Abstract
We report a comprehensive statistical analysis of data on 58 DNA markers (mitochondrial [mt], Y-chromosomal, and autosomal) and sequence data of the mtHVS1 from a large number of ethnically diverse populations of India. Our results provide genomic evidence that (1) there is an underlying unity of female lineages in India, indicating that the initial number of female settlers may have been small; (2) the tribal and the caste populations are highly differentiated; (3) the Austro-Asiatic tribals are the earliest settlers in India, providing support to one anthropological hypothesis while refuting some others; (4) a major wave of humans entered India through the northeast; (5) the Tibeto-Burman tribals share considerable genetic commonalities with the Austro-Asiatic tribals, supporting the hypothesis that they may have shared a common habitat in southern China, but the two groups of tribals can be differentiated on the basis of Y-chromosomal haplotypes; (6) the Dravidian tribals were possibly widespread throughout India before the arrival of the Indo-European-speaking nomads, but retreated to southern India to avoid dominance; (7) formation of populations by fission that resulted in founder and drift effects have left their imprints on the genetic structures of contemporary populations; (8) the upper castes show closer genetic affinities with Central Asian populations, although those of southern India are more distant than those of northern India; (9) historical gene flow into India has contributed to a considerable obliteration of genetic histories of contemporary populations so that there is at present no clear congruence of genetic and geographical or sociocultural affinities.
India has served as a major corridor for the dispersal of modern humans (Cann 2001). The date of entry of modern humans into India remains uncertain. By the middle Paleolithic period (50,000-20,000 years before present [ybp]), humans appear to have spread to many parts of India (Misra 1992). The migration routes of modern humans into India remain enigmatic, and whether there were also returns to Africa from India/Asia is unclear (Maca-Meyer et al. 2001; Roychoudhury et al. 2001; Cruciani et al. 2002). Contemporary ethnic India is a land of enormous genetic, cultural, and linguistic diversity (Karve 1961; Beteille 1998; Majumder 1998). The people of India are culturally stratified as tribals, who constitute 8.08% of the total population (1991 Census of India), and nontribals. There are ∼450 tribal communities in India (Singh 1992), who speak ∼750 dialects (Kosambi 1991) that can be classified into one of the following three language families: Austro-Asiatic (AA), Dravidian (DR), and Tibeto-Burman (TB). Most contemporary nontribal populations of India belong to the Hindu religious fold and are hierarchically arranged in four main caste classes, namely, Brahmin (priestly class), Kshatriya (warrior class), Vysya (business class), and Sudra (menial labor class). In addition, there are several religious communities, who practice different religions, namely, Islam, Christianity, Sikhism, Judaism, and so on. The nontribals predominantly speak languages that belong to the Indo-European (IE) or Dravidian families. The IE and DR groups have been the major contributors to the development of Indian culture and society (Meenakshi 1995). Indian culture and society are also known to have been affected by multiple waves of migration and gene flow that took place in historic and prehistoric times (Ratnagar 1995; Thapar 1995). In a recent study conducted on ranked caste populations sampled from one southern Indian State (Andhra Pradesh), Bamshad et al. (2001) have found that the genomic affinity to Europeans is proportionate to caste rank—the upper castes being most similar to Europeans, particularly East Europeans, whereas the lower castes are more similar to Asians. These findings are consistent with the migration of IE groups into India, the establishment of the caste system, and subsequent recruitment of indigenous people into the caste fold. Because the Indian samples for this study were drawn from one geographical area, whether we can safely generalize these findings needs to be investigated.
The tribals are possibly the original inhabitants of India (Thapar 1966; Ray 1973), although their evolutionary histories and biological contributions to the nontribal populations have been debated (Risley 1915; Guha 1935; Sarkar 1958). Therefore, it is crucial to carry out genetic investigations in geographically and culturally disparate, but ethnically well-defined, populations, using data on a uniform set of mitochondrial (mt), Y-chromosomal, and autosomal DNA markers. Unfortunately, the vast majority of earlier studies on Indian populations have been conducted on ethnically ill-defined populations or have been restricted to a single geographical area or a single set of markers—primarily either mitochondrial or Y-chromosomal (e.g.,Kivisild et al. 1999a; Bamshad et al. 2001). The objectives of the present study are to (1) provide a comprehensive view of genomic diversity and differentiation in India, and (2) to draw inferences on the peopling of India, and the origins of the ethnic populations, specifically in relation to the various competing hypotheses, such as whether the Austro-Asiatic or the Dravidian-speaking tribal groups were the original inhabitants of India (Risley 1915; Guha 1935; Sarkar 1958).
We analyzed genetic variation in 44 geographically, linguistically, and socially disparate ethnic populations of India (Table 1). These include 10 restriction site polymorphisms (RSPs), one insertion/deletion (InDel) polymorphism, and hypervariable segment 1 (HVS1) sequences on mtDNA; 11 RSPs, 1 InDel, and 10 short tandem repeat (STR) loci on Y-chromosomal DNA; and 8 InDel and 17 RSPs on autosomal DNA.
Table 1.
Names of Study Populations, Sample Sizes, Linguistic, and Ethnological Information
|
Sample size
|
||||||
|---|---|---|---|---|---|---|
|
mt
|
||||||
| Population namea[code] | RSP | HVS1 seq | Y | Autosomal | Linguistic affiliation | Social category |
| 1. Agharia [AGH]3 | 24 | 10 | 9 | 24 | Indo-European | Middle caste |
| 2. Ambalakarer [AMB]4 | 30 | 10 | 18 | 50 | Dravidian | Middle caste |
| 3. Bagdi [BAG]3 | 31 | 10 | 11 | 31 | Indo-European | Lower caste |
| 4. Chakma [CHK]2 | 10 | 10 | 4 | 10 | Tibeto-Burman | Tribe |
| 5. Chamar [CHA]1 | 25 | 10 | 18 | 25 | Indo-European | Lower caste |
| 6. Gaud [GAU]3 | 13 | 10 | 4 | 15 | Indo-European | Middle caste |
| 7. Gond [GND]6 | 51 | 10 | Dravidian—Gondi dialect | Tribe | ||
| 8. Halba [HAL]6 | 47 | 20 | 20 | 48 | Indo-European Primarily Marathi | Tribe |
| 9. Ho [HO]3 | 54 | 10 | 20 | 54 | Austro-Asiatic | Tribe |
| 10. Irula [ILA]4 | 30 | 14 | 18 | 50 | Dravidian | Tribe |
| 11. lyengar [IYN]4 | 30 | 10 | 20 | 51 | Dravidian | Upper caste |
| 12. lyer [IYR]4 | 30 | 10 | 20 | 50 | Dravidian | Upper caste |
| 13. Jamatiya [JAM]2 | 55 | 10 | 16 | 55 | Tibeto-Burman | Tribe |
| 14. Jat Sikh [JSK]1 | 48 | 15 | Indo-European | Middle caste | ||
| 15. Kamar [KMR]6 | 54 | 10 | 19 | 57 | Dravidian | Tribe |
| 16. Khatris [KHT]1 | 48 | 15 | Indo-European | Middle caste | ||
| 17. Konkan Brahmins [KBR]5 | 31 | 10 | Indo-European | Upper caste | ||
| 18. Kota [KOT]4 | 30 | 25 | 15 | 45 | Dravidian | Tribe |
| 19. Kurumba [KUR]4 | 30 | 10 | 18 | 54 | Dravidian | Tribe |
| 20. Lodha [LOD]3 | 32 | 14 | 17 | 32 | Austro-Asiatic | Tribe |
| 21. Mahishya [MAH]3 | 33 | 10 | 9 | 34 | Indo-European | Middle caste |
| 22. Manipuri (Meitei) [MNP]2 | 11 | 9 | Tibeto-Burman | Upper caste | ||
| 23. Maratha [MRT]5 | 41 | 10 | Indo-European | Middle caste | ||
| 24. Mizo [MZO]2 | 29 | 14 | 20 | 29 | Tibeto-Burman | Tribe |
| 25. Mog [MOG]2 | 25 | 10 | 6 | 25 | Tibeto-Burman | Tribe |
| 26. Munda [MUN]3 | 7 | 6 | 49 | Austro-Asiatic | Tribe | |
| 27. Muria [MUR]6 | 30 | 12 | 8 | 28 | Dravidian—Gondi dialect | Tribe |
| 28. Muslim [MUS]1 | 28 | 10 | 19 | Indo-European | Islamic religious group | |
| 29. Naba-Baudh [NBH]5 | 40 | 10 | Indo-European | Lower caste (recently adopted Buddhism) | ||
| 30. Pallan [PLN]4 | 30 | 10 | 15 | 50 | Dravidian | Lower caste |
| 31. Punjab Brahmins [PBR]1 | 48 | 12 | Indo-European | Upper caste | ||
| 32. Rajput [RAJ]1 | 51 | 10 | 35 | 52 | Indo-European | Middle caste |
| 33. Riang [RIA]2 | 51 | 12 | 17 | 50 | Tibeto-Burman | Tribe |
| 34. Santal [SAN]3 | 20 | 14 | 15 | 24 | Austro-Asiatic | Tribe |
| 35. Saryupari Brahmins [SBR]6 | 26 | 19 | Indo-European | Upper caste | ||
| 36. Scheduled caste-Punjab [SCH]1 | 48 | 15 | Indo-European | Lower caste | ||
| 37. Tanti [TAN]3 | 16 | 10 | 6 | 16 | Indo-European | Lower caste |
| 38. Tripperah (Tripuri) [TRI]2 | 51 | 20 | 17 | 50 | Tibeto-Burman | Tribe |
| 39. Toda [TOD]4 | 50 | 10 | 8 | 50 | Dravidian | Tribe |
| 40. Toto [TTO]3 | 30 | 20 | 12 | 30 | Tibeto-Burman | Tribe |
| 41. Uttar Pradesh Brahmins [UBR]1 | 27 | 10 | 17 | 27 | Indo-European | Upper caste |
| 42. Vanniyar [VAN]4 | 30 | 10 | 14 | 50 | Dravidian | Middle caste |
| 43. Vellala [VLR]4 | 43 | 10 | 16 | 43 | Dravidian | Middle caste |
| 44. West Bengal Brahmins [WBR]3 | 22 | 10 | 13 | 23 | Indo-European | Upper caste |
Geographical location: 1 = North, 2 = Northeast, 3 = East, 4 = South, 5 = West, and 6 = Central.
RESULTS
Distribution of mtDNA Lineages Indicates a Small Founding Group of Females
The HpaI np 3592 mtDNA restriction site locus was monomorphic in all populations. We observed 32 distinct 10-locus mtDNA RSP haplotypes (maximum number within a population = 13, for Rajput and Tipperah; minimum = 2, for Kota and Toda) among the 1490 individuals studied from the 44 populations, five (15.7%) of which were exclusive to the northeastern populations. (Haplotype frequencies are provided in Supplemental Table 1, available online at www.genome.org.) Although the frequency distributions of haplotypes among populations differed significantly (p < 0.05), one haplotype, belonging to haplogroup (HG) M, accounted for 46.4% of all mtDNA molecules. This modal haplotype in the pooled data set was also the modal haplotype in 34 (77%) of the 44 study populations. The 10 populations in which this haplotype was not the most frequent primarily comprised ethnic groups of the northern (Uttar Pradesh Brahmins, Punjab Brahmins, Rajput, Muslim) and the northeastern regions (Chakma, Jamatiya, Mog, Toto). In these populations, the modal haplotypes belong to non-M HGs, especially HG-U. However, even in these populations, the haplotype that was modal in the overall data set was generally the second most frequent haplotype. Thus, the distribution of mtDNA haplotypes shows that there is a strong uniformity of female lineages in India.
Among the 528 individuals for whom mtDNA HVS1 sequences were determined, the total number of distinct sequences observed was 323, of which 91 (28.2%) were shared by multiple individuals. In all, 298 (56.4%) individuals shared HVS1 sequences. Thus, there was a significant sharing both in the number of sequences and in the proportion of individuals sharing these sequences. This reinforces our earlier conclusion of uniformity of female lineages. The most parsimonious explanation of these findings is that there was a small number of founding female lineages in Indian populations. The small number of founding female lineages may have either resulted from a founder effect caused by a small number of women entering India or, possibly more likely, caused by the group of founding females, irrespective of the size of the group, being drawn from an ancestral population with a relatively homogeneous pool of mitochondrial haplotypes.
Haplogroup Distributions Are Largely Concordant With Gene Flow Into India and Provide Insights Into Population Structure
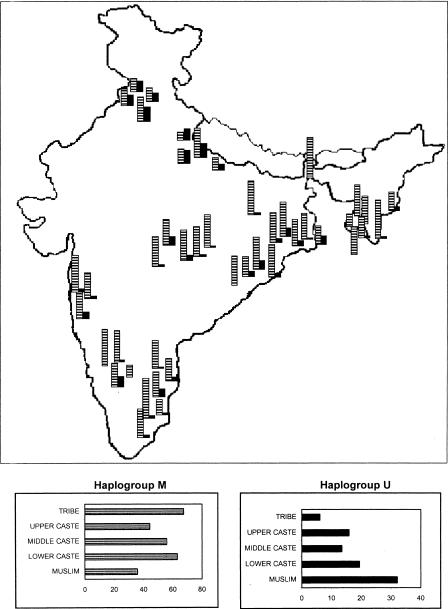
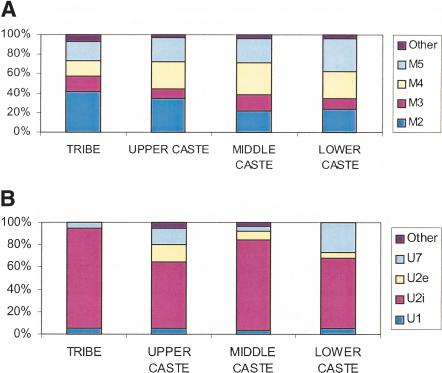
The frequencies of the most predominant mtDNA HGs in India, M and U, are roughly inversely correlated (Fig. 1). HG-M frequency is very high (overall 59.9%: range 18.5% [Brahmins of Uttar Pradesh] to 96.7% [Kota]), confirming that it is an ancient marker in India. HG-M frequency is the highest among tribal groups, particularly in the AA tribals. Among HG-M individuals, 98.22% belong to subHG-M*, defined by the presence of T at np 16,223. Figure 2A presents the frequencies of various known (Bamshad et al. 2001) subHGs of M* among different sociocultural categories. (Detailed data are given in Suppl. Tables 2-4.) Individuals belonging to subHG-M2 had the highest nucleotide diversity in HVS1, indicating that M2 may be the most ancient in India. It occurs in significantly higher (p < 0.05) frequencies among tribals (28%), particularly among the AA tribals (32%), than among castes (8.8%). Furthermore, the coalescent time of M2 found in India was estimated to be greater than most east Asian and Papuan branches of HG-M (Forster et al. 2001), indicating that India was settled early after humankind came out of Africa (Kivisild et al. 1999b). These findings imply that the contemporary tribals are descendants of the initial settlers. HG-U is a complex mtDNA lineage, whose age was estimated from our data to be 45,000 ± 25,000 years, not significantly different from an earlier estimate (Torroni et al. 1996). Its frequency is significantly (p < 0.001) higher among the IE-speaking caste groups, compared with other caste or tribal groups. Of particular interest are the frequencies of subHGs U2i (Indian-specific cluster of subHG U2 that predated the arrival of IE speakers from Central and West Asia into India; Kivisild et al. 1999a), U2e (Western-Eurasian cluster of U2), and U7 (an ancient Indian subHG). The frequencies of the subHGs of U are presented in Figure 2B. It is striking that the tribals do not possess U2e, and have the highest frequency of U2i. The gradations in frequencies (Figs. 1 and 2) of HG-M, particularly of subHG-M2, and also of HG-U, notably the absence of U2e among tribals, indicate that (1) tribals are more ancient than the castes,(2) there has been considerable admixture with Central and West Asians during the formation of the caste system, and (3) many new female lineages were introduced by the IE speakers.
Figure 1.
Frequencies (%) of mitochondrial haplogroups M (hatched) and U (solid black) in 44 ethnic populations, and among sociocultural groups of populations (insets).
Figure 2.
Frequencies (%) of subhaplogroups of (A) M and (B) U among tribal and ranked caste populations.
Because U2i appears to be the indigenous subHG of U in India, we sought to find phylogenetic relationships among the distinct HVS1 sequences within this subHG and their observed frequencies in various linguistic groups. The phylogenetic network (Bandelt et al. 1999) is complex (Suppl. Fig. 1). There is no major starlike cluster to indicate sudden population expansions, nor is there any clear sociolinguistic clustering of related sequences.
The frequency differences among ethnic groups of the YHGs are dramatic (Fig. 3). The tribals, irrespective of linguistic group, possess significantly lower (p < 0.0001) frequencies of HG-P*, which probably arose in Central Asia (Zerjal et al. 2002), than castes (Table 2). The distribution of HG-BR* frequencies, which is the most ancestral lineage in Europe (Rosser et al. 2000), is rather inconsistent with IE admixture. The tribals possess higher frequencies than castes. However, there are significant differences (p < 0.0001) in frequencies among tribals belonging to the TB (8%), AA (27%), and DR (65%) tribals. These inconsistencies and differences may be due to the fact that HG-BR* probably contains a heterogeneous set of chromosomes that are not closely related (Zerjal et al. 2002). We could not find phylogenetic clades within this HG. Dramatically higher frequencies of HG-K* were observed among TB (72%) and AA (52%) tribals, compared with other tribal or caste groups (Table 2). The age of this HG (∼20,000 yr) is 5000-15,000 yr higher than other HGs (Table 3). Contrary to Quintana-Murci et al.'s (2001) suggestion of a cline of HG-J frequencies from the Middle East (where this HG has high frequencies) into India, resulting from diffusion of people with agriculture, we do not find any clear cline with the addition of new data.
Figure 3.
Frequencies (%) of Y-chromosomal haplogroups among ethnic populations. (Population codes are given in Table 1.)
Table 2.
Frequencies of Y Haplogroups in Ethnic Populations of India Belonging to Various Sociolinguistic Groups
|
Haplogroup (%)
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Linguistic group | Social group | Sample size | P* | BR* | R1a | J | K* | L |
| Austro-Asiatic | Tribe | 52 | 7.7 | 28.8 | 0.0 | 13.5 | 50.0 | 0.0 |
| Dravidian | Tribe | 84 | 4.8 | 67.9 | 3.6 | 9.5 | 6.0 | 8.2 |
| Caste | 103 | 19.4 | 32.2 | 11.6 | 22.3 | 4.8 | 9.7 | |
| Tibeto-Burman | Tribe | 87 | 4.7 | 10.3 | 0.0 | 14.9 | 70.1 | 0.0 |
| Indo-European | Tribe | 19 | 0.0 | 36.8 | 15.8 | 21.1 | 26.3 | 0.0 |
| Caste | 122 | 22.1 | 32.8 | 23.8 | 12.3 | 6.6 | 2.4 | |
Table 3.
Estimated Ages of Various Y-Chromosomal Haplogroups
| Haplogroup | ||||||
|---|---|---|---|---|---|---|
| Estimate (yr) | P* | BR* | R1a | J | K* | L |
| Age | 16,322 | 12,508 | 5416 | 15,300 | 20,049 | 14,290 |
| Lower 95% Cl | 9477 | 7263 | 3145 | 8884 | 11,641 | 8297 |
| Upper 95% Cl | 29,979 | 22,974 | 9947 | 28,103 | 36,824 | 26,246 |
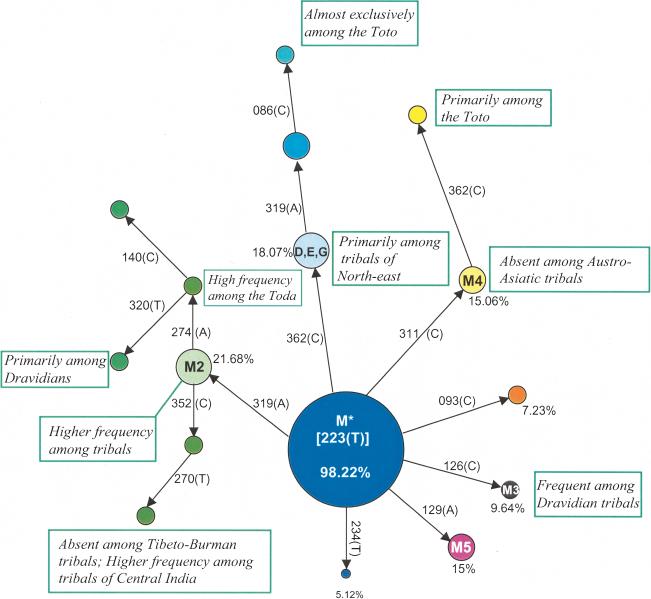
Anthropologists and historians have contended (Karve 1961; Kosambi 1991) that fission as a process has been very widespread in the formation of ethnic subgroups in India, resulting not only from pressures on natural resources but also because of social regulations. The common genetic consequences of fission are founder effect and drift, which are expected to result in high frequencies of haplotypes or nucleotide motifs in the daughter populations that are infrequent in the parental populations. Evidence of this is clearly seen from an evolutionary network, considerably simplified to highlight the principal features, of HVS1 sequences of individuals belonging to HG-M (Fig. 4). Many motifs occur exclusively or in high frequencies in some populations or groups or regions, which is consistent with fission and founder effect. Similarly, a motif GCGC at nps 16,051, 16,206, 16,230, and 16,311, respectively, was found in 16% of individuals belonging to subHG U2i. This motif occurs exclusively among tribal, middle-, and lower-caste populations, but not among the upper-caste populations. Evidence of fission and founder effect are also discernible from Y-STRP haplotype data: One 10-site haplotype is shared by several Kamar and Kota tribals, whereas another is shared by several Kurumbas. There are also large differences in the numbers of Y-STRP haplotypes and haplotype diversities among populations. (Detailed data are presented in Suppl. Tables 5 and 6.)
Figure 4.
Phylogenetic network of mtHVS1 sequences belonging to subhaplogroup M*, with frequency distributions of motifs in populations.
Austro-Asiatic-Speaking Tribals May Be the Earliest Inhabitants of India
Sociocultural and linguistic evidence indicates (Risley 1915; Thapar 1966; Pattanayak 1998) that the AA tribals are the original inhabitants of India. Some other scholars have, however, argued that tribal groups speaking DR and AA languages have evolved from an older original substrate of proto-Australoids (Keith 1936), whereas the TB tribals are later immigrants from Tibet and Myanmar (Guha 1935). Our findings strongly support the hypothesis that AA tribals are the earliest inhabitants of India. They possess the highest frequencies of the ancient east-Asian mtDNA HG-M and exhibit the highest HVS1 nucleotide diversity (Table 4). They also have the highest frequency of subHG M2 (19%), which had the highest HVS1 nucleotide diversity compared with other subHGs and therefore possibly the earliest settlers (the estimated coalescence time is 63,000 ± 6000 ybp; Kivisild et al. 1999a). Although all sociolinguistic groups seem to have undergone significant population expansions as evidenced by the unimodality of the HVS1 mismatch distributions (data not shown) and by the values of the relevant statistics (small values of the “raggedness” statistic and significantly large negative values of Fu's Fs statistic; Table 4), the AA tribals show the highest value of the estimated expansion time, ∼55,000 years, which is ∼15,000 years larger than the estimates for the other groups. Although we cannot be sure that this expansion took place in India, in conjunction with the other findings, it appears that this group of tribals may be the earliest inhabitants of India. A young subclade M4, with an estimated coalescence time of 32,000 ± 7500 ybp (Kivisild et al. 1999a), whose overall frequency is ∼15% in India, is completely absent among them. It is, therefore, likely that M4 arose after the expansion of the AA tribals and their entry into India.
Table 4.
Numbers of Polymorphic Sites, Nucleotide Diversities, Mismatch Statistics, Estimated Expansion Times, and Other Statistics Pertaining to Population Expansion Based on HVS1 Sequence Data Classified by Social and Linguistic Affiliation
|
Tribe
|
Caste
|
||||
|---|---|---|---|---|---|
|
Linguistic group
|
Linguistic group
|
||||
| Austro-Asiatic | Dravidian | Tibeto-Burman | Dravidian | Indo-European | |
| No. of sequences | 46 | 94 | 95 | 60 | 195 |
| No. of polymorphic sites | 57 | 55 | 74 | 63 | 115 |
| Nucleotide diversity (π) ± SD | 0.0224 ± 0.0059 | 0.0170 ± 0.0045 | 0.0173 ± 0.0046 | 0.0154 ± 0.0042 | 0.0176 ± 0.0046 |
| Mean no. of mismatches ± SD | 8.00 ± 1.89 | 6.07 ± 1.46 | 6.16 ± 1.48 | 5.51 ± 1.35 | 6.28 ± 1.50 |
| Expansion time (y) (range) | 54,656 (40,243−69,024) | 41,470 (30,488−52,439) | 42,085 (30,976−53,415) | 37,644 (27,439−47,683) | 42,905 (31,707−54,146) |
| Raggedness, r | 0.0199 | 0.0069 | 0.0200 | 0.0094 | 0.0097 |
| Fu's Fs (p value) | − 24.93 (0.0) | − 25.26 (0.0) | − 25.20 (0.0) | − 25.45 (0.0) | − 24.87 (0.0) |
The Northeastern Corridor May Have Served as a Major Passage of Entry Into India
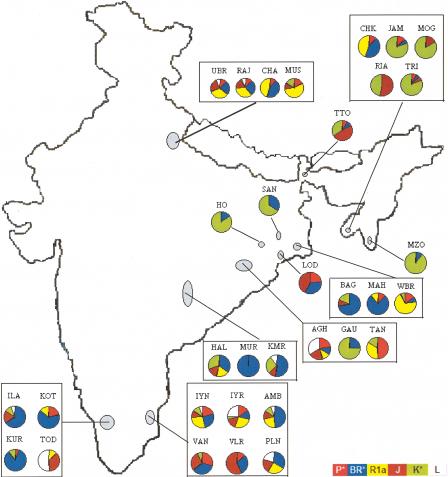
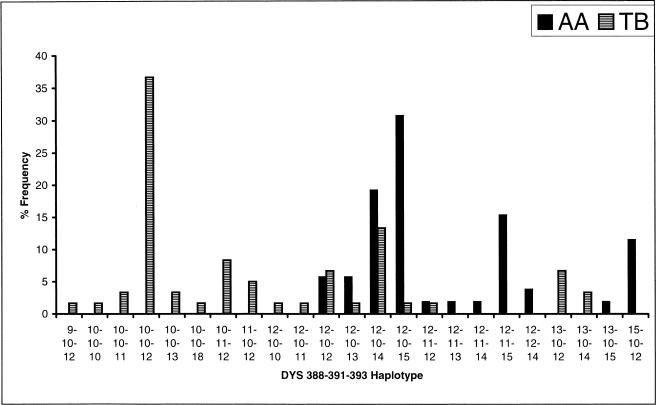
High frequencies of Y-HG K* (Fig. 3) are found among the TB populations, mainly confined to northeast India, and also among the Han Chinese (Su et al. 2000). The TB subfamily of the Sino-Tibetan language family has been subdivided (Grimes 1999) into four branches: Baric, Bodic, Burmese-Lolo, and Karen. Based on a study of Y-chromosomal haplotypes, Su et al. (2000) have contended that after the proto-Tibeto-Burman people left their homeland in the Yellow River basin, the Baric branch moved southward and peopled the northeastern Indian region after crossing the Himalayas. This branch did not possess the YAP insertion element, which has also not been found in any of the TB populations of India. Our findings, therefore, are consistent with Su et al.'s (2000) inference and indicate that the TB speakers entered India from the northeastern corridor. It is, however, surprising that the AA tribals, who primarily inhabit the eastern and central Indian regions, also possess high frequencies of Y-HG K* (Fig. 3). There is one major tribal group (Khasi) of northeastern India, who speak a dialect that belongs to the AA subfamily. Besides India, AA languages are spoken in south-east Asia. Thus, it is likely that a fraction of the AA tribals also entered India through the northeastern corridor. However, it does not seem that all of them have entered through this corridor. The expansion time of AA tribals was estimated to be ∼55,000 yr (Table 4), which is ∼13,000 yr greater than that estimated for TB tribals. But the age of the Y-HG K* estimated from the pooled variance of repeat numbers at the STRP loci among AA tribals (8500 yr) is about half of that estimated for TB tribals (15,000 yr). The AA tribals can also be clearly distinguished (estimated probability of correct identification = 0.8) from the TB tribals on the basis of haplotype frequencies at three Y-STRP loci (Fig. 5). We, therefore, believe that the ancestors of the present-day AA tribals in India initially entered India either through the northwestern corridor or through a southern exit route from Africa, and then later through the northeastern corridor during the Austronesian diaspora from southern China through southeast Asia to Papua New Guinea (Diamond 1997). It is possible that the ancestors of the AA speakers entered India through the northwest from out-of-Africa as they moved south of the Himalayas, and another ancestral group moved north of the Himalayas, settled in southern China, and then entered later through the northeast.
Figure 5.
Frequency distributions of Y-chromosomal STR haplotypes that best discriminate between the Austro-Asiatic (AA) and Tibeto-Burman (TB) population groups.
Genetic Differentiation and Affinities
Estimation of FST statistics and AMOVA were carried out among populations grouped by well-defined criteria, separately for the different sets of markers, mt DNA (RSP and HVS1), Y-chromosomal (RSP and STRP), and autosomal. (Detailed allele and haplotype frequency data for autosomal loci are provided in Suppl. Tables 7 and 8.) The results for the different sets of markers are qualitatively similar. The FST values are highest for tribal populations inhabiting different geographical regions or belonging to different linguistic groups (Table 5). We note that there is some degree of confounding between geographical region of habitat and linguistic groups (see Methods). Genomic differentiation among the caste groups is substantially smaller than among the tribal groups. With respect to Y STRPs, the upper castes of different geographical regions show stronger differentiation compared with middle or lower castes, possibly reflecting historical male gene flow. The FST values based on autosomal markers are less than those for mitochondrial and Y-chromosomal markers, which is expected because of stronger drift effects arising from the fact that the effective size of a population with respect to mt and Y markers is a quarter of that for autosomal markers. FST values for Y markers are higher than for mtDNA markers possibly because of social practices that enhance female mobility compared with male mobility (Bamshad et al. 1998; Bhattacharyya et al. 1999). The consistency of the results of our FST and AMOVA analyses is very reassuring. The AMOVA results (Table 5) indicate that the extent of variation is the highest among individuals within population groups. Genetic differentiation with respect to Y-chromosomal markers is generally high, particularly geographical differentiation with respect to YRSPs (11.29%), possibly because of the social practices mentioned above. The extent of variation in mt-RSP haplotype frequencies among upper castes of different geographical regions is also high (11.1%). This implies that there is stronger geographical substructuring of upper-caste populations, compared with populations of other ranks. Sociocultural effects on genomic substructuring seem minimal as the proportions of variance attributable to caste-tribal group differences or linguistic differences are quite low. However, there is high genetic differentiation among populations belonging to both caste and tribal groupings, particularly the tribal populations, implying that neither of these two broad groups is genetically homogeneous.
Table 5.
Estimates of Fst and AMOVA Results Based on Mitochondrial, Y-Chromosomal, and Autosomal Polymorphisms for Different Groupings of the Populations Studied
|
% variation attributable toa
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Fst
|
Between groups
|
Between populations within groups
|
|||||||||||||
|
mt
|
Y
|
mt
|
Y
|
mt
|
Y
|
||||||||||
| Grouping | RSP | HVS1 | RSP | STRP | Auto | RSP | HVS1 | RSP | STRP | Auto | RSP | HVS1 | RSP | STRP | Auto |
| 2 groups: caste and tribe | 0.112 | 0.102 | 0.233 | 0.211 | 0.053 | 2.38 | 0.62 | 7.87 | 0.42 | 0.99 | 8.81 | 9.56 | 14.97 | 10.04 | 4.25 |
| 6 groups: geographical regions | 0.108 | 0.099 | 0.241 | 0.229 | 0.052 | 4.32 | 1.01 | 11.29 | 3.16 | 1.88 | 6.51 | 8.99 | 12.49 | 8.1 | 3.32 |
| 4 groups: linguistic groups | 0.109 | 0.101 | 0.243 | 0.232 | 0.055 | 2.64 | 1.29 | 8.51 | 1.86 | 1.89 | 8.29 | 8.90 | 15.30 | 8.97 | 3.46 |
| 3 groups: ranked castes—upper, middle and lower | 0.056 | 0.033 | 0.144 | 0.119 | 0.020 | 0.37 | ≈0 | 5.39 | ≈0 | 0.11 | 5.23 | 3.83 | 8.38 | 4.67 | 1.78 |
| Upper castes of different geographical regions | 0.101 | 0.045 | 0.089 | 0.249 | 0.008 | 11.10 | 1.71 | 4.02 | 3.67 | 0.98 | ≈0 | 2.78 | 4.83 | 1.24 | ≈0 |
| Middle castes of different geographical regions | 0.053 | 0.047 | 0.050 | 0.115 | 0.022 | 2.46 | 0.66 | ≈0 | 0.65 | ≈0 | 2.88 | 4.05 | 10.51 | 3.88 | 2.29 |
| Lower castes of four different geographical regions | 0.010 | 0.016 | 0.192 | 0.121 | 0.023 | 0.84 | ≈0 | ≈0 | ≈0 | ≈0 | 0.16 | 1.83 | 32.24 | 9.17 | 4.77 |
| 4 groups: tribes of four different geographical regions (Northeast, East, South, and Central) | 0.138 | 0.158 | 0.239 | 0.272 | 0.069 | 1.88 | 1.90 | 5.25 | 1.37 | 2.12 | 11.94 | 13.90 | 18.70 | 10.03 | 4.47 |
| 4 groups: tribes of four different linguistic groups | 0.136 | 0.157 | 0.243 | 0.275 | 0.069 | 0.40 | 0.06 | 6.86 | 0.82 | 1.80 | 13.15 | 15.39 | 17.54 | 10.52 | 4.84 |
The percentages of variation attributable to “between individuals within groups” are not shown. These are obtainable by subtracting from 100 the sum of the percentages of total variation attributable to the other two sources of variation that are shown.
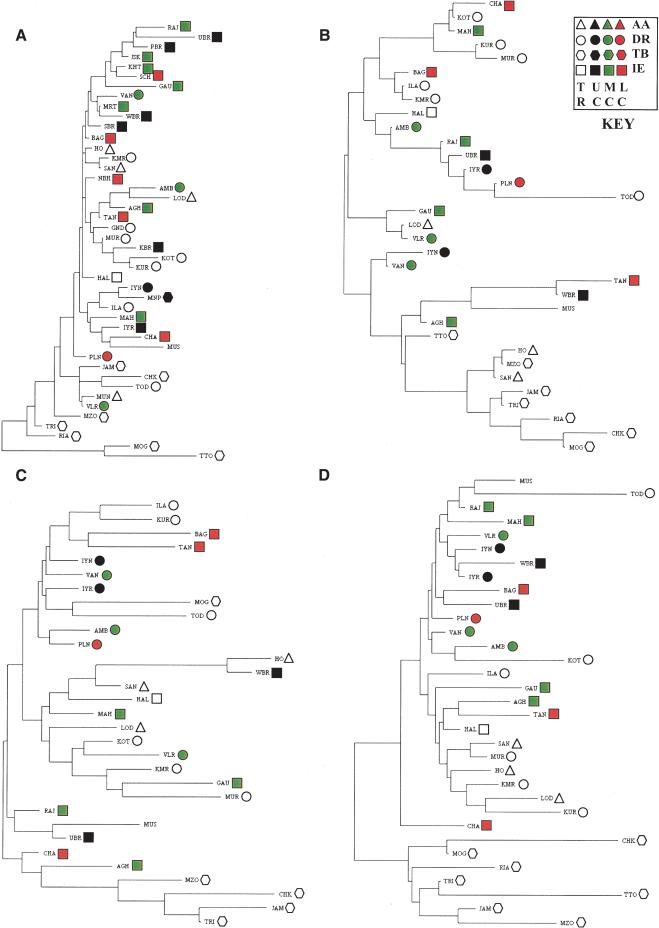
Genetic affinities were analyzed on the basis of data on different sets of markers (Fig. 6). No strong clustering of populations belonging to the same social, geographical, or linguistic group is observed, except that the TB speakers of northeast India form a separate cluster irrespective of the set of markers used. With respect to mtDNA-RSP haplotype frequencies (top of Fig. 6A), a small cluster comprising several populations of the north is also observed. This cluster reflects the high frequencies of haplotypes belonging to HG-U in these populations. Using the 323 distinct HVS1 sequences, a neighbor-joining (NJ) tree was also constructed (data not shown). There was again no clustering of the distinct sequences by geographical region, social rank, or linguistic group. However, most of the sequences belonging to HG-U formed a separate cluster. We have also formally tested whether there is any association between geographic and genetic distances by using the nonparametric Mantel test. No statistically significant association was found, irrespective of the set of marker loci used for computing the genetic distances.
Figure 6.
Neighbor-joining tree depicting genetic affinities among Indian ethnic populations based on (A) mitochondrial RSP haplotype frequencies, (B) Y-haplogroup frequencies, (C) Y-STRP frequencies, and (D) autosomal markers. The social [(UC) upper caste, (MC) middle caste, (LC) lower caste, (TR) tribal] and linguistic [(AA) Austro-Asiatic, (DR) Dravidian, (TB) Tibeto-Burman, (IE) Indo-European] background of each population is color-coded; the key to the color codes is given on the top right-hand corner.
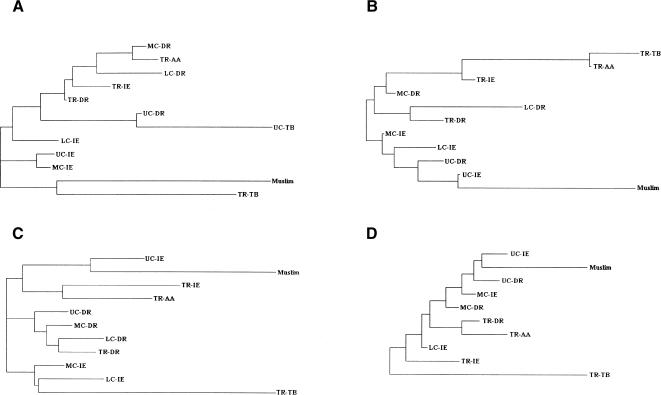
To discover broader patterns of genetic variation, we have pooled populations into social × linguistic subgroups, and have carried out a phylogenetic analysis. The results are presented in Figure 7. The genetic affinities do not reveal any significant additional patterns, except that (1) the Muslims and Tibeto-Burman-speaking tribals are each differentiated from all other categories, irrespective of the genetic system; and (2) with respect to the Y-STRP markers, the Dravidian speakers, irrespective of their position in the social hierarchy, show close affinities (Fig. 7C). The close affinity of the Muslims of Uttar Pradesh with the Indo-European upper-caste groups with respect to Y-chromosomal and autosomal markers (Fig. 7B-D) is easily explained from historical evidence of conversions to Islam in north India (Thapar 1966).
Figure 7.
Neighbor-joining tree depicting genetic affinities among categories of populations cross-classified by social rank [(UC) upper caste, (MC) middle caste, (LC) lower caste, (TR) tribal] and linguistic group [(AA) Austro-Asiatic, (DR) Dravidian, (TB) Tibeto-Burman, (IE) Indo-European] based on (A) mitochondrial RSP haplotype frequencies, (B) Y-haplogroup frequencies, (C) Y-STRP frequencies, and (D) autosomal markers.
Dravidian Speakers, Now Confined to Southern India, May Have Earlier Been Widespread Throughout India: Genetic Signatures of “Elite Dominance”?
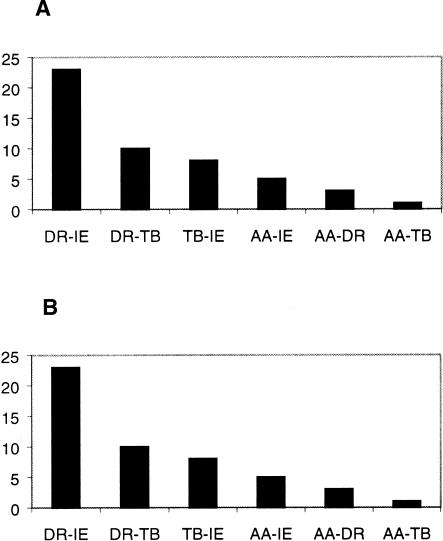
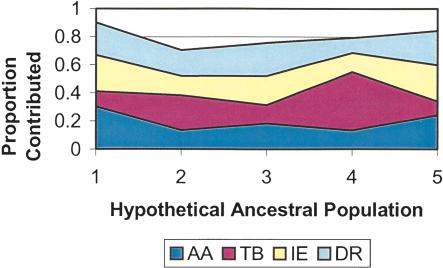
The IE and DR speakers share a significantly larger number of HVS1 sequences (Fig. 8A) compared with those between other groups. The number of individuals sharing these sequences is also the largest between IE and DR groups (Fig. 8B). These facts are striking, especially because the geographical regions presently inhabited by them are virtually disjoint. To explore in further detail the presence of cryptic population structure and the relationships among the various subgroups of populations, we have carried out a “population structure” analysis (Pritchard et al. 2000). In this analysis, an unknown number (K) of hypothetical ancestral populations is assumed to have contributed to the genetic profiles of contemporary populations. The number of hypothetical ancestral populations and their relative genetic contributions are statistically estimated from allele frequency data of contemporary populations. The results of population structure analysis based on our autosomal data also show (Fig. 9) that the DR and IE speakers are the most similar, in the sense that the proportional contributions of the five estimated hypothetical ancestral populations to these two groups are the most similar. These findings are consistent with the historical view that the DR speakers were possibly widespread throughout India (Thapar 2003). When the ranked caste system was formed after the arrival of the IE speakers ∼3500 ybp, many indigenous people of India, who were possibly DR speakers, embraced (or were forced to embrace) the caste system, together with the IE language and admixture. In fact, Renfrew (1992) has suggested that the elite dominance model, which envisages the intrusion of a relatively small but well-organized group that takes over an existing system by the use of force, may be appropriate to explain the distribution of the IE languages in north India and Pakistan. As the IE speakers, who entered India primarily through the northwest corridor, advanced into the Indo-Gangetic plain, indigenous people, especially the DR speakers, may have retreated southward to avoid linguistic dominance, after an initial period of admixture and adoption of the caste system. As evidenced by their strong genetic similarities (data not shown), the IE-speaking Halba tribals were most probably a DR-speaking tribal group, which is consistent with IE dominance over DR tribals.
Figure 8.
Frequency distributions of mtHVS1 sequences shared between groups of populations—Austro-Asiatic (AA), Dravidian (DR), Tibeto-Burman (TB), and Indo-European (IE). (A) Number of sequences shared and (B) number of individuals sharing sequences.
Figure 9.
Results of STRUCTURE analysis: Proportional contributions of five hypothetical ancestral populations to Austro-Asiatic (AA), Dravidian (DR), Tibeto-Burman (TB), and Indo-European (IE) groups.
Central Asian Populations Have Contributed to the Genetic Profiles of Upper Castes, More in the North Than in the South
Central Asia is supposed to have been a major contributor to the Indian gene pool, particularly to the north Indian gene pool, and the migrants had supposedly moved to India through Afghanistan and Pakistan. From mtHVS1 data, we have estimated FST values between the populations of the Central Asia and Pakistan regions (data were collated from Calafell et al. 1996; Comas et al. 1998; Kivisild et al. 1999a) and those belonging to various geographical regions of India. Populations of Central Asia and Pakistan show the lowest (0.017) coefficient of genetic differentiation with the north Indian populations, higher (0.042) with the south Indian populations, and the highest (0.047) with the northeast Indian populations. The Central Asian populations are genetically closer to the upper-caste populations than to the middle- or lower-caste populations, which is in agreement with Bamshad et al.'s (2001) findings. Among the upper-caste populations, those of north India are, however, genetically much closer (FST = 0.016) than those of south India (FST = 0.031). Phylogenetic analysis of Y-HG data collated from various sources (Hammer et al. 2000; Nebel et al. 2000; Rosser et al. 2000; Qamar et al. 2002) and with those generated in the present study also showed a similar picture (data not shown). One explanation, consistent with those of the previous section, is that even after the DR speakers retreated to the south to avoid elite dominance, there has been admixture between Central and West Asians and northern Indian populations.
METHODS
Populations
Blood samples were drawn with informed consent from individuals, unrelated to the first cousin level, belonging to 44 populations, chosen to represent ethnic groups of all geographical regions and sociocultural and linguistic categories. A list of populations, with sample sizes and brief notes on their sociocultural backgrounds, is provided in Table 1. We note that (1) population groups of the north are IE speakers, and those of the south are DR speakers; (2) the AA speakers are all tribals and are primarily confined to the central, eastern, and northeastern regions; (3) the TB speakers are confined to the northeastern region; and (4) the number of IE-speaking tribal groups is very few. Thus, there is an extent of confounding of geography, culture, and language in the distribution of ethnic groups of India, which is reflected in the nature of our statistical analyses and inferences. Because of various reasons, including lack of adequate amounts of DNA, not all markers could be examined in all populations.
Loci and Protocols
Ten mtDNA restriction site polymorphisms (HaeIII np 663, HpaI np 3592, AluI np 5176, AluI np 7025, DdeI np 10,394, AluI np 10,397, HinfI np 12,308, HincII np 13,259, AluI np 13,262, HaeIII np 16517) and 1 Insertion/Deletion polymorphism (IDP; COII/tRNALys intergenic 9-bp deletion) were screened using standard primers and protocols (Torroni et al. 1993,1996). Sequencing of the mtDNA Hyperveriable Segment-1 (HVS1; np 16,024-16,380) was carried out on an ABI-3100 sequencer, after PCR amplification using standard primers in both directions (Vigilant et al. 1991). HVS1 sequencing was carried out in a subset (528 of 1490) of individuals, randomly selecting at least 10 individuals from each ethnic group.
We screened 22 Y-chromosomal markers; 12 were binary (YAP, 92r7, SRY 4064, sY81, SRY + 465, TAT, M9, M13, M17, M20, SRY10831, and p12f2) and 10 were short tandem repeats (STR; DYS19, DYS388, DYS 389I and II, DYS390, DYS391, DYS392, DYS393, DYS425, DYS426), using primer sequences and protocols described before (Casanova et al. 1985; Thomas et al. 1999; Rosser et al. 2000; Mukherjee et al. 2001). For STRs, an ABI-3100 DNA sequencer and Genescan version 3.1 and Genotyper version 2.1 software packages were used. Haplogroup definitions as given in Y Chromosome Consortium (2002) were followed. However, for the lineages P*(×R1b8, R1a, Q3), BR*(×B2b, CE, F1, H, JK), and K*(×K1, LN, O2b, O3c, P), we have used the abbreviated HG symbols P*, BR*, and K*.
We screened 25 polymorphic autosomal loci. Eight were IDPs, and the remaining 17 were RSPs. The names and GDB (http://gdbwww.gdb.org) accession nos. or ALFRED UID (http://alfred.med.yale.edu) of the RSP loci are: ESR1 (GDB: 185229); NAT (GDB: 187676); CYP1A (GDB: 9956062)-MspI; PSCR (GDB: 182305); T2 (GDB: 196856); LPL (GDB: 285016); ALB (GDB: 178648); ALAD-MspI (GDB: 155925); ALAD-RsaI (GDB: 155924); HB ψβ-HincII (GDB: 56084); HB 3′ψβ-HincII; HB 5′β-HinfI; HoxB4-MspI (UID: SI0001670); DRD2 (UID: SI000191L)-TaqIB,-TaqID,-Taq1A; and ADH2-RsaI (UID: SI000002C). The names of the IDPs and primers and protocols used are as given in Majumder et al. (1999a) and Tishkoff et al. (1996). RSPs were screened using primers and protocols of Jorde et al. (1995), Majumder et al. (1999b), and K. Kidd (pers. comm.).
Statistical Methods
Maximum likelihood estimates of haplotype frequencies at linked autosomal loci were obtained via the EM algorithm using the HAPLOFREQ package (Majumdar and Majumder 2000). DNA sequences were aligned using CLUSTALW (http://www2.ebi.ac.uk/clustalw/). The Cambridge sequence was used as reference during alignment.
Tests of significance in sparse cross-classified tables were carried out by a bootstrap test procedure devised by us. Standard statistical analyses were carried out using SPSS. Population genetic analyses were performed using ARLEQUIN (Schneider et al. 2000; http://lgb.unige.ch/arlequin/) and DnaSP (Rozas and Rozas 1999; http://www.ub.es/dnasp/). Mantel tests were carried out using MANTEL, version 2.0 (http://www.sci.qut.edu.au/NRS/mantel.htm). Expansion times using HVS1 data were estimated (Slatkin and Hudson 1991) assuming a mutation rate of 20.5% per site per million years (Bonatto and Salzano 1997). For phylogenetic analyses using HVS1 data, DNA distances were calculated using NETWORK (http://www.fluxus-engineering.com/sharenet.htm) and the maximum likelihood method as implemented in PHYLIP version 3.5 (http://evolution.genetics.washington.edu/phylip.html) assuming a 30:1 transition:transversion ratio (Lundstrom et al. 1992). For phylogenetic reconstruction from data on autosomal markers, mtDNA-RSPs, and Y-chromosomal markers, Nei's (1987) DA distance measure and the neighbor-joining method (Saitou and Nei 1987) were used, as implemented in DISPAN (http://oat.bio.indiana.edu:7580/) and PHYLIP version 3.5c packages. The age (A) of a Y-HG was estimated as A = g × s2/μ, where g is the generation time (assumed to be 30 yr); s2 is the variance of the STR repeat number among haplotypes belonging to the HG, averaged over all STR loci; and μ is the mutation rate per generation at an STR locus, taken to be 0.18% (Quintana-Murci et al. 2001). A Markov Chain Monte Carlo analysis of population structure (Pritchard et al. 2000) was carried out using the program STRUCTURE (http://pritch.bsd.uchicago.edu/software.html).
Acknowledgments
We thank our collaborators C.S. Chakraborty, R. Lalthantluanga, Mitashree Mitra, A. Ramesh, N.K. Sengupta, Samir K. Sil, Jai Rup Singh, Chitra Mahadik Thakur, and M.V. Usha Rani for help in collecting samples and at various other stages of this work. We thank Sujit Maiti for help with data management and in preparing the figures. We also thank Lynn Jorde, Ken Kidd, Andy Merriwether, Antonio Torroni, and Chris Tyler-Smith for providing advice and for sharing laboratory protocols with us during the initial stages of this work. Hans Bandelt pointed out some errors in our sequence data; we are grateful to him. This work was partially supported by grants from the Department of Biotechnology, Government of India.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
[Supplemental Material is available online at www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: C.S. Chakraborty, R. Lalthantluanga, M. Mitra, A. Ramesh, N.K. Sengupta, S.K. Sil, J.R. Singh, C.M. Thakur, M.V. Usha Rani, L. Jorde, K. Kidd, A. Merriwether, A. Torroni, and C. Tyler-Smith.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1413403.
References
- Bamshad, M.J., Watkins, W.S., Dixon, M.E., Jorde, L.B., Rao, B.B., Naidu, J.M., Prasad, B.V.R., Rasanayagam, A., and Hammer, M.F. 1998. Female gene flow stratifies Hindu castes. Nature 395: 851-852. [DOI] [PubMed] [Google Scholar]
- Bamshad, M.J., Kivisild, T., Watkins, W.S., Dixon, M.P., Ricker, L.E., Rao, B.B., Naidu, M., Prasad, B.V.R., Reddy, P.G., Rasanayagam, A., et al. 2001. Genetic evidence on the origins of Indian caste populations. Genome Res. 11: 994-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt, H.J., Forster, P., and Rohl, A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16: 37-48. [DOI] [PubMed] [Google Scholar]
- Beteille, A. 1998. The Indian heritage—A sociological perspective. In The Indian human heritage (eds. D. Balasubramian and N. Appaji Rao), pp. 87-94. University Press, Hyderabad, India.
- Bhattacharyya, N., Basu, P., Das, M., Pramanik, S., Banerjee, R., Roy, B., Roychoudhury, S., and Majumder, P.P. 1999. Negligible gene-flow across ethnic boundaries in India, revealed by analysis of Y-chromosomal DNA polymorphisms. Genome Res. 9: 711-719. [PubMed] [Google Scholar]
- Bonatto, S.L. and Salzano, F.M. 1997. A single and early origin for the peopling of the Americas supported by mitochondrial DNA sequence data. Proc. Natl. Acad. Sci. 94: 1866-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calafell, F., Underhill, P., Tolun, A., Angelicheva, D., and Kalaydjieva, L. 1996. From Asia to Europe: Mitochondrial DNA sequence variability in Bulgarians and Turks. Ann. Hum. Genet. 60: 35-49. [DOI] [PubMed] [Google Scholar]
- Cann, R.L. 2001. Genetic clues to dispersal of human populations: Retracing the past from the present. Science 291: 1742-1748. [DOI] [PubMed] [Google Scholar]
- Casanova, M., Leroy, P., Boucekkine, C., Weissenbach, J., Bishop, C., Fellous, M., Purrello, M., Fiori, G., and Siniscalco, M. 1985. A human Y-linked DNA polymorphism and its potential for estimating genetic and evolutionary distance. Science 230: 1403-1406. [DOI] [PubMed] [Google Scholar]
- Comas, D., Calafell, F., Mateu, E., Perez-Lezaun, A., Bosch, E., Martinez-Arias, R., Clarimon, J., Facchini, F., Fiori, G., Luiselli, D., et al. 1998. Trading genes along the silk road: mtDNA sequences and the origin of Central Asian populations. Am. J. Hum. Genet. 63: 1824-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciani, F., Santolamazza, P., Shen, P., Macaulay, V., Moral, P., Olckers, A., Modiano, D., Holmes, S., Destro-Bisol, G., Coia, V., et al. 2002. A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am. J. Hum. Genet. 70: 1197-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond, J. 1997. Guns, germs and steel: The fates of human societies. Jonathan Cape, London.
- Forster, P., Torroni, A., Renfrew, C., and Rohl, A. 2001. Phylogenetic star contraction applied to Asian and Papuan mtDNA evolution. Mol. Biol. Evol. 18: 1864-1881. [DOI] [PubMed] [Google Scholar]
- Grimes, B.F. 1999. The ethnologue: Languages of the world. Summer Institute of Linguistics, California.
- Guha, B.S. 1935. The racial affinities of the people of India. In Census of India, 1931, Part III—Ethnographical. Government of India Press, Simla, India.
- Hammer, M.F., Redd, A.J., Wood, E.T., Bonner, M.R., Jarjanazi, H., Karafet, T., Santachiara-Benerecetti, S., Oppenheim, A., Jobling, M.A., Jenkins, T., et al. 2000. Jewish and middle eastern non-Jewish populations share a common pool of Y-chromosome biallelic haplotypes. Proc. Natl. Acad. Sci. 97: 6769-6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorde, L.B., Bamshad, M.J., Watkins, W.S., Zenger, R., Fraley, A.E., Krakowiak, P.A., Carpenter, K.D., Soodyall, H., Jenkins, T., and Rogers, A.R. 1995. Origins and affinities of modern humans: A comparison of mitochondrial and nuclear genetic data. Am. J. Hum. Genet. 57: 523-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karve, I. 1961. Hindu society: An interepretation. Deshmukh Prakashan, Poona, India.
- Keith, A. 1936. Review of B.S. Guha's “Racial affinities of the peoples of India.” Man 29: 37. [Google Scholar]
- Kivisild, T., Bamshad, M.J., Kaldma, K., Metspalu, M., Metspalu, E., Reidla, M., Laos, S., Parik, J., Watkins, W.S., Dixon, M.E., et al. 1999a. Deep common ancestry of Indian and western-Eurasian mitochondrial DNA lineages. Curr. Biol. 9: 1331-1334. [DOI] [PubMed] [Google Scholar]
- Kivisild, T., Kaldma, K., Metspalu, M., Parik, J., Papiha, S., and Villems, R. 1999b. The place of the Indian mitochondrial DNA variants in the global network of the maternal lineages and the peopling of the old world. In Genome diversity: Applications in human population genetics (eds. S.S. Papiha et al.), pp. 135-152. Kluwer, New York.
- Kosambi, D.D. 1991. The culture and civilisation of ancient India in historical outline. Vikas Publishing House, New Delhi, India.
- Lundstrom, R.S., Tavare, S., and Ward, R.H. 1992. Estimating substitution rates from molecular data using the coalescent. Proc. Natl. Acad. Sci. 89: 5961-5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maca-Meyer, N., Gonzalez, A.M., Larruga, J.M., Flores, C., and Cabrera, V.M. 2001. Major genomic mitochondrial lineages delineate early human expansions. BMC Genetics 2: 13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar, P. and Majumder, P.P. 2000. HAPLOFREQ: A computer program package for maximum-likelihood estimation of haplotype frequencies in a population via the EM algorithm. Tech Rep AHGU-1/2000. Indian Statistical Institute, Calcutta, India.
- Majumder, P.P. 1998. People of India: Biological diversity and affinities. Evol. Anthrop. 6: 100-110. [Google Scholar]
- Majumder, P.P., Roy, B., Banerjee, S., Chakraborty, M., Dey, B., Mukherjee, N., Roy, M., Guha Thakurta, P., and Sil, S.K. 1999a. Human-specific insertion/deletion polymorphisms in Indian populations and their possible evolutionary implications. Eur. J. Hum. Genet. 7: 435-446. [DOI] [PubMed] [Google Scholar]
- Majumder, P.P., Roy, B., Balgir, R.S., and Dash, B.P. 1999b. Polymorphisms in the β-globin gene cluster in some ethnic populations of India and their implications on disease. In Molecular intervention in disease (eds. S. Gupta and O.P. Sood), pp. 75-83. Ranbaxy Science Foundation, New Delhi.
- Meenakshi, K. 1995. Linguistics and the study of early Indian history. In Recent perspectives of early Indian history (ed. R. Thapar), pp. 53-79. Popular Prakashan, Bombay, India.
- Misra, V.N. 1992. Stone age in India: An ecological perspective. Man and Env. 14: 17-64. [Google Scholar]
- Mukherjee, N., Nebel, A., Oppenheim, A., and Majumder, P.P. 2001. High-resolution analysis of Y-chromosomal polymorphisms reveals signatures of population movements from Central Asia and West Asia into India. J. Genet. 80: 125-135. [DOI] [PubMed] [Google Scholar]
- Nebel, A., Filon, D., Weiss, D.A., Weale, M., Faerman, M., Oppenheim, A., and Thomas, M.G. 2000. High-resolution Y chromosome haplotypes of Israeli and Palestinian Arabs reveal geographic substructure and substantial overlap with haplotypes of Jews. Hum. Genet. 107: 630-641. [DOI] [PubMed] [Google Scholar]
- Nei, M. 1987. Molecular evolutionary genetics. Columbia University Press, New York.
- Pattanayak, D.P. 1998. The language heritage of India. In The Indian human heritage (eds. D. Balasubramanian and N.A. Rao), pp. 95-99. Universities Press, Hyderabad, India.
- Pritchard, J.K., Stephens, M., and Donnelly, P. 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qamar, R., Ayub, Q., Mohyuddin, A., Helgason, A., Mazhar, K., Mansoor, A., Zerjal, T., Tyler-Smith, C., and Mehdi, S.Q. 2002. Y-chromosomal DNA variation in Pakistan. Am. J. Hum. Genet. 70: 1107-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci, L., Krausz, C., Zerjal, T., Sayar, S.H., Hammer, M.F., Mehdi, S.Q., Ayub, Q., Qamar, R., Mohyuddin, A., Radhakrishna, U., et al. 2001. Y-chromosome lineages trace diffusion of people and languages in southwestern Asia. Am. J. Hum. Genet. 68: 537-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnagar, S. 1995. Archaeological perspectives of early Indian societies. In Recent perspectives of early Indian history (ed. R. Thapar), pp. 1-52. Popular Prakashan, Bombay, India.
- Ray, N. 1973. Nationalism in India. Aligarh Muslim University, Aligarh, India.
- Renfrew, C. 1992. Archaeology, genetics and linguistic diversity. Man 27: 445-478. [Google Scholar]
- Risley, H.H. 1915. The people of India. Thacker Spink, Calcutta, India.
- Rosser, Z.H., Zerjal, T., Hurles, M.E., Adojaan, M., Alavantic, D., Amorim, A., Amos, W., Armenteros, M., Arroyo, E., Barbujani, G., et al. 2000. Y-chromosomal diversity in Europe is clinal and is influenced primarily by geography, rather than by language. Am. J. Hum. Genet. 67: 1526-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychoudhury, S., Roy, S., Basu, A., Banerjee, R., Vishwanathan, H., Usha Rani, M.V., Sil, S.K., Mitra, M., and Majumder, P.P. 2001. Genomic structures and population histories of linguistically distinct tribal groups of India. Hum. Genet. 109: 339-350. [DOI] [PubMed] [Google Scholar]
- Rozas, J. and Rozas, R. 1999. DnaSP version 3: An integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15: 174-175. [DOI] [PubMed] [Google Scholar]
- Saitou, N. and Nei, M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406-425. [DOI] [PubMed] [Google Scholar]
- Sarkar, S.S. 1958. Race and race movements in India. In The cultural heritage of India (ed. S.K. Chatterjee), Vol. 1, pp. 17-32. The Ramakrishna Mission Institute of Culture, Calcutta, India. [Google Scholar]
- Schneider, S., Kueffer, J.-M., Roessli, D., and Excoffier, L. 2000. ARLEQUIN: A software for population genetic data analysis. University of Geneva, Geneva, Switzerland.
- Singh, K.S. 1992. People of India: An introduction. Anthropological Survey of India, Calcutta, India.
- Slatkin, M. and Hudson, R.R. 1991. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129: 555-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, B., Xiao, C., Deka, R., Seielstad, M.T., Kangwanpong, D., Xiao, J., Lu, D., Underhill, P., Cavalli-Sforza, L.L., Chakraborty, R., et al. 2000. Y chromosome haplotypes reveal prehistorical migrations to the Himalayas. Hum. Genet. 107: 582-590. [DOI] [PubMed] [Google Scholar]
- Thapar, R. 1966. A history of India, Volume 1. Penguin, Middlesex, UK.
- ____. 1995. The first millennium B.C. in northern India (up to the end of Mauryan period). In Recent perspectives of early Indian history (ed. R. Thapar), pp. 80-141. Popular Prakashan, Bombay, India.
- ____. 2003. Early India: From the origins to AD 1300. University of California Press, Berkeley, CA.
- Thomas, M., Bradman, N., and Flinn, H. 1999. High throughput analysis of 10 microsatellite and 11 diallelic polymorphisms on the human Y-chromosome. Hum. Genet. 105: 577-581. [DOI] [PubMed] [Google Scholar]
- Tishkoff, S.A., Ruano, G., Kidd, J.R., and Kidd, K.K. 1996. Distribution and frequency of a polymorphic Alu insertion at the plasminogen activator locus in humans. Hum. Genet. 97: 759-764. [DOI] [PubMed] [Google Scholar]
- Torroni, A., Schurr, T.B., Cabell, M.F., Brown, M.D., Neel, J.V., Larsen, M., and Smith, D.G. 1993. Asian affinities and continental radiation of the four founding Native American mtDNAs. Am. J. Hum. Genet. 53: 563-590. [PMC free article] [PubMed] [Google Scholar]
- Torroni, A., Huoponen, K., Francalacci, P., Petrozzi, M., Morelli, L., Scozzari, R., Obinu, D., Savontaus, M.-L., and Wallace, D.C. 1996. Classification of European mtDNAs from an analysis of three European populations. Genetics 144: 1835-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigilant, L.A., Wilson, A.C., and Harpending, H. 1991. African populations and the evolution of human mitochondrial DNA. Science 253: 1503-1507. [DOI] [PubMed] [Google Scholar]
- Y Chromosome Consortium. 2002. A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 12: 339-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerjal, T., Wells, R.S., Yuldasheva, N., Ruzibakiev, R., and Tyler-Smith, C. 2002. A genetic landscape reshaped by recent events: Y-chromosomal insights into Central Asia. Am. J. Hum. Genet. 71: 466-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://alfred.med.yale.edu; ALFRED.
- http://evolution.genetics.washington.edu/phylip.html; PHYLIP.
- http://gdbwww.gdb.org; GDB.
- http://lgb.unige.ch/arlequin/; ARLEQUIN.
- http://oat.bio.indiana.edu:7580/; DISPAN.
- http://pritch.bsd.uchicago.edu/software.html; STRUCTURE.
- http://www.fluxus-engineering.com/sharenet.htm; NETWORK.
- http://www2.ebi.ac.uk/clustalw/; CLUSTALW.
- http://www.sci.qut.edu.au/NRS/mantel.htm; MANTEL.
- http://www.ub.es/dnasp/; DnaSP.