al. (2001), the gene is renamed sideroflexin 1 (Sfxn1), for the siderocytic anemia and flexed tail phenotypes observed in mutant mice (see Fig. 4B). (B) Current representation of the Sfxn1 gene record in MGI and LocusLink, demonstrating the types of information integrated with sequences at the two resources. Wide arrows indicate data types shared between MGI and LocusLink, and the direction of transfer. MGI and LocusLink also exchange gene name synonyms and corresponding gene record identifiers. Hypertext links to various annotations and data are provided at both resources: official mouse gene nomenclature (MGI provides to LocusLink; A), mapping information (reconciled between MGI and LocusLink; B), allele and phenotype information (MGI; C), polymorphisms (LocusLink provides links to dbSNP, data not shown; D), gene ontology (MGI provides to LocusLink; E), homology information (MGI provides curated mammalian orthology data; F), expression (MGI; G), UniGene (H), LocusLink/MGI reciprocal links (I), mouse genome annotations (J), protein domains (also at LocusLink, data not shown; K), Database of Transcribed Sequences (DoTS, MGI; L), TIGR Mouse Gene Index (MGI; M), mRNA-genome alignments (LocusLink; N), references (O), RefSeqs (LocusLink provides to MGI; P), and sequences (exchanged between MGI and LocusLink; Q).
Figure 5.
Representation of a novel FANTOM2 gene in MGI. Detail pages for the gene and molecular segment objects and for the sequence summary report are shown. Novel FANTOM2 gene nomenclature incorporates the RIKEN clone IDs of representative sequences from clusters. Clusters of sequences for the same gene are represented by associating the sequence identifiers and molecular segment records of all cluster members to the gene record (of the 37 molecular segments of type cDNA for this gene shown, five are FANTOM2 clones; the rest are IMAGE cDNAs associated with gene 0910001B06Rik via UniGene cluster 28470). Molecular segment records for FANTOM2 clones contain clone library source information, and they link to the FANTOM2 annotation pages for the corresponding sequences.
As of September 28, 2002, there were 24,838 curated associations between MGI and LocusLink gene records, 9199 of which had transcript-based RefSeqs9 (sequence accession designator, NM_123456, NP_123456 for the corresponding protein). The list of associations among MGI gene accession identification numbers, LocusLink identifiers, and RefSeq sequence accessions is updated daily and can be downloaded from the MGI (ftp://ftp.informatics.jax.org/pub/informatics/reports) or LocusLink (ftp://ftp.ncbi.nih.gov/refseq/LocusLink/) FTP sites. Reciprocal hypertext links are provided by MGI and LocusLink to corresponding gene records. MGI also provides hypertext links to RIKEN functional annotation details for all RIKEN clones (Fig. 5).
Mouse Genome Sequence Draft Assembly
Of the 22,444 predicted mouse genes reported for the initial analysis of the mouse genome10 (Waterston et al. 2002), 11,254 represent known genes that could be associated with a gene in MGI and LocusLink through RefSeq or SWISS-PROT sequences.11 Links from the computational gene predictions for known genes in the mouse genome to gene records in MGI and LocusLink are provided using genome browsers such as NCBI's Map Viewer (http://www.ncbi.nih.gov/mapview/), the EMBL-EBI and Sanger Institute's Ensembl (http://www.ensembl.org; Hubbard et al. 2002), and the University of California at Santa Cruz's Genome Browser (http://www.genome.ucsc.edu; Kent et al. 2002). The mouse genome sequence is being used extensively in ongoing efforts to curate and integrate the FANTOM2 data at MGI and LocusLink.
DISCUSSION
The FANTOM2 data set contributes significantly to our understanding of the mouse genome and transcriptome. A high novelty rate for genes in MGI was observed (57%), excluding the 18,794 clones loaded into MGI without gene associations (Table 3). When resolved to genes, these remaining sequences certainly will contribute to the novel genes from this set. This high novelty rate reflects the successful efforts of the RIKEN Genome Exploration Research Group to target novel mouse transcripts. The high rate of FANTOM2 singletons (Table 3) is largely due to this pursuit of novelty (Carninci et al. 2003). The number of MGI genes associated with FANTOM2 data (19,980) is clearly an underestimate of the total genes represented by these data. There remains some redundancy between MGI genes and the 18,794 FANTOM2 clones loaded with no MGI gene association. As this redundancy is resolved, we will associate the remaining FANTOM2 clones with MGI genes. A total gene number estimate for the FANTOM2 set can be obtained from the FANTOM2 RTPS6.3. Construction of RTPS6.3 combined representative sequences of the FANTOM2 clusters with all public mouse cDNAs, and then followed ordered steps to reduce redundancy and separate distinct sequences according to the computational definition of a TU (Okazaki et al. 2002; see “Genes versus TUs”). The total number of RTPS6.3 TUs that incorporate FANTOM2 sequences is 33,409 (Table 5). Comparing the number of MGI genes (19,980) versus TUs (17,123) associated with the same set of FANTOM2 sequences (Table 5) provides upper and lower limits of the genes represented. This difference in gene number is due to differences in how TUs and MGI genes are defined (see “Genes versus TUs”), and to the TU merges that took place after mapping to the mouse genome assembly, a final step in construction of RTPS6.3. The number of gene records in MGI and LocusLink represented by FANTOM2 data will change as we continue to address issues of remaining redundancy and overclustering, and assimilate additional public cDNA sequences and updates to the genome assembly.
The tools and strategies used for large-scale annotation and integration of FANTOM2 sequences were rooted in the experience gained from annotation of the 21,076 FANTOM1 cDNAs (Kawai et al. 2001). Since the release of those data, LocusLink and MGI have improved gene-to-sequences connections, identifying >3000 cases for merges and improved annotation. Our experience with the gene redundancy introduced from the FANTOM1 set largely influenced the decision to postpone creating gene records for 16,129 FANTOM2-new singletons, until additional redundancy analyses are completed. The FANTOM2 data posed even greater challenges of redundancy detection and tracking. These challenges drove the development of tools for enriched graphical displays of multiple sequence alignments, and for annotation of sequence groups based on cluster analyses. The cluster-based approach increased curation efficiency and effectiveness, because the clusters reduced the number of alignments to analyze. The global views of clusters placed alignments in biologically relevant contexts, making detection of splice variants, truncated clones, and problem sequences much easier. Integration of this large data set required a coordinated union of automated and manual curation methods. Lessons learned from this effort will influence efficient integration of large sequence data sets from the genome assembly, the MGC, and further contributions from the FANTOM Consortium.
The integration of FANTOM2 cDNAs into MGI, and subsequently into LocusLink and mouse RefSeq, is part of a broader system of data integration that includes sequences associated with published literature, gene models from genome annotation pipelines, and additional high-quality contributed data sets such as the full-length cDNAs of the MGC.
This integration is the result of a continuing collaboration between MGI, LocusLink, SWISS-PROT, and RIKEN, with significant contributions from the Human Genome Organization (HUGO) Gene Nomenclature Committee (HGNC). Curators from these groups work together to ensure that data represented by the individual databases are linked by common gene objects (Gasteiger et al. 2001; Pruitt and Maglott 2001; Ringwald et al. 2001; Blake et al. 2002; Okazaki et al. 2002). The cooperative curation across different databases allows high-confidence data inheritance among these resources, consequently enhancing computational analyses of new data. These collaborations provide a model for the integration of biological knowledge represented in model organism databases with other genome informatics resources for which genome sequence data are available or rapidly emerging.
METHODS
Connecting the RIKEN Mouse Genome Encyclopedia to MGI and LocusLink
Overview
Although steps were taken to reduce clone redundancy, many of the sequences from the FANTOM2 effort were from the same gene (Carninci et al. 2003). A combination of computational and manual inspection approaches was necessary to associate the FANTOM2 sequences to genes represented in the MGI and LocusLink databases. The sequences were clustered to identify groups of clones that potentially represent the same gene, and to reduce the number of alignments requiring manual inspection. Clusters containing sequences in MGI were identified (MGI-associated clusters), and then sequence comparison and graphical cluster views were used to establish cluster-to-gene relationships. Once cluster-to-gene relationships were established, groups of clustered clones could be represented consistently in the MGI and LocusLink databases (Fig. 1). Sequence clustering and cluster triage methods are described below. Computational analyses of FANTOM2 data, including sequence clustering, were performed in January 2002. Curatorial analyses of FANTOM2 clusters and functional annotations were performed prior to public release of FANTOM2-new sequences.
Sequence Clustering
Redundancy in the FANTOM2 data set was detected by three independent sequence clustering builds generated by the RIKEN Genome Exploration group, the NCBI UniGene group (UniGene Build Mm.98), and The Institute for Genomic Research (TIGR), respectively. The RIKEN Genome Exploration group generated a cluster build using only the 60,770 FANTOM2 cDNA sequences with their unpublished clustering algorithm. The NCBI UniGene and TIGR Gene Index groups clustered the FANTOM2 clone sequences together with all mouse cDNAs and ESTs available from public sequence databases. The clustering methods are described elsewhere (Okazaki et al. 2002.) Identifiers were assigned to the clusters from each build (including singletons), and a cluster union ID was generated by computing the union of clones from all three groups (Figs. 2, 3). FANTOM2 clusters that incorporated at least one sequence associated with an MGI gene (including FANTOM1 sequences or non-RIKEN sequences) were considered MGI-associated clusters, and cluster unions that contained any MGI-associated clusters were considered MGI-associated cluster unions (Table 1).
Cluster Triage
The process of cluster triage involved the evaluation of cluster integrity (i.e., the likelihood that all members of the cluster are derived from the same gene). Because the clustering parameters and starting sequence sets were different for each build, a given clone could be grouped with different sequences by the three builds (although each clone is included in only one cluster from each build). Our curation strategy was based on the assumption that clusters of FANTOM2 cDNA clones grouped consistently by the three independent cluster builds have high cluster integrity and should require comparatively less curatorial analysis than do more complex clusters. An extension of this idea is that clusters that consistently represent the same MGI gene over the three independent cluster builds (by incorporation of sequences that are associated with that MGI gene), or even the same set of MGI genes, also have higher integrity. To use cross-cluster consistency of either FANTOM2 clone or MGI gene representation for targeting higher confidence clusters, we developed a system to label (or code) clones according to this consistency (Table 2). Figure 2 shows an example of how these consistency codes were assigned to clones.
RIKEN and MGI Status Codes
Status codes are the indicators of consistency across clusters from the different cluster builds, and they convey a measure of sequence grouping confidence. The codes are three-character identifiers assigned to each FANTOM2 sequence and indicate either the coincidence of FANTOM2 clone representation over the three different clusters containing a given clone (RIKEN status code), or the coincidence of MGI gene representation over the three cluster views for that clone (MGI status code). If the number and identities of all FANTOM2 sequences were the same in all three cluster views containing a given FANTOM2 sequence, then the value of the RIKEN status code was set to RNT (equal representation in RIKEN, NCBI's UniGene, and TIGR clusters containing that sequence). If the cluster views for a sequence were identical in two of the three cluster builds with respect to FANTOM2 clone number and identity, then the RIKEN status code values were either -NT, RN-, or R-T (equal representation in the two clusters abbreviated). If all three cluster views for a given sequence were different with respect to FANTOM2 clone number and/or identity, then the value of the RIKEN status codes for that sequence was set to “---” (Fig. 2). MGI gene representation was determined for each cluster from sequence-to-gene associations in MGI for sequences in the clusters, and MGI gene relationships for both RIKEN and non-RIKEN sequences in the clusters were considered. The five values for MGI status codes are identical to those for the RIKEN status codes in structure and in meaning, except that they refer to MGI gene representation in the clusters (Figs. 2, 3). If no MGI genes are represented across clusters containing the same clone, this is not considered equivalent for the MGI Status Code.
Number Codes
Number codes were established to communicate the total number of either FANTOM2 clones (RIKEN number code) or MGI genes represented (MGI number code) in each cluster view (RIKEN, NCBI UniGene, and TIGR) for a given clone. Number codes contain three digits, where the first, second, and third digits indicate the total number of FANTOM2 clones (for RIKEN number) or MGI genes represented (for MGI number) in the RIKEN, UniGene, and TIGR cluster views, respectively (Figs. 2, 3). In some cases, the cluster builds failed to group all sequences for the same gene into the same cluster, because the overlapping regions were short or flanked by introns. To find relationships between FANTOM2 sequences and MGI genes not detected by clustering, BLAST was performed between the 60,770 FANTOM2 cDNAs and all DDBJ/EMBL/GenBank sequences associated with a single gene in MGI (RepeatMasker was used to mask the FANTOM2 sequences [options: -mus -xsmall -nolow], and WU-BLAST 2.0 BLASTN was used for the alignments [options: M = 1 N = -2 Q = 2 R = 2 S = 100 s2 = 25 -filter=none -lcmask]). To relate FANTOM2 sequences that had sequence similarity to overlapping public sequence database sequences from the BLAST results, but were not part of the same cluster union, we encoded BLAST (BLASTN) results such that all clones that hit any combination of overlapping DDBJ/EMBL/GenBank IDs (P score ≤ 1e - 50) were assigned the same BLAST group ID.
Visualizing Clusters and Annotations of FANTOM2 Clones
Web-based graphical displays of clone annotations and sequence clusters were developed by RIKEN (Okazaki et al. 2002). To support cluster triage, manual cluster curation, and MGI integration, a software system was developed by the MGI software group to track and query information required for and derived from our curatorial process. The system had two components: a relational database (implemented in Sybase) that was used to store information about the clones, and a graphical user interface (implemented in TeleUse, a motif-based user interface management system) that was used to display, edit, and query the database. The database supported the association of each sequence with the following attributes: sequences e.g., length, quality), clusters (cluster IDs, status, and number codes), MGI gene associations, curated cluster grouping, and nomenclature. Data from all 60,770 FANTOM2 clones were entered into the database. To view the context of these sequences with other mouse cDNAs and ESTs, additional records were created for 16,557 public database sequences that were members of UniGene and TIGR Mouse Gene Index clusters that had at least one FANTOM2 clone sequence. MGI gene associations for sequences in the table were updated weekly.
Sequence Cluster Curation
To evaluate the quality of sequence clusters and to determine if each cluster represented a single gene, curators evaluated sequence alignments of the clustered clones, as well as BLAST results of FANTOM2 sequences searched against public sequence databases and the publicly available mouse genome draft assembly (Mouse Genome Sequencing Consortium versions 2 and 3). The BLAST results of the clone sequences searched against the draft mouse genome were helpful in identifying partially processed mRNAs and were often essential for the resolution of clusters with complicated sequence alignment patterns.
Clone orientation, repetitive sequence, transcript processing, and the distributions of sequence mismatches were among the factors considered in evaluating the quality of each cluster. FANTOM2 cDNAs from the same unit of transcription that varied due to alternative, partial, or intron-containing transcripts were considered to represent the same gene. Occasionally, sequences from different genes were grouped together into a computed cluster. This occurred for several reasons, including (1) a limitation of the clustering parameters (as in the case of clustered paralogs), (2) a consequence of legitimate biological overlap among transcripts (as in the case of some closely linked genes), (3) read-through transcripts, or (4) artifact (chimeric clones). To record the curated associations between sequences in manually inspected clusters, the same “final cluster” identifier was associated with all legitimate cluster members. To maintain the curated associations between clones and existing MGI genes, the appropriate existing MGI gene ID was used as the final cluster identifier.
Curators selected a single FANTOM2 sequence to serve as the representative sequence for each resolved cluster. This selection was based on the length of the predicted coding sequence (CDS), sequence quality (measured by Phred score), and sequence length. If the best sequence in the cluster were a non-FANTOM2 sequence, then curators recorded the best overall sequence in addition to selecting the “best” FANTOM2 sequence. Information about the most-representative sequence in a cluster was used by the FANTOM2 group in construction of a mouse RTPS (Okazaki et al. 2002), as well as feedback for their clustering algorithm, which in addition to automated sequence similarity grouping also predicted a representative sequence for each cluster. Sequences found to be problematic in some way (for example, chimeric, intron-containing, frame-shifted CDS, poor quality sequence, uncharacterizable) were recorded by the MGI curators. Sequences with the annotated status of uncharacterizable were usually part of complicated alignments, in which the sequence relationship between cluster members was unclear.
Functional Annotation of FANTOM2 Clones
The RIKEN group developed a Web interface for clone annotation (Okazaki et al. 2002). This interface allowed a worldwide team of annotators to login and visualize the results of the FANTOM2 clone annotation pipeline, and to register curated confirmations or changes to these automated annotations. The three objectives for clone annotation were (1) to choose a functionally relevant name for each FANTOM2 clone, (2) to select the most likely coding sequence (CDS) region for each protein-coding clone and confirm that automated GO annotations associated with the sequence were consistent with the CDS region selected, and (3) to annotate the status of each clone with respect to various features of sequence and clone quality in the context of relative biological orientation (such as frame shifts, truncations, intron presence).
Sequence-to-Gene Associations
Because the FANTOM2 data set contains all FANTOM1 clones (which were associated with MGI genes previously) and because some new FANTOM2 clones represent known mouse genes, there is extensive redundancy between the FANTOM2 data and MGI genes. For MGI-curated FANTOM2 clusters, both cluster integrity and redundancy with existing MGI genes were considered. FANTOM2 sequences were associated to MGI genes, either by direct sequence-gene associations or, if the sequence is a “Problem Sequence”, by molecular segment-gene relationships (see next section). New sequence information often leads to reinterpretation of gene models in MGI, which are corrected by changes in data representation (i.e., gene merges, splits and renames, or reassociations of data from one gene to another). When MGI curators confirmed the need for updates to existing MGI data, changes were made as part of the load of FANTOM2 data into MGI. Given the volume of FANTOM2 data and the time required for manual curation, only half of the MGI-associated clusters were evaluated manually at the time the FANTOM2 data were released. For clusters not evaluated by MGI curators, sequence-to-MGI gene associations followed the clone-to-cluster relationships established either by an unpublished RIKEN clustering algorithm or by other FANTOM2 annotators, although MGI gene record merges and splits were not processed for these clusters.
Prior to loading the FANTOM2 data, additional sequence similarity searches were performed against mouse EST sequences in MGI to avoid creating gene record redundancy. In addition, to synchronize NCBI gene model annotations with LocusLink and MGI gene records, curators at LocusLink reviewed inconsistencies involving FANTOM2 sequences from their ongoing mouse genome annotation efforts. Following the load, NCBI computed reannotations of its mouse genome assembly.
Data Integration and Representation
Molecular Segments
Clones are represented in the MGI database as unique database objects called molecular segments. When possible, relationships between molecular segments and gene records are established. Attributes of molecular segment objects include molecular source information (e.g., clone library name, mouse strain, tissue, age), sequence accession IDs, other foreign database accession IDs, and references. Molecular Segment objects were created in MGI for all 60,770 FANTOM2 cDNA clones (21,076 of which were created with the load of FANTOM1 data; Kawai et al. 2001) and associated with the corresponding molecular source information, DDBJ seqids, and references (Fig. 5).
Relationships were established between each FANTOM2 molecular segment object and its appropriate MGI gene object. With few exceptions, FANTOM2 molecular segments are associated with a single MGI gene. The exceptions come from FANTOM2 clones that overlap more than one gene, as a consequence of either a normal cellular phenomenon (e.g., an antisense transcript) or an artifact (e.g., chimeric clones or unprocessed read-through transcripts). For sequences judged to have a problem (“problem sequences” e.g., chimeric clones, intron-containing clones, clones with poor quality sequence information), a note was attached to the molecular segment records of these FANTOM2 clones (Fig. 3). Because short FANTOM2 clones (<300 bp) are less likely to represent full-length cDNAs, a note was attached to the molecular segment records of such clones to make this information accessible to users (Fig. 3).
Chromosome Assignments
FANTOM2 cDNAs were compared to the mouse whole genome assembly (Okazaki et al. 2002) to place the genes represented by the cDNAs on the appropriate chromosome. For MGI gene records associated with FANTOM2 data, this map position was accepted when no mapping ambiguity was evident. In cases of mapping conflict between members of the same multiple sequence FANTOM2 cluster, or if the sequence(s) for the novel gene did not map to the assembly, a chromosome value of “unknown” was used for novel FANTOM2 genes. For FANTOM2 sequences associated with existing MGI genes that have a map position, the chromosome value derived from assembly mapping of FANTOM2 sequences was incorporated only if it did not conflict with the chromosome values in MGI for those genes. LocusLink used a similar approach with alignments to the MGSC assembly (version 3) or to finished BACs computed at NCBI.
GO Annotation
MGI gene records associated with FANTOM2 sequences inherited GO annotations electronically if the associated FANTOM2 sequences (1) encode InterPro domains with GO term relationships, (2) show significant sequence similarity to SWISS-PROT/TrEMBL (SPTR) proteins with GO term relationships, or (3) show significant similarity to paralogous sequences associated with other MGI gene records annotated to GO terms. All GO annotations derived from expert curation were loaded.
Nomenclature and Orthology
MGI is the authoritative source of official genetic nomenclature for the mouse. The LocusLink and RefSeq groups at NCBI incorporate official nomenclature when available. It is the policy of mouse, rat, and human international gene nomenclature committees to coordinate the names of orthologous mammalian genes (Maltais et al. 2002; Wain et al. 2002). When possible, official nomenclature for novel MGI gene records from the FANTOM2 load, and for existing gene records with uninformative names, was mined from informative official nomenclature from orthologs (usually human). Otherwise, the nomenclature for new FANTOM2 gene records followed the structure used for novel FANTOM1 genes (gene symbol: RIKEN cloneID”Rik”, gene name: “RIKEN cDNA” RIKEN cloneID “gene”, e.g., symbol: 0910001B06Rik, name: RIKEN cDNA 0910001B06 gene; Kawai et al. 2001). If the nomenclature of a novel or existing MGI gene were influenced by the nomenclature of a mammalian ortholog, then an ortholog record was established in MGI (mammalian homology record). Some curated ortholog records were established in MGI without nomenclature updates (Table 4).
Genes Versus TUs
The FANTOM2 Consortium adopted an algorithmic definition for the sequences of a transcribed genomic region, which was designated a TU (Okazaki et al. 2002). A TU is intended to encompass all overlapping transcripts derived from the same strand of a transcribed region. This definition is similar to the definition used in MGI for genes that have sequence information; however, there are a few notable differences. TU boundaries are defined by transcript boundaries; thus, TUs do not include regulatory sequences that lie outside of the transcribed region. In addition, if two closely linked genes are transcribed from the same strand, and if an occasional unprocessed transcript from the upstream gene reads through into the downstream gene, then these two genes are grouped into the same TU. In MGI, although genes can be defined in the classical genetic sense, typically they are defined as the region necessary and sufficient to express the complete set of products derived from a unit of transcription. In general, this definition is similar to that of the HUGO Gene Nomenclature Committee (HGNC; Wain et al, 2002), who consider a unit of transcription to be a transcribed region of the genome in which transcription products share at least part of one exon, and if the shared exon encodes protein, then the transcription products share a reading frame in this exon. Automatic identification of gene features via the NCBI annotation pipeline also combines evidence from multiple transcripts into one feature when exon or intron boundaries are shared. There are strong similarities between the definitions of an MGI gene and a FANTOM2 TU, and most cases of alternative overlapping, FANTOM2 transcripts are represented equivalently in MGI and in the FANTOM2 RTPS. An important implementation difference between RTPS TUs and MGI genes is in the interpretation of valid antisense transcripts. Separate MGI gene records and RTPS TUs are recognized for overlapping transcripts that initiate from opposite strands. For the RTPS, overlapping FANTOM2 clones with opposing sequence orientation were split into separate “antisense” TUs. High confidence in the cloning orientation of FANTOM2 cDNA clones validated this step. The criteria for MGI to create separate gene records for overlapping sequences with opposing orientations is more strict (i.e., MGI requires supporting evidence that a transcript has originated from the opposite strand, e.g., the lack of shared transcript processing junctions and termini between sense and antisense candidates, or published accounts of antisense function).
Coordination Between MGI and LocusLink/RefSeq
MGI and LocusLink coordinate their respective representations of mouse genes through daily and weekly data exchanges and personal communications. From MGI, LocusLink downloads MGI gene accession identifiers, official mouse gene nomenclature, alternative names, gene-to-sequence associations, chromosome assignments for mouse genes, homology reports, marker reports, links to the Gene Expression Database, and functional annotation based on GO terms. Some of these data are used to create Entrez LinkOut files for MGI, to integrate MGI homology data into HomoloGene, and to support Web links from UniGene, marker, and map resources back to MGI. The LocusLink/RefSeq group assimilates the nomenclature and sequence data and uses them as frames of reference for analysis of new records from DDBJ/EMBL/GenBank, of current UniGene clusters, for placement on the genomic assembly and for comparison to nomenclature or gene structure in other genomes. If conflicts are identified, MGI and LocusLink staff work cooperatively to resolve them. Representative transcript and predicted protein sequences then are selected from each gene to be instantiated as RefSeq accessions. Whenever possible, C57BL/6J sequences are selected as the source for these records.
From LocusLink, MGI receives LocusIDs, RefSeq, and DDBJ/EMBL/GenBank sequence identifiers, and proposed nomenclature for novel genes and additional gene-to-sequence associations for genes already in the MGI database. MGI also receives official human gene nomenclature for human genes from LocusLink. A system of data integrity checks identifies data from either resource that would result in inconsistencies, such as a transcript-derived nucleotide accession being associated with more than one gene. Frequent personal correspondence between MGI and LocusLink staff resolve conflicting data representations as they arise and help maintain high levels of coordination and data quality. MGI participates in a similarly robust exchange of mouse protein sequence data with SWISS-PROT to provide curated associations of gene records in MGI to SWISS-PROT, InterPro protein domains, and additional gene-to-sequence associations. The continuous reconciliation of information among MGI, LocusLink/RefSeq, and SWISS-PROT is a foundation for the connections between MGI gene records and the records of other MGI collaborators, such as UniGene and Ensembl (Fig. 4B).
Linking the Mouse Genome Sequence to MGI and LocusLink
One of the primary objectives of the MGI and NCBI groups relative to the Mouse Genome Sequencing initiative is to make it easy for researchers to navigate from computationally annotated views of the mouse genome sequence to manually annotated and curated annotations about expression, function, phenotype, homology, etc., that are available in the MGI database. Because the RefSeq (Pruitt and Maglott 2001) resource at NCBI is used as one of the primary data sets in different genome annotation pipelines and because RefSeq sequence identifiers have curated associations with gene records in both LocusLink and MGI, computationally based gene models associated with a RefSeq sequence are associated easily with equivalent gene objects in both the LocusLink and MGI resources. Because the MGI group also works collaboratively with SWISS-PROT curators to validate protein sequence and gene associations, gene models from genome annotation pipelines that use mouse protein sequences from SWISS-PROT can also be associated easily with the appropriate records in LocusLink and MGI.
Tab-delimited files of the curated relationships between accessioned objects from DDBJ/EMBL/GenBank, RefSeq, LocusLink, SWISS-PROT, and MGI are available from the MGI FTP site (ftp://ftp.informatics.jax.org/pub/informatics/reports). These files are updated nightly. After automated integration, LocusLink/RefSeq generates similar tab-delimited files (ftp://ftp.ncbi.nih.gov/RefSeq/LocusLink) each morning. In addition to the curated records, these files also include the accessions (predicted mRNAs and proteins) from NCBI's annotation of the mouse genome. These association files can then be used by the mouse genome annotation groups to compare and connect computational gene models that represent known genes to the detailed biological knowledge of the curated genes represented in LocusLink and MGI. Links from the computational gene models to LocusLink and MGI are currently available from the genome browsers at NCBI, the EMBL-EBI and Sanger Institute, and the University of California at Santa Cruz.
Acknowledgments
We wish to acknowledge all members of the MGI group. The integration of mouse biological data in MGI is possible only through their cooperative and dedicated efforts. We thank Leah Rae Donahue and Stanton Short for access to the Sfxn1f mutant mouse and mouse photography, respectively. The Mouse Genome Sequencing (MGS) Project, The Mouse Genome Database (MGD), the Gene Expression Database (GXD), and The GO project are components of the MGI database system. M.G.S. is supported by DOE grant FG02-99ER62850. M.G.D. is supported by National Human Genome Research Institute grant HG-00330. G.X.D. is supported by National Institute of Child Health and Human Development grant HD-33745. The GO project is supported by National Human Genome Research Institute grant HG-002273.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.991003.
Footnotes
The Mouse Genome Informatics group at The Jackson Laboratory is a consortium of multiple investigators who work cooperatively to provide a comprehensive information resource on the genetics, genomics, and biology of the laboratory mouse.
Since that time, RefSeq has added two other accession types for mouse, NR_123456 for noncoding RNAs and NG_123456 for genomic segments (primarily for pseudogenes).
We note that the total number of genes annotated by NCBI's computed annotation of the mouse assembly sequence (Build 2) is 36,976.
In 228 cases, RefSeq or SWISS-PROT sequences that correspond to single curated genes in LocusLink and MGI were associated with more than one gene model from the first Ensembl annotation of the assembly. These may represent instances of paralogs, and are being targeted for manual curation.
References
- Ashburner, M., Ball, C.A., Blake, J.A., Botstein, D., Butler, H., Cherry, J.M., Davis, A.P., Dolinski, K., Dwight, S.S., Eppig, J.T., et al. d2000. Gene ontology: Tool for the unification of biology: The Gene Ontology Consortium. Nat Genet. 25: 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake, J.A., Richardson, J.E., Bult, C.J., Kadin, J.A., Eppig, J.T., and the Mouse Genome Database Group. 2002. The Mouse Genome Database (MGD): The model organism database for the laboratory muse. Nucleic Acids Res. 30: 113-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult, C.J., Richardson, J.E., Blake, J.A., Kadin, J.A., Ringwald, M., Eppig, J.T., and the Mouse Genome Database Group. 2000. Mouse genome informatics in a new age of biological inquiry. Proceedings of the IEEE International Symposium on Bio-Informatics and Biomedical Engineering pp. 29-32. IEEE Computer Society, Los Alamitos, California.
- Carninci, P., Waki, K., Shiraki, T., Konno, H., Shibata, K., Itoh, M., Aizawa, K., Arakawa, T., Ishii, Y., Sasaki. D., et al. 2003. Targeting a complex transcriptome: The construction of the mouse full-length cDNA encyclopedia. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Fleming, M.D., Campagna, D.R., Haslett, J.N., Trenor III, C.C., and Andrews, N.C. 2001. A mutation in a mitochondrial transmembrane protein is responsible for the pleiotropic hematological and skeletal phenotype of flexed-tail (f/f) mice. Genes & Dev. 15: 652-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger, E., Jung, E., and Bairoch, A. 2001. SWISS-PROT: Connecting biological knowledge via a protein database. Curr. Issues Mol. Biol. 3: 47-55. [PubMed] [Google Scholar]
- Hubbard, T., Barker, D., Birney, E., Cameron, G., Chen, Y., Clark, L., Cox, T., Cuff, J., Curwen, V., Down, T., et al. 2002. The Ensembl database genome database project. Nucleic Acids Res. 30: 38-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, H.R., Mixter, R., and Permar, D. 1933. Flexed tail in the mouse, Mus musculus. Genetics 18: 335-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai, J., Shinagawa, A., Shibata, K., Yoshino, M., Itoh, M., Ishii, Y., Arakawa, T., Hara, A., Fukunishi, Y., Konno, H., et al. 2001. Functional annotation of a full-length mouse cDNA collection. Nature 409: 685-690. [DOI] [PubMed] [Google Scholar]
- Kent, J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., and Haussler, D. 2002. The human genome browser at UCSC. Genome Res. 12: 996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Maltais, L.J., Blake, J.A., Chu, T., Lutz, C.M., Eppig, J.T., and Jackson, I. 2002. Rules and guidelines for mouse gene, allele, and mutation nomenclature: A condensed version. Genomics 79: 471-474. [DOI] [PubMed] [Google Scholar]
- Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., Nikaido, I., Osato N., Saito, R., Suzuki, H., et al. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563-573. [DOI] [PubMed] [Google Scholar]
- Pruitt, K.D. and Maglott, D.R. 2001. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 29: 137-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringwald, M., Eppig, J.T., Begley, D.A., Corradi, J.P., McCright, I.J., Hayamizu, T.F., Hill, D.P., Kadin, J.A., and Richardson, J.E. 2001. The Mouse Gene Expression Database (GXD). Nucleic Acids Res. 29: 98-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausberg, R.L., Feingold, E.A., Klausner, R.D., and Collins, F.S. 1999. The Mammalian Gene Collection. Science 286: 455-457. [DOI] [PubMed] [Google Scholar]
- Wain, H.M., Lovering, R.C., Bruford, E.A., Lush, M.J., Wright, M.W., and Povey, S. 2002. Guidelines for human gene nomenclature. Genomics 79: 464-470. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.ncbi.nih.gov/; National Center for Biotechnology Information.
- ftp://ftp.informatics.jax.org/pub/reports/MGI_ProblemSequence.rpt; Mouse Genome Informatics FTP site.
- ftp://ftp.informatics.jax.org/pub/informatics/reports; Mouse Genome Informatics FTP site.
- ftp://ftp.ncbi.nih.gov/refseq/LocusLink/; LocusLink FTP site.
- http://www.ncbi.nih.gov/mapview/; National Center for Biotechnology Information Map Viewer.
- http://www.ensembl.org; EMBL-EBI and Sanger Institute's Ensembl.
- http://www.genome.ucsc.edu; University of California at Santa Cruz's genome browser.