Abstract
FACTS (Functional Association/Annotation of cDNA Clones from Text/Sequence Sources) is a semiautomated knowledge discovery and annotation system that integrates molecular function information derived from sequence analysis results (sequence inferred) with functional information extracted from text. Text-inferred information was extracted from keyword-based retrievals of MEDLINE abstracts and by matching of gene or protein names to OMIM, BIND, and DIP database entries. Using FACTS, we found that 47.5% of the 60,770 RIKEN mouse cDNA FANTOM2 clone annotations were informative for text searches. MEDLINE queries yielded molecular interaction-containing sentences for 23.1% of the clones. When disease MeSH and GO terms were matched with retrieved abstracts, 22.7% of clones were associated with potential diseases, and 32.5% with GO identifiers. A significant number (23.5%) of disease MeSH-associated clones were also found to have a hereditary disease association (OMIM Morbidmap). Inferred neoplastic and nervous system disease represented 49.6% and 36.0% of disease MeSH-associated clones, respectively. A comparison of sequence-based GO assignments with informative text-based GO assignments revealed that for 78.2% of clones, identical GO assignments were provided for that clone by either method, whereas for 21.8% of clones, the assignments differed. In contrast, for OMIM assignments, only 28.5% of clones had identical sequence-based and text-based OMIM assignments. Sequence, sentence, and term-based functional associations are included in the FACTS database (http://facts.gsc.riken.go.jp/), which permits results to be annotated and explored through web-accessible keyword and sequence search interfaces. The FACTS database will be a critical tool for investigating the functional complexity of the mouse transcriptome, cDNA-inferred interactome (molecular interactions), and pathome (pathologies).
In large-scale sequence annotation, efficient identification of relevant text information and integration of this information with biomolecular data is often a limiting factor in inferring new functions for genes, transcripts, or proteins. Publishing (and patenting) of novel molecular findings increasingly depends on finding relevant information buried in a vast mass of text and biomolecular data, akin to finding a needle in a haystack. Much biological knowledge can be gleaned from MEDLINE records and their references. Although the knowledge discovery process could be dramatically enhanced by integrating information retrieval with natural language-processing techniques, surprisingly little progress has been made in this area. The ideal, therefore, is to create text-mining tools that can deal with complex, context-dependent biological relationships of genes, transcripts, and their products.
ENTREZ (Schuler et al. 1996), one of the most widely used keyword-based biological information retrieval tools that links sequence database entries with literature, is restricted to the textual presentation of retrieved abstracts with their Medical Subject Headings (MeSH) (Nelson et al. 2001) and cross-referenced sequence identifiers, if available. Filtering functions that could aid the summarization of results by extracting sentences from the retrieved abstracts are not provided in ENTREZ. PubGene (Jenssen et al. 2001), through its graphical network output provides a broad view of biological relationships extracted from abstracts. From a gene name query, PubGene generates a complex graphical network of co-occurring gene names in abstracts, which are hypothesized to have some biological relationship. Both ENTREZ and PubGene-extracted results contain significant noise that requires the biologist to refine queries and manually pursue a large number of retrieved links and abstracts to find the sought-after answer. Further, PubGene results depend on cooccurrence of gene names or symbols. Therefore, the ambiguity of automatically collected gene symbols or short gene names in abstract retrieval without disambiguation or filtering may amplify erroneous associations.
A number of text retrieval and mining tools (see also Links page of FACTS Web site) that cannot be addressed for reasons of space, allow the computational identification and/or extraction of query-relevant biological relationships (e.g., protein interactions). For example, PreBIND (http://deep.mshri.on.ca/prebind/) classifies abstracts retrieved by user-specified yeast protein names, identifies abstracts containing likely interaction information, and extracts the candidate protein names. The success of retrieval depends on the presence of yeast protein names or alternate names in an internal name and synonym dictionary. Although PreBIND and other textual protein interaction extraction tools (e.g., Ono's PPI extractor [Ono et al. 2001]) are useful to identify interaction candidates, functions that would aid further computational exploration of biological functions and pathways associated with interaction candidates are not provided.
Text-mining tools for identifying gene-disease relations from abstracts are important in the process of testing a disease-association hypothesis and providing support in interpreting results. XplorMed (Perez-Iratxeta et al. 2001) permits the step-wise exploration of user-provided or keyword-retrieved abstracts by quantitative word dependencies and categorization by keywords representing a concept, for example, disease MeSH. However, the upper input limit is 500 abstracts. Further, the output (keyword chains linked to abstracts) may contain rather general keywords (e.g., cell or patient) that are not linked with a gene. In contrast, G2D (Perez-Iratxeta et al. 2002) extracts disease and substance Medical Subject Heading (MeSH) from MEDLINE articles associated with a mapped human gene and computationally infers gene ontology (GO) terms (Ashburner et al. 2000) based on substance MeSH. The combined GO and substance MeSH concept mapping provide useful context information on potential biological roles of gene-disease candidates. Because the G2D database can be queried only by accessions and genome mapping positions, its application is limited.
Each of the existing text-mining tools has its own unique strengths and limitations in respect to information retrieval and extraction of relevant biological information. Input restrictions and lack of integrated biological context information (e.g., tissue and expression) limit many of the existing tools to the casual exploration of small data sets related to one topic (e.g., only protein interactions or only disease associations). At the time of annotating and analyzing 60,000 RIKEN mouse FANTOM2 cDNA clones (The FANTOM Consortium and The RIKEN Genome Exploration Research Group Phase I & II Team 2002; Mouse Genome Sequencing Consortium 2002), none of the tools available to us suited the large-scale textual information retrieval on the basis of clone annotations, followed by the extraction of molecular interaction, gene ontology, and disease association information. Further, we required annotation capability of the results, because this is critical to prevent massive error propagation when computationally inferred functional information is incorporated into curated databases. In light of these issues, we sought to construct a system that is (1) transcript focused (FANTOM2 clone set), (2) supports large-scale data retrieval, (3) interrelates basic gene-name annotations with sequence and MEDLINE abstract-inferred molecular interaction-containing sentences, disease associations, gene ontologies, and other sequence-related information (e.g., cDNA library source and protein motifs) and external data (e.g., gene expression), (4) produces results that can be both annotated and mined and, (5) generates intuitive search reports from traversing the integrated data by keywords, concept-containing keywords, or sequences.
RESULTS AND DISCUSSION
FACTS System
The FACTS system (version 1.0) contains 12 core programs (see also Supplement 1) for keyword-based retrieval, filtering, and processing, sequence similarity searches, and a result database that can be queried through 10 user interfaces. Two types of keyword-based queries need be distinguished, that are, command-line and web-based queries. Command-line queries or batch queries of MEDLINE or sequence databases to retrieve information are available to FACTS developers or expert users who downloaded the core programs. Web-based dynamic querying of MEDLINE, followed by the processing of retrieved abstracts in FACTS is not supported in this version. Data that were processed (e.g., disease MeSH associated with a FANTOM cDNA clone) by the FACTS system and integrated into its database (version 1) can be queried by keywords or sequence through Web-based interfaces. These queries permit users to retrieve and explore functional information of a FANTOM2 cDNA clone that was associated by FACTS.
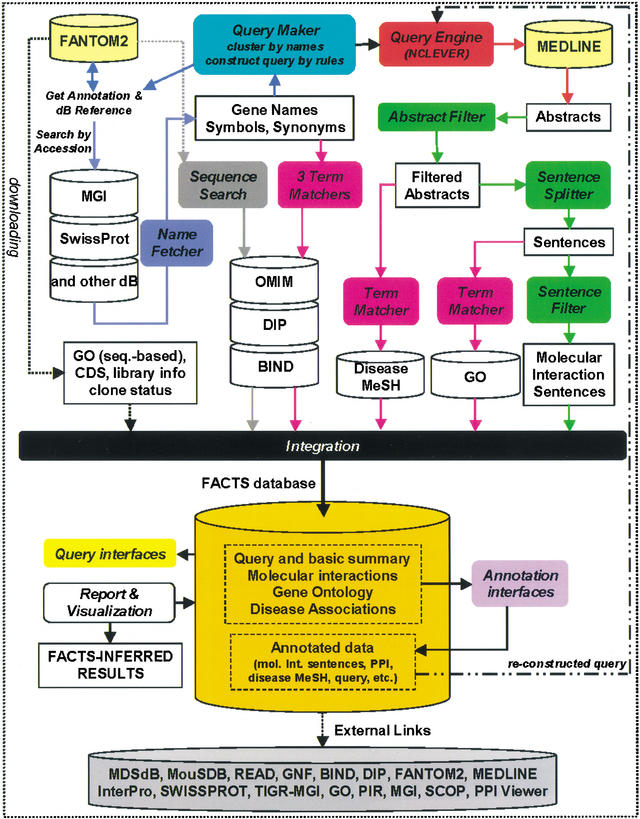
A schema of how the FACTS system works is shown in Figure 1. Implementation details of the PERL programs are shown as flowcharts in Supplement 1. The program NameFetcher uses CloneIDs and the database accessions of their annotation sources (e.g., MGI database) to extract gene/protein name, symbol, or synonym accession from specified fields in the source databases (Supplement 2). The combination of source database name and accession yields the query ID for the extracted information. If multiple CloneIDs have the same source accession, they are clustered and receive only one query ID.
Figure 1.
The FACTS System consists of 12 core programs (round-shaped boxes). External databases are FANTOM2 and MEDLINE. Other databases were installed locally. Arrows indicate the flow between programs and databases. Details are described in the text.
QueryMaker reads the query IDs of the NameFetcher to construct, according to query rules, from the gene names, symbols, and synonyms MEDLINE queries that are input for NCLEVER (Rioux et al. 1994). NCLEVER emulates ENTREZ, but allows batch queries to MEDLINE. The NCLEVER-retrieved abstracts are processed by AbstractFilter, which removes abstracts related to plants, and abstracts with gene names referring to cell line names. The filtered abstracts are split into sentences by the SentenceSplitter and filtered for molecular interaction-containing sentences by the SentenceFilter. The sentences and their associated query identifier, clone accession, MEDLINE identifiers are stored in the FACTS database for retrieval and annotation.
Three TermMatcher programs read in names, symbols, and synonyms derived from the NameFetcher output and matches them to name fields in OMIM (Hamosh et al. 2002), Biomolecular Interaction Network Database (BIND) (Bader and Hogue 2000), or Database of Interaction Proteins (DIP) (Xenarios et al. 2001). Two TermMatcher programs match disease MeSH to all MeSH fields in AbstractFilter-derived abstracts and GO terms to SentenceSplitter-processed sentences. SequenceSearch (FASTY and TBLASTN) searches FANTOM2 sequences against DIP, BIND, and OMIM to obtain sequence similarity search-based assignments protein-interaction candidates and OMIM disease information.
Other sequence-inferred information such as clone status (e.g., truncated), coding sequence (CDS) information, BLASTN (Altschul et al. 1997), or FASTY (Pearson et al. 1997) derived evidence (e.g., percent identity, match length, and clone length) for annotation categories of FANTOM2 cDNA clones and curated, sequence similarity search-based GO mappings were integrated from the FANTOM2 database. The text-based (keyword-retrieved molecular interaction sentences, GO terms that matched to sentences, and disease MeSH that matched to MH field of abstracts), sequence-inferred, and external data (e.g., splice variant information) were integrated and loaded into the relational PostgreSQL FACTS database.
Functionality of FACTS System
The uniqueness of the FACTS system lies in its rule-based, large-scale retrieval of textual information on molecular interactions, gene ontology GO, and potential disease associations, together with its biologically oriented and comparative integration of sequence-based data into the FACTS database for data mining and annotation.
The FACTS database can be searched from 10 interfaces by various keyword, accession or predefined queries, and sequences. The interface `Search by Gene Name' permits the keyword-based retrieval of FACTS entries restricted by gene name, symbol, and synonym of the RIKEN clone. To address the different needs of biologists, we implemented partial and exact matching of keywords plus optional target restrictions on database references of clone annotations, annotation categories (homolog, similar to, etc.) derived from the FANTOM2 MATRICS (Mouse Annotation Teleconference for RIKEN cDNA sequences) annotation pipeline, and/or tissue source of the cDNA library. For example, a user with interest in exploring functional information on receptors expressed in the brain can query with the keyword `receptor' in partial matching mode, restricted to clones derived from brain cDNA libraries. `Search by keyword' allows the keyword-based retrieval of molecular interaction containing sentences, BIND protein names, OMIM, disease MeSH, and GO terms associated with a clone annotation. A search with `Inflammation' restricted to disease MeSH and clones derived from skin cDNA libraries will retrieve clones associated with immune response and inflammatory pathways, together with their curated gene names, corresponding hyperlinked MEDLINE identifiers, and FACTS reports.
The interface `Data and Result Sources' contains predefined queries on 14 data sources, including computational extracted molecular interactions, annotated PPI sentences, term-matching or sequence similarity search-inferred OMIM titles, disease MeSH, and external data, for example, MDS (Kawaji et al. 2002), MousDB (Zavolan et al. 2003), and InterPro (Apweiler et al. 2000). The items can be queried alone or in any combination and restricted by cDNA library source and annotation category to obtain a hyperlinked statistic of associated functions for each clone.
Another entry point to the FACTS database is the BLAST search interface. Sequence searches of the FANTOM2 or RTPS (Representative Transcript Protein Set) sequence sets (The FANTOMConsortium and The RIKEN Genome Exploration Research Group Phase I & II Team 2002; Mouse Genome Sequencing Consortium 2002; Baldarelli et al. 2003) result in BLAST outputs with hyperlinks to the FACTS report of entries. Search by accession permits queries with multiple CloneID, DDBJ accessions, MEDLINE identifiers of abstracts or FANTOM2 cluster ID, or RTPS accessions to retrieve FACTS-inferred functional information.
The association query interfaces `Infer mol. interaction associations' and `Infer disease associations' facilitate the extraction of potential interaction partners and their disease associations on the basis of shared disease MeSH terms, OMIM titles, or molecular interaction-containing sentences. Combinatorial FACTS database searches with text or sequence-inferred GO, disease MeSH, OMIM title, and/or molecular interaction sentences are facilitated by the interface `Infer by term'. The search target can be restricted by annotation category (e.g., homolog, similar to, etc.) and/or molecular interaction-associated clones. The search report includes clone ID, gene name, hyperlinks to the MeSH, GO, OMIM of the clone, and information on whether the clone is an alternative splice variant. Users can thereby identify candidate transcripts that are directly and indirectly associated by shared functional concepts for microarray construction, signaling, or disease gene pathway studies.
FACTS database query results are linked to a functional report containing seven tables as follows: (1) basic information, (2) summary of search and extraction results, (3) molecular interactions from PubMed abstracts, (4) protein interaction pairs derived from BIND and DIP databases, (5) disease MeSH terms of MEDLINE abstracts, (6) OMIM and Morbid Map titles, and (7) gene ontology terms.
The basic information table provides users with a summary of computational assigned and curated gene names, symbols, and synonyms. The integrated information on CDS status (e.g., immature, 3′UTR, 5′UTR, etc.), clone status information (e.g., length, truncated, antisense, and immature), and annotation category (e.g., homolog, similar to, etc.) aids query prioritization and annotation selection. A clone that contains only the 3′UTR of a gene is obsolete for exploring protein–protein interactions, whereas it is still useful for inferring and annotating potential protein–RNA interactions of UTR site regulatory elements. InterPro-derived domain information may provide hints for localizing sequence regions that are critical for protein interaction.
The summary table of query and extraction results displays simple statistics of query matches in MEDLINE abstracts, downloaded, and processed abstracts, followed by the number of query word-containing sentences, interaction word, and query word-containing sentences and disease MeSH terms. For results extracted from GO, OMIM, and BIND, we compared text and sequence-based counts and reported the overlap between both methods. A button `Annotate' opens an interface for the annotation of computationally extracted molecular interaction sentences, disease MeSH, OMIM, and text-based GO assignments.
The comparison of constructed query strings and hits in abstracts with gene or protein names (query), symbols, and synonyms (N&S) of the data sources enables the detection of erroneous queries and improvement of the query construction rules. A query reconstruction function in the FACTS report interface lets users correct the query string, request requerying, and updating of the results from the FACTS administration. This feature allows the cyclic improvement of information retrieval.
Query Construction for Text Retrieval
The FACTS system was applied to both the computational and human annotated FANTOM2 cDNA clone set (Suppl. 3). Here, we report only the results for human curated clones. The coverage and specificity of abstract retrieval from MEDLINE using gene or protein names depends on how queries are formulated. Synonym and symbol usage affects the sensitivity, whereas the specificity is influenced by common English words (e.g., protein), ambiguous words and symbols, or phrases that are unlikely to occur in abstracts. We addressed those issues by defining query rules to distinguish informative from uninformative names and decreased the number of unsuccessful retrievals by missed alternate names or overly specific words. The rules were empirically derived from the knowledge of molecular biologists plus systematic gathering and visual inspection of names, alternate names, and symbols from various databases. From the corrected queries, we are able to derive new rules. During the development of FACTS, 24 rules were added through query reconstruction.
Depending on the data sources, symbols or words were not always suitable for querying MEDLINE abstracts because the annotations contained (1) uninformative names such as hypothetical protein or similar to SIMILAR TO KIAA0266 GENE PRODUCT, (2) phrases that are unlikely to occur in an abstract (e.g., SIMILAR TO MANNOSIDASE, ALPHA, CLASS 2C, and MEMBER 1 and 3) symbols or synonyms with one or two letters or alpha-numeric characters. Those symbols or synonyms were deleted from the query.
We derived 205 query construction rules (Suppl. 4A) that are specific for FANTOM2 cDNA clone annotations and their data sources (Suppl. 2). The majority of rules targeted the removal of annotations that would result in excessive MEDLINE abstract retrieval (e.g., membrane protein) and the deletion of uninformative prefixes, and terms (e.g., similar to, structure containing, -pending, member 1 and 3, class 2C) derived from the FANTOM2 MATRICS annotation pipeline or data sources. A few rules determined the generation of spelling-variants for symbols (e.g., Pax6 and Pax-6) and the use of the Boolean operators “and” and “or” For example, the SWISSPROT name GLUTATHIONE S-TRANSFERASE, MU TYPE 3 was converted to Glutathione S-transferase AND “mu” AND “3”.
Because the query construction process by NameFetcher and QueryMaker led to a clustering of FANTOM2 gene name annotations by their database source accessions, we reduced the number of queries significantly. By querying with the gene name of the database reference rather than the clone annotation itself, we imply that a clone annotated as `similar to Abca1' or `weakly similar to Abca1' may have functions related to Abca1 of the data source. The annotation prefixes were based on sequence identity, match length thresholds calculated from BLASTN, or FASTY outputs as described in detail by The FANTOMConsortium and The RIKEN Genome Exploration Research Group Phase I & II Team (2002) and The Mouse Genome Sequencing Consortium (2002). As the clustering by annotation source accession may result in irrelevant functional associations, we integrated this sequence search-based evidence (e.g., FASTY, 77% ID, 100% length, match = 835) from the FANTOM2 database field `evidence' to provide users a confidence measure for FACTS assigned molecular functions.
Before the FANTOM2 MATRICS, we constructed 14,210 queries from the computational gene-name annotations of 60,770 clones (Suppl. 2). Curators subsequently changed 42.7% of the annotations, which prompted 9895 (69.6%) changes in FACTS queries. A total of 8662 (60.9%) resulted in new FACTS query constructions or reassignments of existing queries; 985 (6.8%) queries became uninformative and were removed. Curation reduced the number of queries to 13,245. These comprised curated gene names in addition to 34,158 symbols, synonyms, and alternate names of 28,843 (47.5%) clones. A total of 31,927 (52.5%) of human-curated clone annotations were uninformative for MEDLINE queries, but still informative for functional inference from their sequences.
Information Retrieval and Extraction From MEDLINE and Other Biomolecular Databases
NCLEVER retrieved for 9873 (74.5%) MEDLINE queries more than 1.2 million abstracts (Table 1A). The number of retrieved abstracts for each query varied from 1 to 1907 abstracts. Among the top 20 query retrievals were C-protein, LINE protein, Ran oncogene, Rad51 one misconstructed query (DiGeorge syndrome-related protein FKSG5 or protein), and also queries with ambiguous symbols (e.g., Gpr106 or GREAT and alcoholsulfotransferase or STD). We addressed the frequently occurring ambiguity problem of gene symbols with three characters by systematic filtering of the abstracts in the context of full name or all available synonyms. Abstracts containing only the three-letter symbol were removed. Ambiguity of four or five character symbols (e.g., GREAT) occurred less frequently and was dealt with on a case-by-case basis when reported through the query reconstruction function.
Table 1A.
Summary of MEDLINE Abstract Retrieval and Extraction of Molecular Interaction-Containing Sentences, Disease MeSH and GO Terms
| MEDLINE | Clones | % | Query | % | Abstract | % | Sentence | % | Term |
|---|---|---|---|---|---|---|---|---|---|
| Queried | 28,843 | 100.0 | 13,245 | 100.0 | n/a | n/a | n/a | n/a | n/a |
| w/o abstract | 7,619 | 26.4 | 3,372 | 25.5 | n/a | n/a | n/a | n/a | n/a |
| Abs. retrieved | 21,779 | 75.5 | 9,873 | 74.5 | 1,201,630 | 100.0 | 10,744,757 | 100.0 | n/a |
| Abs. removed | 11,351 | 39.4 | 4,933 | 37.2 | 461,194 | 38.4 | 4,093,616 | 38.1 | n/a |
| Abs. remain | 20,611 | 71.5 | 9,356 | 70.6 | 740,436 | 61.6 | 6,651,141 | 61.9 | n/a |
| Mol. interact. | 14,021 | 48.6 | 6,362 | 48.0 | 156,879 | 13.1 | 261,043 | 2.4 | n/a |
| Disease MeSH | 13,789 | 47.8 | 6,304 | 47.6 | 201,925 | 16.8 | n/a | n/a | 3,672 |
| GO | 19,720 | 68.4 | 8,973 | 67.7 | 418,714 | 34.8 | 3,831,845 | 35.7 | 4,765 |
(Abs) Abstract, (n/a) not applicable, (w/o abstract) without abstract, (mol. interact) molecular interaction information-containing sentences. Term: refers to number of non-redundant GO or disease MeSH terms. All numbers are nonredundant. Note that one clone can be associated with multiple abstracts. Cleaning of abstracts affected 11,351 transcripts, of which 1,168 had only one abstract. Therefore, the number of clones with remaining abstracts is 20,611
A total of 3372 (25.5%) of queries did not match any word in MEDLINE abstracts at the time of querying (Table 1A). Queries without retrieved abstracts were often derived from domain- and structure-containing, or SWISS-PROT (Bairoch and Apweiler 2000) and PIR-inferred annotations. Abstracts rarely contain domain or fold names unless they are from review articles or associated with the discovery or characterization of functionally important domains or folds. Multiple word-containing protein names without available symbols (e.g. [ALR-like protein]; AK077567) or classifications of proteins (SWISS-PROT) that are informative but too specific to appear in the abstract (e.g., Atp11a or [ATPase and class VI and type 11A]; AK006628) were another source of unsuccessful queries. We avoided total removal of classifications (e.g., class or type) or combinations of all query words by the Boolean operator “or” to prevent generating large numbers of false positive abstracts.
The downloaded abstracts served as a basis for this analysis. We did not construct Boolean queries with terms specifying a particular function (e.g., cyclin E and interact) to avoid having to re-query all of MEDLINE when the focus changes. We were interested in molecular interactions, particularly protein interactions, disease associations, and gene ontology. Before extracting the above information, we cleaned the downloaded abstracts by rules (Suppl. 4B–D) developed from human curation of abstracts retrieved by 40 randomly chosen queries and biological knowledge. For example, abstracts containing plant, but not transplant or implant, were removed. Furthermore, we used rules to delete abstracts containing variations of cell or cell line preceding or following a gene symbol (e.g., IL-4 dependent cell line and CCR3 cells). For example, CCR3 cells, which refers to cells transfected with CCR3, were detected in 3 of 521 abstracts retrieved with a CCR3 query (http://facts.gsc.riken.go.jp/CCR3/). In total, the disambiguation or filtering and cleaning steps reduced the number of downloaded abstracts by 38% and affected 34% of queries.
Molecular Interactions Inferred From Abstracts
At present, the FACTS database is loaded with 740,436 abstracts related to 20,611 clones (9356 queries). MEDLINE abstract-inferred molecular interactions were derived from 261,043 predicted molecular interaction sentences associated with 14,021 (48.6%) clones (6362 queries). A total of 9148 (65%) of the molecular interaction sentences-associated clones have an InterPro domain, whereas 4351 (30.9%) clones are also members of 12,841 splice variation clusters in MouSDB. These clones may therefore reveal important clues on the effect of alternative splicing on predicted protein functions.
Molecular interactions in abstracts are generally explicit and confined to one sentence. If the interaction encompasses two sentences, the neighboring sentence may implicitly refer to “it” or “which”, meaning an interaction described in the preceding sentence. We decided, therefore, to extract a single sentence, rather than sentence pairs, using 10 sentence delimiter rules (Suppl. 4B). Sentences containing molecular interactions were defined by the presence of at least one of the 75 interaction indicator words (e.g., bind or inhibit) and the query word(s). Interaction indicators (Suppl. 4C) comprised stemmed words (e.g., bind* equals bind, binds, and binding), complete words, and phrases expressing protein–protein, protein–DNA, protein–RNA, protein–small molecule (e.g., drug) interactions. Because receptors and kinases constitute the largest protein families involved in signal transduction and are potential drug targets, we included combinations of phrases containing `receptor' or `kinase' (kinase xxx associat*). Sentences containing one or more of 101 false positive interaction indicator words (e.g., fluorescence activated cell sorter [FACS], cell interaction, activation curve; see Supplement 4D) were removed.
This strategy resulted in a significant data size reduction and concentration of interaction sentences (see Table 1A). For example, molecular interactions associated with CMKBR3 or CCR3 or chemokine (C-C) receptor 3 or MIP-1 αRL2 were summarized in 174 sentences extracted from 123 abstracts (A530083H05; AK041106, and Web site reference FACTS CCR3). To obtain similar molecular interaction information on CMKB3 from ENTREZ-retrieved abstracts would require significant longer reading time. The advantage of our sentence-based approach is that context information can be captured. For example, FACTS extracted from abstract 11396683 (Marone et al. 2001) two sentences: (1) Human basophils and mast cells express the chemokine receptor CCR3, which binds the chemokines eotaxin and RANTES, and (2) by interacting with the CCR3 receptor on Fc epsilonRI+ cells, HIV-I Tat protein is a potent chemo-attractant for human basophils and lung mast cells. Besides three CMKRB3-interacting proteins, the sentences contain cell type-specific expression information and the suggestion of a role in HIV infection. Even sentences of the following type: “The objectives of this study were to investigate CCR3-mediated activation of the mitogen-activated protein (MAP) kinases... and c-jun N-terminal kinase (JNK) in eosinophils...” (Kampen et al. 2000) are useful for biologists, because they provide pointers to intracellular signaling information and potential cellular or disease context. We also considered molecular interactions that definitely do not occur as informative. FACTS extracted from MEDLINE abstract 11404385 four sentences that summarize the interactions of CCR1, CCR3, CCR4, and CCR5, with the ligands RANTES and MIP1-α. The sentence “MIP-1α has similar binding characteristics to RANTES except that it does not bind to CCR3” is important, as it implies that MIP-1 α and RANTES have different receptor ligand-binding sites.
Although molecular interaction-containing sentences represent a fuzzy concept, they provide useful biological and functional information on direct and indirect interactions for exploration and annotation. A FACTS database search using the interface `Infer Molecular Interactions' with the keyword `Tumor necrosis factor receptor-associated factor 6' restricted to annotation category MGI (known gene) retrieves 47 potential molecular interactions, of which 24 are nonredundant. When selecting the TRAF6 interacting candidate, tumor necrosis factor receptor superfamily member 1b (TNFRSF1B), three molecular interaction sentences are displayed as evidence. Nerve growth factor-dependent TRAF6-TNFRSF1B interaction was shown in Schwann cells (Khursigara et al. 1999). The predicted shared disease MeSH for Traf6 and Tnfrsf1b is Autoimmune disease.
We purposely did not narrow the computationally extracted text information to a particular interaction type or interaction pairs (e.g., protein names). (1) Molecular interaction-containing sentences provide richer information, potentially on the context surrounding the interaction. (2) Blaschke et al. (2001) showed, in a small case study, only 30% extraction overlap between interacting proteins of DIP and MEDLINE abstract-derived sentences describing those interactions. (3) Extraction of particular interaction pairs or complexes from sentences that are reliable and biologically applicable can only be achieved by a combination of human curation and sequence comparison of the inferred interacting molecules with their sources.
Annotation
The computationally inferred molecular interaction sentences have a broad coverage and contain potentially interesting functional information in compressed form for biological interpretation and annotation. The computationally derived molecular interaction sentences can be curated for protein–protein, protein–DNA, protein complexes, or protein–small molecule interactions after completing an annotator registration form. The registration and annotation mode applies also for the other computational inferred information (e.g., GO, OMIM, and disease MeSH). All annotations are rechecked before being added to the FACTS annotation results or transferred to external databases as curated entries.
Annotation of Molecular Interaction-Containing Sentences
When matching (TermMatcher) gene/protein names, symbols, or synonyms derived from FANTOM cDNA clone annotations to the name fields of BIND and DIP entries, 10.2% (2099) of clones overlapped with 7.2% (1611) entries of the combined nonredundant BIND and DIP-derived experimental protein–protein interactions. Because 1589 (75.7%) of those clones were also associated with computationally extracted molecular interaction sentences, this number constitutes only 11.3% of all molecular interaction candidates (14,021 clones). Therefore, it is important to augment experimental mouse protein–protein interactions (PPI) stored in public databases, including the FANTOM2 PPI Viewer (Suzuki et al. 2003), with reliably inferred PPI information from the literature. As a consequence, we applied a multiple-step strategy to annotate PPI-containing sentences and to assign sequence candidates to the names of the interacting proteins.
Because PPI represent a subset of molecular interactions, we performed on the existing FANTOM clone query-derived sentences exact and case-insensitive term-matching with feature table descriptions “product” and “gene” of mouse, human, rat, and chimpanzee entries of GenBank (Release 127.0) to generate a PPI candidate-enriched sentence set for human curation. We limited the set to 431,234 candidate sentences of 166,375 abstracts derived from 4,728 queries of the FANTOM2 clone categories `homolog,' `similar to,' `inferred,' and `weakly similar to.' To reduce the number of potential false positives, we removed 87,389 sentences derived from spurious and ambiguous queries such as LINE protein, and C protein among others. For the remaining sentences, we deleted another 34,566 sentences containing the molecular interaction indicator words at the beginning or end of the sentence. The sentences in this set were often found to contain uninformative or unrelated interaction. For example, sentences beginning with `Binding was inhibited' or `Binding was determined' did not specify what was bound. Similarly, sentences with words `DNA binding,' `after binding,' `tissue binding,' or `sperm-egg binding' at the end were frequently found to be unrelated to PPI. To further enrich the sentences for interaction statements concerning two different proteins, we deleted 284,170, because the predicted protein names or symbols were identical. Another 16,205 sentences containing the interaction indicator words activat*, inactivat*, or inhibit* were removed, because visual inspection of sentences showed that the majority referred to interactions other than PPI. At the end of the filtering, we obtained 8904 sentences with 9233 computational binary PPI associated with 2850 clones.
Despite the computational preprocessing, the resulting sentences still contained other molecular interactions in addition to PPI (e.g., protein–DNA, protein–RNA, protein complexes, and protein–drug). The deleted and remaining sentences are valuable sets for refining our filtering strategy to minimize information loss and noise. We curated 5458 sentences associated with 1199 clones (414 queries). Annotation and double checking yielded 402 PPI interaction pairs derived from 845 PPI candidate sentences relating to 223 clones (96 queries) and 179 GenPept derived sequences. In addition, we obtained 18 protein–DNA and seven protein–small molecule interactions. We transferred 402 PPI interaction pairs to the FANTOM2 PPI viewer. Because the GenPept-derived sequences are associated with less information than FANTOM2 sequences, we compared them against the FANTOM2 cDNA set using TFASTY (protein sequence against translated database; no frame-shift, no unexpected stop codon). A total of 58 GenPept sequences that matched to FANTOM2 cDNA sequences with greater than 95% identity over more than 95% CDS length were manually inspected and replaced with FANTOM2 sequences. To reduce redundancy among the sequences of interacting proteins, we compared them against each other using BLASTP (>95% identity, >95% length) and selected one representative. Finally, we obtained from 402 protein interactions pairs, 90 PPI networks comprising 39 non-FANTOM2 mouse, 19 rat, 63 human, and 276 PPI curated FANTOM2-derived predicted proteins.
The small overlap (4.3%) of 402 annotated, double checked, and sequence post-processed, and rechecked PPI pairs and 9233 computationally predicted PPI pairs in our rigorous multistep PPI extraction procedure shows the limitations of text-based extraction of protein names or symbols that can be automatically associated with sequence accessions and species information. Even with improved protein name dictionaries such as PNAD (Yoshida et al. 2000) we do not expect significant improvement, because the automatic association of text-derived protein names with the correct species and sequences that are not frame-shifted or immature is problematic.
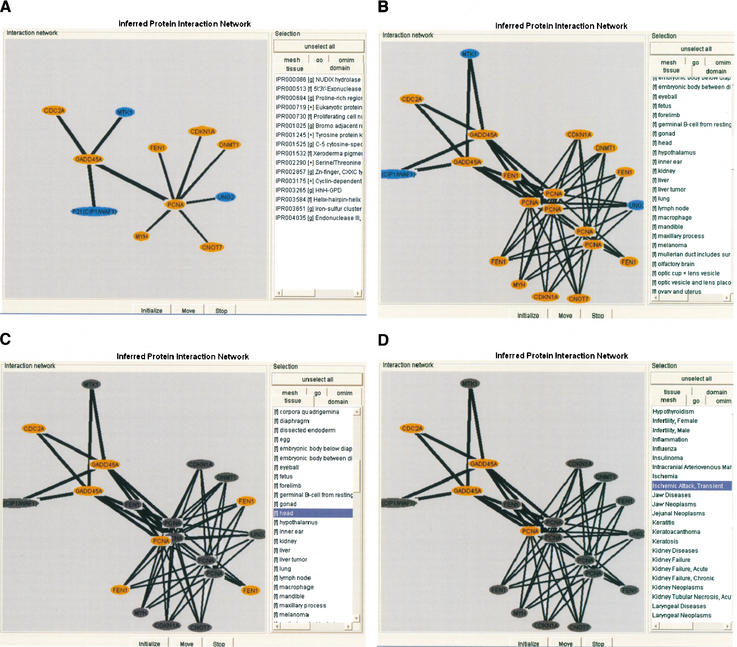
The protein–protein interaction networks constructed from the curated interactions were visualized in a static summary view and in a dynamic JAVA applet viewer in context of tissue, OMIM Morbidmap, disease MeSH, InterPro (Apweiler et al. 2000), and GO information. A network may contain identical protein names if the gene name annotation refers to clones of different library origin (see http://facts.gsc.riken.go.jp/viewer/InteractionViewer.php?CloneID=E230016M11 for clone ID E230016M11). The network shown in Figure 2A consists of 11 proteins involved in DNA repair, cell death, and cell cycle control. The transcript-based view (Fig. 2B) allows the observer to predict PPIs in context of expression as inferred from dBEST tissue distributions using BLASTN and FANTOMcDNA library information. Growth arrest and DNA-damage-inducible 45 α (Gadd45a; AK029473 and AK054076) was cloned from neonate day 0 head and oviduct libraries. Selection of head (Fig. 2C) resulted in the subnet CDC2A— GADD45A—PCNA—FEN1, which is supported by expression data. When adding the disease MeSH term `ischemic attack' (Fig. 2D) to the head expression criteria, the FEN1–PCNA interaction disappears, suggesting that FEN1 does not play a role in ischemia. Gadd45a and Pcna are induced by ischemic damage of the adult and fetal brain and are important in suppressing apoptosis and cell survival (Li et al. 1997; Charriaut-Marlangue et al. 1999). Cdc2a is the mouse homolog of human Cdc2, which was shown to be up-regulated in ischemic heart muscle and is implicated in cell survival after infarction (Reiss et al. 1996). To our knowledge, there are no reports on the cell survival-related role of CDC2 in the ischemic brain. This example shows that context-inferred PPI networks may result in interesting experimental targets, such as the role of the mouse CDC2A and other interacting proteins in the ischemic brain.
Figure 2.
(A) MEDLINE abstract-derived PPI network for curated PPI of 11 proteins: CDC2A, CDKN1A, CNOT7, FEN1, GADD45A, PCNA, MTK1, P21(CIP1/WAF1), UNG2, MYH, and DNMT1 (see also http://facts.gsc.riken.go.jp/viewer/InteractionViewer.php?CloneID=2810049I05 for protein interaction viewer of clone ID 2810049I05). Detailed information on data source accessions and inferred functions (OMIM Morbidmap, disease MeSH, tissue distribution, gene ontology assignment, and InterPro domains) appears after clicking on the circles. Orange-colored circles symbolize mouse proteins. Blue circles show potential interacting proteins of human origin. (B) The PPI network shown in A is derived from 64 interaction pairs inferred from 14 FANTOM2 and 5 GenPept sequences. The clone-based display of inferred PPI facilitates the visualization of context information. (C) Selection of head shows CDC2A—GADD45A—PCNA—FEN1 PPI associated with transcripts that are expressed in the head. Symbols preceding the tissue names, (f) FANTOM2 READ database-derived EST tissue information; (g) dbEST-derived EST tissue information. (l) FANTOM2 cDNA library source-derived tissue information. (D) Additional selection of disease MeSH Ischemic attack, transient removes FEN1 from the PPI network.
BIND and DIP-Inferred Protein Interactions
BIND and DIP databases were queried by gene name and/or symbol plus all FANTOM2 cDNA sequences. To assign protein interactions recorded in the BIND and DIP databases, we used a word-matching and a sequence similarity-based search strategy. Gene/protein names, symbols, or synonyms derived from FANTOMcDNA clone annotations to the name were matched (TermMatcher) to the name fields of BIND and DIP entries. Matching names and/or symbols were extracted together with the accessions of the entry. We obtained 1346 molecular interactions for 1509 (7.3%) clones (636 queries). Sequence-based assignments of BIND (http://facts.gsc.riken.go.jp/pi_seq_base.html) and DIP protein interactions were performed by comparing all DNA sequences of FANTOM clones in the FACTS database against the protein sequences of the BIND and DIP databases using the FASTY program with BLOSUM80 matrix. The best match with greater than 95% identity over more than 95% length to the FANTOM2 query sequence was selected as a protein interaction candidate. For BIND-derived candidates, we extracted the interacting protein names from the BIND name fields and integrated them into FACTS database to facilitate an integrated display with molecular interaction sentences. Due to restrictions in integrating data from DIP, we hyperlinked candidates to their original DIP entry.
Of note is the small overlap (BIND 2.2%, DIP 4.9%) of inferred PPI by term-matching and sequence searching (Table 3A, below) and less than 5% of source database coverage. We can largely exclude term-matching problems, as the target symbols or names are from identical data sources (e.g., MGI) and do not occur in free text format.
Table 3A.
Clone-Based Comparison of Inferred Functions
| All | Text | (%) | Overlap | (%) | Sequence (%) | ||
|---|---|---|---|---|---|---|---|
| GO | 20,338 | 19,720 | 97.0 | 15,900 | 78.2 | 16,518 | 81.2 |
| Component | 19,649 | 18,731 | 95.3 | 10,765 | 54.8 | 11,683 | 59.5 |
| Function | 19,582 | 17,947 | 91.7 | 12,954 | 66.2 | 14,589 | 74.5 |
| Process | 19,295 | 17,544 | 90.9 | 11,792 | 61.1 | 13,543 | 70.2 |
| OMIM | 11,732 | 10,931 | 93.2 | 1,814 | 15.5 | 2,615 | 22.3 |
| Morbid map | 3,950 | 2,585 | 65.4 | 1,124 | 28.5 | 2,489 | 63.0 |
| BIND | 1,634 | 1,509 | 92.4 | 36 | 2.2 | 161 | 9.9 |
| DIP | 973 | 639 | 65.7 | 48 | 4.9 | 382 | 39.3 |
Note: Numbers are based on 20,611 clones
However, term-matching is blind to alternative splice forms, truncated or partial sequences that were not extracted by FASTY. Because PPI can be abolished by substitution of one amino acid residue as demonstrated for PDZ domain ligands (Gee et al. 2000), all text-extracted PPI need to be carefully scrutinized by either sequence similarity search of the data source sequence or reading the full-text article before proceeding with experimental validation. The computational BIND sequence-inferred networks partially overlap with mouse experimentally derived mouse PPI as reported by Suzuki et al. (2001).
Gene Ontology-Inferred Functions
The GO-controlled vocabulary describes functionality of gene products. The assignment of GO codes from free text has been performed recently for a small set of GO terms to evaluate three different document classification methods (Raychaudhuri et al. 2002). The best method achieved 72% accuracy. We opted for simple term-matching using manually modified GO terms. The modifications included splitting phrases or compound words (e.g., GO 0004791 thioredoxin reductase [NADPH] becomes thioredoxin reductase “and” NADPH) and removal of words (e.g., in sensu), which are unlikely to occur in abstracts. Term-matching was performed to both query-word containing sentences and molecular interaction sentences. GO terms with split phrases were assigned if all the words occurred in one sentence, regardless of their combination. Curated, sequence-inferred GO associations were extracted from the FANTOM2 database and compared with the sentence-matched GO terms.
A total of 4765 GO terms were matched to one or more sentences of 418,714 abstracts associated with 19,720 (68.4%) clones (8,973 queries) (Table 1A). The overlap between curated sequence-inferred and text-inferred GO-associated clones is 78.2% (Table 3A, below). Clones associated with sequence or text-inferred GO only amount to 3.0% and 18.8%, respectively (Table 3A, below). If we compare the text and curated, sequence-based GO inference on GO term level, 50.1% of GO terms were associated by term-matching only, 16.4% by sequence, and 33.4% by both methods (Table 3B, below). A total of 1778 sequence-inferred GO assignments covered 21.4% (8,612 of 40,159) clones that were not considered informative for text queries or whose query failed to retrieve abstracts whose abstracts were removed during the cleaning procedure (Table 2, below).
Table 3B.
Term-Based Comparison of Inferred Functions
| All | Text | (%) | Overlap | (%) | Sequence | (%) | |
|---|---|---|---|---|---|---|---|
| GO | 5,702 | 4,765 | 83.6 | 1,907 | 33.4 | 2,844 | 49.9 |
| Component | 621 | 558 | 89.9 | 215 | 34.6 | 278 | 44.8 |
| Function | 2,747 | 2,137 | 77.8 | 1,059 | 38.6 | 1,669 | 60.8 |
| Process | 2,333 | 2,069 | 88.7 | 633 | 27.1 | 897 | 38.4 |
| OMIM | 4,725 | 4,532 | 95.9 | 564 | 11.9 | 757 | 16.0 |
| Morbid map | 1,147 | 961 | 83.8 | 552 | 48.1 | 738 | 64.3 |
| BIND | 1,445 | 1,346 | 93.1 | 33 | 2.3 | 132 | 9.1 |
| DIP | 877 | 554 | 63.2 | 90 | 10.3 | 413 | 47.1 |
Note: Numbers are based on 20,611 clones
Table 2.
Summary of Sequence-Based Functions for Clones Without Abstract
| Category | Clones | (%) | Term | Source dB coverage |
|---|---|---|---|---|
| w/o abstract | 40,159 | 100.0 | n/a | n/a |
| GO | 8,612 | 21.4 | 1,778 | 15.4 |
| OMIM | 661 | 1.6 | 269 | 1.9 |
| Morbid map | 566 | 1.4 | 261 | 12.3 |
| BIND | 101 | 0.3 | 65 | 0.6 |
| DIP | 48 | 0.1 | 49 | 0.4 |
A total of 40,159 clones comprise 7,619 (19.0%) without retrieved abstract, 1,168 (2.9%) with retrieved abstract(s) that were removed during filtering, and 31,372 (78.1%) clones with annotations that were uninformative for query construction
GO inference by term-matching shows high coverage (Table 1B), because the terms can be associated with query and co-occurrence with other gene names. For example, SCYA27 (small inducible cytokine 27, AK005520) is described by two sequence-inferred GO IDs for molecular functions (chemokine and cytokine) and one for cellular role (extracellular) that were also captured from the sentences. Additional sentence matching-derived GO terms that increase the functional information for SCYA27 include `homeostasis,' `necrosis,' `receptor,' and `chemokine receptor.' SCYA27 interacts with chemokine receptor GPR2. The interaction is involved in regulation of T cell-mediated skin inflammation, autoimmune skin diseases, and homeostasis (Homey et al. 2000). The GO term necrosis is associated with TNF α, which induces SCYA27 expression. The GO terms `extracelluar matrix' (SCYA27 binds to the extracellular matrix) and `chromosome' (Scya27 maps to chromosome 4) are false positives. The detection of false positive GO term associations caused by cooccurrence of unrelated genes or spurious hits (e.g., kinase in the phrase kinase inhibitor genistein) is assisted by a hyperlinked table of sequence and sentence-inferred GO comparisons and MEDLINE ID references for the sentence-inferred GO terms.
Table 1B.
Summary of Text and Sequence-Based Functional Associations
| Category | Clones | (%) | Query | (%) | Term | Source DB coverage |
|---|---|---|---|---|---|---|
| TEXT | ||||||
| Disease MeSH | 13,789 | 66.9 | 6,304 | 47.6 | 3,672 | 93.3 |
| GO | 19,720 | 95.7 | 8,973 | 67.7 | 4,765 | 41.4 |
| OMIM | 10,931 | 53.0 | 5,248 | 39.6 | 4,532 | 31.7 |
| Morbid map | 2,585 | 12.5 | 1,218 | 9.2 | 961 | 45.4 |
| BIND | 1,509 | 7.3 | 636 | 4.8 | 1,346 | 12.0 |
| DIP | 639 | 3.1 | 304 | 2.3 | 554 | 4.9 |
| SEQUENCE | ||||||
| GO | 16,518 | 80.1 | n/a | n/a | 2,844 | 24.7 |
| OMIM | 2,615 | 12.7 | n/a | n/a | 757 | 5.3 |
| Morbid map | 2,489 | 12.1 | n/a | n/a | 738 | 34.9 |
| BIND | 161 | 0.8 | 161 | 0.8 | 132 | 1.2 |
| DIP | 382 | 1.9 | 382 | 1.9 | 413 | 3.7 |
Comparisons among text and sequence-derived categories are based on 20,611 clones
All numbers are nonredundant
The data mining function “infer by terms” alleviates false positives and captures indirect functional associations by combination of the Boolean `and' with multiple GO terms or combination of GO with disease MeSH terms. A search of the sentence-inferred GO and abstract-inferred disease MeSH in the clone annotation categories `MGI' (identical to known gene), `homolog,' and `similar to' with GO terms `chemokine receptor' and `homeostasis' and `necrosis' and disease MeSH term `Inflammation' retrieves 279 clones, representing 143 gene product candidates directly and indirectly related to the above concepts. The output includes hyperlinks to a comparison of text and curated sequence-based GO assignments, disease MeSH terms, and information on potential alternative splicing. Seventy-four candidates are potential splice variants.
The same search on the curated sequence-inferred GO and disease MeSH failed to retrieve any candidates, because the combination of the three GO did not occur. Another reason was that sequence-inferred GO terms were deleted by the FANTOM2 MATRICS curator because of premature stop codons or partial sequence status (e.g., JAK3 homolog, AK043429). In some cases, GO terms were not assigned. One of the candidates, stromal cell-derived factor 1 (AK045092), plays a role in chemokine, cytokine signaling, and homeostasis of thyroid tissues (Aust et al. 2001) and hematopoiesis. The curated sequence-inferred GO terms `cytokine,' `extracellular,' `chemotaxis,' `immune response,' and `chemokine' were also present in the text-inferred GO terms, whereas `homeostasis,' a term for cell growth and/or maintenance, was only found in text-inferred GO. This data mining function is a powerful tool to infer shared pathways or disease associations that would not be possible from sequence-inferred information only, or with disease and GO information retrieval tools known to us.
OMIM-Inferred Human Disease Associations
OMIM is a database of inherited disease associations (Hamosh et al. 2002). OMIM titles were extracted from the OMIM Genemap and Morbidmap files by matching the associated gene symbols to the FANTOM clone annotations. OMIM Morbidmap is a subset of OMIM that contains inherited or heritable disease genes with cytogenetic map location. Matching of FACTS queries to gene name and symbol in Morbidmap inferred 961 potential disease associations (Table 1B) for 2585 clones (1218 queries). The noncurated TBLASTN (protein query against translated sequence database) search results (E- 50 e-value threshold) of FANTOM2 cDNA sequences against 1022 human disease-associated sequences (Schriml et al. 2003) yielded 738 inferred disease associations for 2489 clones with abstracts and 261 for clones without abstracts (Table 2). The overlap between text and sequence-based methods was only 28.5% for clone and 48.1% for the Morbidmap disease association (Tables 3A and 3B). Visual inspection of the disease candidate clones that were only associated with human disease by TBLASTN (34.6%) revealed isoforms and clones with hypothetical domain-containing gene names that could not be identified by term-matching. False positive TBLASTN-predicted disease associations were detected by term-matching. For example, Gabrg1 (AK032128, OMIM 137166) was wrongly associated with the Greig cephalopolysyndactyly syndrome related to Gli3 (OMIM 165240) and CCR2 or CCR5 OMIM entries (601267, 601373) were assigned to CCR3 (A530083H05, AK041106). False positive disease associations inferred by term-matching were caused by ambiguous query symbols or words. Thus, the use of both methods together leads to an increased confidence if computational assignments overlap. Further, it enhances the detection of inconsistencies that can then be corrected by human curation.
MeSH-Inferred Human Disease Associations
MeSH and GO terms comprise a curated, controlled vocabulary that link related concepts in a hierarchical structure. MeSH are applied in indexing abstracts and establishing concept relationships among them. We used the human disease-specific MeSH Tree (2001 Release) for term-matching to MeSH Headings (MH) of abstracts. MH is one of the MEDLINE record descriptors that follow the abstract text. If identical disease MeSH occurred in different abstracts associated with the same query (e.g., MH1:abstract 1; MH1:abstract 2), the redundant MeSH terms were removed (e.g., MH1:abstract 1; MH1:abstract 2) and the nonredundant MeSH were assigned to the corresponding MEDLINE identifiers. We inferred 3672 disease MeSH for 66.9% (13,789) of clones (6,304 queries) with cleaned abstracts. Of those, 23.5% (3244 clones) overlapped with 82% of predicted hereditary disease associations from Morbidmap. The computationally derived disease MeSH-clone distribution (Supplement 5A) shows a high frequency of clone associations with neoplasms (49.5%), pathological conditions, signs and symptoms (48.1%), nervous system diseases (35.9%), immunological diseases (30.1%), and neonatal diseases and abnormalities (28.8%). Notably, one clone can be associated with multiple disease MeSH categories. Breast neoplasm is associated with a clone (AK007298) derived from a gene similar to tissue kallikrein or glandular kallikrein that belongs to MeSH categories `Neoplasm' and `Endocrine Diseases,' because in human breast cancer, glandular kallikrein is differentially regulated by steroid hormones (Magklara et al. 2000). Symptoms and pathologies are a very broad category with often spurious associations. For example, the MeSH term, `Acute disease,' was assigned, on average, to every sixth clone.
MeSH Annotation and Disease Associations
Clones that were annotated as similar to (>70% and <85% identity over >70% of length) are good targets to discover new disease associations. FACTS predicted 708 clones (522 queries) with disease MeSH associations and 591 text and sequence-inferred OMIM association out of a total of 2578 clones (1114 queries) of the category `similar to.' As MeSH-inferred disease associations are important in the identification of nonhereditary disease associations, we evaluated the performance of FACTS by comparing the results of computational and medical expert annotations.
From 708 clones, we assigned the curator 333 clones (234 queries) selected randomly by clone IDs. The curator confirmed that of these 333 clones, 234 (70%) had relevant disease-MeSH-inferred disease associations. Ninety nine clones (62 queries) were deleted because they represented indirect or nonspecific (e.g., acute disease), associations or were incorrect associations caused by ambiguous gene names or queries irrelevant to disease associations (e.g., similar to SIMILAR ENIGMA, similar to serine-rich protein, etc.). On average, disease MeSH annotation (see Suppl. 5B,C) reduced the clone and term coverage in the 23 disease MeSH categories by more than 50% and mostly affected disease MeSH in the branches of the MeSH tree hierarchy, whereas the MeSH distribution among the top hierarchies did not change. Only 273 (24.2%) of 1127 nonredundant disease MeSH terms attributed by FACTS to the 333 human curated clones were confirmed upon curation. For example, in a clone annotated as similar to squamous cell carcinoma antigen 2 (AK003650, cloneID 1110023A16), 6 of 16 MeSH associations were deleted. Among the deleted MeSH associations, was Psoriasis, which originated from a MeSH-indexed abstract containing a description on the sequence similarity of human hurpin to SCCA2 and the overexpression of hurpin in psoriatic skin lesions (Abts et al. 1999). AK003650 and hurpin are both serin protease inhibitors (IPR000215). However, AK003650 does not contain the putative reactive sites Thr 356 and Ser 357, characteristic for the ovalbumin-serpin family member hurpin. In addition, AK003650 showed only 49% identity to human hurpin, but 82.5% identity to mouse SCCA2.
The disease association data mining function `Infer disease association' can be used if the focus is on targeting human disease-associated FANTOM2 cDNA clones that share the same signaling pathway. A query with Psoriasis restricted to the annotation category of known genes (MGI) and database reference MGD, retrieves 373 entries (clone ID, gene name) representing 171 nonredundant transcripts that are potentially associated with psoriasis. Selection of soluble acid phosphatase 1 retrieves a list of 14 potential associated molecules with their molecular interaction sentences and disease MeSH. Among six nonredundant candidates are STAT1 and STAT3, which both share Psoriasis and 11 other disease MeSH terms with soluble acid phosphatase 1. Psoriasis is associated with the STAT pathway through interferon gamma (Komine et al. 1996).
In rule-based systems, it is important to know the extent of information loss. The comparison of manual and FACTS-retrieved disease associations (OMIM Morbidmap and disease MeSH) for clones of the category `similar to' showed that 27% and 20% of them were detected by only one of the two methods, respectively. The overlap between both methods was 53% (data not shown). Hence, FACTS-inferred human disease associations are applicable for gross classification and decision support of biomedical experts, and should assist in rapidly expanding manually extracted potential human disease genes for further experimental studies.
Conclusions
The FACTS strategy of combining computationally inferred functional associations from sentences, controlled vocabularies, and sequence sources produces a summarization effect. The integrated display of interrelated information on molecular interaction, disease association, and gene ontology enables users to explore unexpected relations and to test them experimentally. To make the system accessible to researchers, we have deliberately implemented a simple annotation system that allows remote curation via the internet. Existing data mining or natural language processing systems did not satisfy the requirements for output, searching, and in-depth exploration of higher functional relationships of mouse transcripts. In particular, publicly available molecular interaction databases suffer from lack of integrated context information on potential disease associations. The FACTS sequence and text-based functional inference and annotation system provides a useful tool that will be expanded progressively in terms of entry numbers and functionality to analyze the mouse transcriptome and gene expression information.
METHODS
Databases
Query construction depended on mapping database references and accessions to the gene/protein descriptions, symbols, and aliases of computational and human curated RIKEN clones, and we therefore locally installed 17 reference databases (Suppl. 2). In addition to the query-related databases, we downloaded BIND, DIP MeSH tree, GO, OMIM, MDS (Kawaji et al. 2002), MouSDB (Zavolan et al. 2003), RTPS, and VTPS (Variant-based representative Transcript Protein Set) data (Baldarelli et al. 2003) to increase functional information and facilitate query result integration.
Query Construction Rules
The query rules were developed prior to the MATRICS annotation by visual inspection of test query results derived from the computational annotations. Emphasis was placed on detecting the cause of query results with no match or excessive matches to MEDLINE abstracts. Misconstructed queries were corrected and rules derived, as shown in Supplement 4A. Frequently occurring words with no meaning to the subject were deleted from query by using PubMed's stopword list.
Querying MEDLINE and Retrieval of Abstracts
To query MEDLINE, we used three locally installed NCLEVER4.0 MEDLINE search engines (Suppl. 2). All query constructs were formatted into NCLEVER syntax and restricted to MEDLINE entries having an abstract in the English language. To avoid an overload of the NCBI ENTREZ server, we developed a batch query script that sends queries every 1–5 min per search engine. If a query matched to MEDLINE abstracts, we restricted downloading of abstracts to 300 queries per day.
Abstract Processing
From the downloaded MEDLINE-formatted abstracts, we extracted sentences from the AB field (abstract text) and MeSH terms from the MH field. We reduced obvious false positives by deleting abstracts containing the query word in combination with a cell line name, for example, CD34+ cells or cell line MT-1. The latter is an ambiguous symbol for MT-1 cells and metallothionein 1.
To recognize and excise sentences from the text field, we used a sentence delimiter rule that distinguishes the delimiter `.' from the occurrence of `.' in numbers or abbreviations (Suppl. 4B). Sentences containing molecular interaction information were extracted together with the MEDLINE identifiers of the abstract by filtering with interaction indicator words (e.g., bind and coimmunoprecipitate) and the query words (Suppl. 4C). The resulting subset was cleansed from potential false positive interaction indicators (Suppl. 4D) that confer irrelevant interactions. Finally, we assigned to the remaining sentences confidence values for the interactions. For example, sentences describing an interaction with `might' were automatically (Suppl. 4E) given the value (L), for low. Other interactions received the confidence value (M) for medium. High confidence (H) labels were assigned only by annotators.
FACTS Programs
Programs of the FACTS system were written in PERL. Implemention details, including input and output behavior are shown in Supplement 1. The programs NameFetcher, QueryMaker with query construction rules, AbstractFilter, SentenceSplitter with sentence delimiter rules, SentenceFilter, TermMatcher for OMIM, BIND, DIP, GO, and disease MeSH can be downloaded from the FACTS Web site.
Acknowledgments
We thank Mihaela Zavolan for discussions and for providing the accessions of the variant and nonvariant clusters of MouSDB.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1019903.
Footnotes
[Supplemental material is available online at www.genome.org and also at the FACTS Web site http://facts.gsc.riken.go.jp/supplement/.]
References
- Abts, H.F., Welss, T., Mirmohammadsadegh, A., Kohrer, K., Michel, G., and Ruzicka, T. 1999. Cloning and characterization of hurpin (protease inhibitor 13): A new skin-specific, UV-repressible serine proteinase inhibitor of the ovalbumin serpin family. J. Mol. Biol. 293: 29-39. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler, R., Attwood, T.K., Bairoch, A., Bateman, A., Birney, E., Biswas, M., Bucher, P., Cerutti, L., Corpet, F., Croning, M.D., et al. 2000. InterPro—an integrated documentation resource for protein families, domains, and functional sites. Bioinformatics 16: 1145-1150. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., Ball, C.A., Blake, J.A., Botstein, D., Butler, H., Cherry, J.M., Davis, A.P., Dolinski, K., Dwight, S.S., Eppig, J.T., et al. 2000. Gene ontology: Tool for the unification of biologu. The Gene ontology consortium. Nature Genet. 25: 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aust, G., Steinert, M., Kiessling, S., Kamprad, M., and Simchen, C. 2001. Reduced expression of stromal-derived factor 1 in autonomous thyroid adenomas and its regulation in thyroid-derived cells. J. Clin. Endocrinol. Metab. 86: 3368-3376. [DOI] [PubMed] [Google Scholar]
- Bader, G.D. and Hogue, C.W. 2000. BIND—a data specification for storing and describing biomolecular interactions, molecular complexes and pathways. Bioinformatics 16: 465-477. [DOI] [PubMed] [Google Scholar]
- Bairoch, A. and Apweiler, R. 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28: 45-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldarelli, R.M., Hill, D.P., Blake, J.A., Adachi, J., Furuno, M., Bradt, D., Corbani, L.E., Cousins, S., Frazer, K.S., Qi, D., et al. 2003. Connecting sequence and biology in the laboratory mouse. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Blaschke, C., Oliveros, J.C., and Valencia, A. 2001. Mining functional information associated with expression arrays. Funct. Integr. Genomics 1: 256-268. [DOI] [PubMed] [Google Scholar]
- Charriaut-Marlangue, C., Richard, E., and Ben-Ari, Y. 1999. DNA damage and DNA damage-inducible protein Gadd45 following ischemia in the P7 neonatal rat. Brain Res. Dev. Brain Res. 116: 133-140. [DOI] [PubMed] [Google Scholar]
- The FANTOMConsortium and The RIKEN Genome Exploration Research Group Phase I & II Team. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNA. Nature 420: 563-573. [DOI] [PubMed] [Google Scholar]
- Gee, S.H., Quenneville, S., Lombardo, C.R., and Chabot, J. 2000. Single-amino acid substitutions alter the specificity and affinity of PDZ domains for their ligands. Biochemistry 39: 14638-14646. [DOI] [PubMed] [Google Scholar]
- Hamosh, A., Scott, A.F., Amberger, J., Bocchini, C., Valle, D., and McKusick, V.A. 2002. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 30: 52-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homey, B., Wang, W., Soto, H., Buchanan, M.E., Wiesenborn, A., Catron, D., Muller, A., McClanahan, T.K., Dieu-Nosjean, M.C., Orozco, R., et al. 2000. Cutting edge: The orphan chemokine receptor G protein-coupled receptor-2 (GPR-2, CCR10) binds the skin-associated chemokine CCL27 (CTACK/ALP/ILC). J. Immunol. 164: 3465-3470. [DOI] [PubMed] [Google Scholar]
- Jenssen, T.K., Laegreid, A., Komorowski, J., and Hovig, E. 2001. A literature network of human genes for high-throughput analysis of gene expression. Nat. Genet. 28: 21-28. [DOI] [PubMed] [Google Scholar]
- Kampen, G.T., Stafford, S., Adachi, T., Jinquan, T., Quan, S., Grant, J.A., Skov, P.S., Poulsen, L.K., and Alam, R. 2000. Eotaxin induces degranulation and chemotaxis of eosinophils through the activation of ERK2 and p38 mitogen-activated protein kinases. Biochem. Biophys. Res. Commun. 269: 546-552. [PubMed] [Google Scholar]
- Kawaji, H., Schönbach, C., Matsuo, Y., Kawai, J., Okazaki, Y., Hayashizaki, Y., and Matsuda, H. 2002. Exploration of novel motifs derived from mouse cDNA sequences. Genome Res. 12: 367-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khursigara, G., Orlinick, J.R., and Chao, M.V. 1999. Association of the p75 neurotrophin receptor with TRAF6. J. Biol. Chem. 274: 2597-2600. [DOI] [PubMed] [Google Scholar]
- Komine, M., Freedberg, I.M., and Blumenberg, M. 1996. Regulation of epidermal expression of keratin K17 in inflammatory skin diseases. J. Invest. Dermatol. 107: 569-575. [DOI] [PubMed] [Google Scholar]
- Li, Y., Chopp, M., Powers, C., and Jiang, N. 1997. Apoptosis and protein expression after focal cerebral ischemia in rat. Brain Res. 765: 301-312. [DOI] [PubMed] [Google Scholar]
- Magklara, A., Grass, L., and Diamandis, E.P. 2000. Differential steroid hormone regulation of human glandular kallikrein (hK2) and prostate-specific antigen (PSA) in breast cancer cell lines. Breast Cancer Res. Treat. 59: 263-270. [DOI] [PubMed] [Google Scholar]
- Marone, G., Florio, G., Triggiani, M., Petraroli, A., and de Paulis, A. 2001. Mechanisms of IgE elevation in HIV-1 infection. J. Pathol. 194: 239-246. [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-561. [DOI] [PubMed] [Google Scholar]
- Nelson, S.J., Johnston, D., and Humphreys, B.L. 2001. Relationships in medical subject headings. In Relationships in the organization of knowledge. (eds. C.A. Bean and R. Green), pp. 171-184. Kluwer Academic Publishers, New York, NY.
- Ono, T., Hishigaki, H., Tanigami, A., and Takagi, T. 2001. Automated extraction of information on protein-protein interactions from the biological literature. Bioinformatics 17: 155-161. [DOI] [PubMed] [Google Scholar]
- Pearson, W.R., Wood, T., Zhang, Z., and Miller, W. 1997. Comparison of DNA sequences with protein sequences. Genomics 46: 24-36. [DOI] [PubMed] [Google Scholar]
- Perez-Iratxeta, C., Bork, P., and Andrade, M.A. 2001. XplorMed: A tool for exploring MEDLINE abstracts. Trends Biochem. Sci. 26: 573-575. [DOI] [PubMed] [Google Scholar]
- Perez-Iratxeta, C., Bork, P., and Andrade, M.A. 2002. Association of genes to genetically inherited diseases using data mining. Nat. Genet. 31: 316-319. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri, S., Chang, J.T., Sutphin, P.D, and Altman, R.B. 2002. Associating genes with gene ontology codes using a maximum entropy analysis of biomedical literature. Genome Res. 12: 203-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss, K., Cheng, W., Giorando, A., De Luca, A., Li, B., Kajstura, J., and Anversa, P. 1996. Myocardial infarction is coupled with activation of cyclins and cyclin-dependent kinases in myocytes. Exp. Cell. Res. 225: 44-54. [PubMed] [Google Scholar]
- Rioux, P.A., Gilbert, W.A., and Littlejohn, T.G. 1994. A portable search engine and browser for the Entrez database. J. Comp. Biol. 1: 293-295. [DOI] [PubMed] [Google Scholar]
- Schriml, L., Hill, D.P., Blake, J.A., Bono, H., Wynshaw-Boris, A., Pavan, W.J., Ring, B.Z., Beisel, K., Setou, M., RIKEN GER Group and GSL Members, et al. 2003. Human disease genes and their cloned mouse orthologs: Exploration of the FANTOM2 cDNA sequence data set. (this issue). [DOI] [PMC free article] [PubMed]
- Schuler, G.D., Epstein, J.A., Ohkawa, H., and Kans, J.A. 1996. Entrez: Molecular biology database and retrieval system. Methods Enzymol. 266: 141-162. [DOI] [PubMed] [Google Scholar]
- Suzuki, H., Fukunishi, Y., Kagawa, I., Saito, R., Oda, H., Endo, T., Kondo, S., Bono, H., Okazaki, Y., and Hayashizaki, Y. 2001. Protein–protein interaction panel using mouse full-length cDNAs. Genome Res. 11: 1758-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, H., Saito, R., Kanamori, M., Kai, C., Schönbach, C., Nagashima, T., Hosako, J., and Hayashizaki, Y. 2003. The mammalian protein–protein interaction database and its viewing system that is linked to the main FANTOM2 viewer. (this issue). [DOI] [PMC free article] [PubMed]
- Xenarios, I., Fernandez, E., Slawinski, L. Duan, X.J., Thompson, M.J., Marcotte, E.M., and Eisenberg, D. 2001. DIP: The database of interacting proteins: 2001 update. Nucleic Acids Res. 29: 239-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, M., Fukuda, K., and Takagi, T. 2000. PNAD-CSS: A workbench for constructing a protein name abbreviation dictionary. Bioinformatics 6: 169-175. [DOI] [PubMed] [Google Scholar]
- Zavolan, M., Kondo, S., Schonbach, C., Adachi, J., Hume, D.A., RIKEN GER Group and GSL Members, Hayashizaki, Y., and Gaasterland, T. 2003. Impact of alternative initiation, splicing, and termination on the diversity of the mRNA transcripts encoded by the mouse transcriptome. (this issue). [DOI] [PMC free article] [PubMed]
WEB SITE REFERENCES
- http://facts.gsc.riken.go.jp; FACTS home page.
- http://facts.gsc.riken.go.jp/CCR3/; FACTS CCR3.
- http://facts.gsc.riken.go.jp/viewer/InteractionViewer.php?CloneID=2810049I05; FACTS Protein Interaction Viewer for clone 2810049I05.
- http://facts.gsc.riken.go.jp/viewer/InteractionViewer.php?CloneID=E230016M11; FACTS Protein Interaction Viewer for clone E230016M11.
- http://facts.gsc.riken.go.jp/pi_seq_base.html; FACTS sequence-based BIND inferred protein interactions.
- http://deep.mshri.on.ca/prebind/; PreBIND.