Abstract
In the genomic era, one of the fundamental goals is to characterize the function of proteins on a large scale. We describe a method, PANTHER, for relating protein sequence relationships to function relationships in a robust and accurate way. PANTHER is composed of two main components: the PANTHER library (PANTHER/LIB) and the PANTHER index (PANTHER/X). PANTHER/LIB is a collection of “books,” each representing a protein family as a multiple sequence alignment, a Hidden Markov Model (HMM), and a family tree. Functional divergence within the family is represented by dividing the tree into subtrees based on shared function, and by subtree HMMs. PANTHER/X is an abbreviated ontology for summarizing and navigating molecular functions and biological processes associated with the families and subfamilies. We apply PANTHER to three areas of active research. First, we report the size and sequence diversity of the families and subfamilies, characterizing the relationship between sequence divergence and functional divergence across a wide range of protein families. Second, we use the PANTHER/X ontology to give a high-level representation of gene function across the human and mouse genomes. Third, we use the family HMMs to rank missense single nucleotide polymorphisms (SNPs), on a database-wide scale, according to their likelihood of affecting protein function.
The rapid growth in protein sequence databases has led to significant progress in understanding the relationships between protein sequence and function. Hundreds of thousands of protein sequences have been inferred from genomic or complementary DNA sequences derived from >200 different organisms, and recent advances in large-scale direct protein assays, such as protein separation followed by mass spectrometry, promise to further enlarge and refine our knowledge of proteins in vivo. Protein sequence comparison and interpretation of these comparisons have matured to become an extremely useful tool for evolutionary biology. Proteins (from either the same or different organisms) that have related sequences often have related functions. The exceptions to this correlation are as interesting as the rule. Some protein families are relatively restricted in how they are used for different functions, whereas others have been recruited for many different purposes. The evolution of proteins to perform new tasks, either at the molecular level or at the level of broader pathways or processes, is apparently very dependent on the specifics of the individual scaffold. If protein sequence data are to be used to assist in genome-wide functional classification of genes, these functional divergence events must be modeled on a large scale.
Computational algorithms and databases for comparing protein sequences have reached a relatively mature stage of development. In the past few years, profile methods (Gribskov et al. 1987; Henikoff and Henikoff 1991; Attwood et al. 1994), particularly Hidden Markov Models (HMM; Krogh et al. 1994; Eddy 1996) and PSI-BLAST (Altschul et al. 1997), have entered widespread use. The profile has a different amino acid substitution vector at each position in the profile, based on the pattern of amino acids observed in a multiple alignment of related sequences. Profile methods combine algorithms with databases: A group of related sequences is used to build a statistical representation of corresponding positions in the related proteins. The power of these methods therefore increases as new sequences are added to the database of known proteins. Multiple sequence alignments (Dayhoff et al. 1974) and profiles have allowed a systematic study of related sequences. One of the key observations is that some positions are “conserved,” that is, the amino acid is invariant or restricted to a particular property (such as hydrophobicity), across an entire group of related sequences. If the sample of sequences is broad enough, such that we can infer that we are observing the results of mutation and selection at all positions in the protein, these conserved positions are likely to be critical for the function of the protein.
The dependence of profile and pattern-matching approaches (Jongeneel et al. 1989) on sequence databases led to the development of databases of profiles (BLOCKS, Henikoff and Henikoff 1991; PRINTS, Attwood et al. 1994) and patterns (Prosite, Bairoch 1991) that could be searched in much the same way as sequence databases. These approaches typically have better sensitivity and specificity than pairwise sequence comparisons. Even more importantly, these databases also capture human quality assurance (such as sequence correction) and additional expert analysis and interpretation of the grouped sequences. This human intervention makes sequence analysis more accessible to the community of biologists outside the field of computational biology. Today, two of the most widely used protein family databases are Pfam (Sonnhammer et al. 1997; Bateman et al. 2002) and SMART (Schultz et al. 1998; Letunic et al. 2002), which combine expert analysis with the well-developed HMM formalism for statistical modeling of protein families (mostly families of related protein domains).
For some proteins, simply knowing its family membership is enough to predict its function, whereas for others, one must know its subfamily (alternatively referred to as subgroup or subtype; Hannenhalli and Russell 2000) within that family. The detailed task of subfamily-level classification, however, has primarily been carried out as a cottage industry: independent efforts of a large number of labs each focusing on a single family. Phylogenetic trees (representing the evolutionary relationships between sequences) and the related concept of dendrograms (tree structures representing the similarity between sequences) have been used extensively for this purpose. Tree representations are particularly useful for identifying distinct subfamilies (subtrees) of closely related sequences, which tend also to share function (e.g., Chiu et al. 1985; Rollins et al. 1991).
In contrast to protein sequence comparison methods, ontologies to describe protein function are just beginning to enter widespread use. Ontologies define a controlled vocabulary that enables large-scale computational analysis. Early efforts to define biological function ontologies for microbes include EcoCyc (Karp and Riley 1993) and the MIPS classification (Mewes et al. 1997). However, the recent sequencing of large, metazoan genomes such as Drosophila melanogaster and human, demands an ontology that also spans the biological functions of multicellular organisms. The Gene Ontology (GO; Ashburner et al. 2000), still under active development, is emerging as a standard across eukaryotic biology. GO is a very detailed representation of functional relationships, designed as a comprehensive functional annotation vocabulary. Several groups (Lander et al. 2001; Mouse Genome Sequencing Consortium 2002) have selected different samplings of GO terms for illustrating the functional repertoire of genomes. However, there are at present no ontologies having the breadth of GO but designed for high-level browsing and analysis of functions for large numbers of sequences.
Several groups are starting to combine the advantages of ontology terms for functional annotation with the power of Hidden Markov models for statistical, sequence-based inference. Family and domain databases such as Pfam and SMART have associated a number of Hidden Markov Models with GO terms. The TIGRFAMs database (Haft et al. 2003) provides an excellent resource for functional classification of microbial proteins, with >1600 Hidden Markov Models placed into functional categories. The PANTHER database (http://panther.celera.com) was designed as a resource to comprehensively and consistently treat both family and subfamily classification of proteins, focused on metazoans but also covering other organisms.
Rationale
PANTHER Index (PANTHER/X): An Abbreviated Ontology
The goal of PANTHER is to classify proteins by function. Any attempt at classification requires a meaningful set of rules that define the area of study, and how to group objects. Ontologies have been used for some time in computer science for precisely these kinds of applications. In the field of biology, the Gene Ontology (GO) contains >7000 terms to describe molecular function, and almost 5000 terms to describe biological process, arranged as a directed acyclic graph (DAG) up to 12 levels deep. Although this level of detail provides a rich vocabulary for functional annotation of gene products, there are other scientific applications that would benefit from a simpler ontology. We have developed the PANTHER Index (PANTHER/X) ontology to facilitate high-level browsing and analysis of large gene (or protein) lists, such as those generated in whole-genome analysis or in analysis of gene expression array data. PANTHER/X comprises a total of ∼250 categories in each schema (“molecular function” and “biological process”). The ontology borrows heavily from GO, has been fully mapped to GO, and is available on the GO Web site (http://www.geneontology.org). PANTHER/X was designed to be no more than three levels deep, and to be structured such that absolute depth in different parts of the ontology correspond to roughly equivalent levels of functional specificity. This structure was designed for easy navigation, and to partition proteins into biologically meaningful groups. For the first versions of PANTHER/X, we chose to address molecular function and biological process (GO also contains a cellular component ontology).
PANTHER Library (PANTHER/LIB): Subfamilies for Capturing Functional Divergence
If the functions of most proteins were experimentally characterized, assigning proteins to functional categories would be primarily a matter of data entry. Sequencing DNA, however, is a much simpler task than characterizing protein function, thus our present knowledge of protein sequences deduced from DNA far exceeds our knowledge of biological function. Because similar sequences often have similar functions, inferring function from sequence similarity has proved an invaluable tool. However, proteins within a particular family have generally evolved to have different functions, and different protein families show a wide variation in the range of functions they have adopted. It is therefore critical, when predicting protein function from sequence, to allow families to be divided into subfamilies of differing functions. To this end, we developed the PANTHER Subfamily and Family Library (PANTHER/LIB). We adopt the standard definition of subfamilies as subtrees of a family tree built from protein sequence information, but allow subtrees to be defined on a case-by-case basis by biologists who are expert in that particular family or field of biology. After choosing the best “cut” of the tree for predicting function, the biologists associate each subfamily with PANTHER/X terms defining the functions shared by all subfamily members.
Each curator-defined subtree provides a set of “training sequences” for building statistical models (HMMs). Although the present version (3.0) of the PANTHER library has been built using only publicly available sequences as of March 2001, the HMMs can be used to accurately classify novel protein sequences as well. In other words, PANTHER provides not only a controlled vocabulary for protein annotation, but also a means for consistently applying the vocabulary to new proteins. PANTHER/LIB also provides a mechanism to determine whether a new sequence represents a novel subfamily of an existing family. HMMs are built on the family level as well—if a sequence scores more highly against the family HMM than any subfamily HMM, it generally represents a novel subfamily. Family HMMs are associated with only those PANTHER/X terms that are common to all of its subfamilies, ensuring that these predictions are not more specific than justified by the data.
HMMs for Suggesting Functionally Important Residues
HMM libraries, such as PANTHER, Pfam, and SMART, are used primarily to recognize and annotate conserved motifs in protein sequences. However, the position-specific amino acid probabilities in an HMM can also be used to annotate individual positions in a protein as being conserved (or conserving a property such as hydrophobicity) and therefore likely to be required for molecular function. For example, a mutation (or variant) at a conserved position is more likely to impact the function of that protein. In addition, HMMs from different subfamilies of the same family can be compared with each other, to provide hypotheses about which residues may mediate the differences in function or specificity between the subfamilies.
Library Analogy
A useful analogy might be that PANTHER provides a library of information about protein families. Each book in the library (PANTHER/LIB) corresponds to a protein family, including a multiple sequence alignment and a tree to graphically view sequence relationships together with information about each family member. Each book is divided into chapters (subfamilies) by biologist curators, who also assign meaningful names to the book and chapters. The PANTHER/X ontology might then be analogous to an index of the PANTHER library. The curators assign each subfamily, as well as the family, to appropriate ontology categories, in effect indexing sequences by their functions. Statistical models (HMMs) are built from the sequences in each family and subfamily, and these HMMs can then be used to index (classify) novel sequences. PANTHER has an interface for browsing and searching by either function or family/subfamily terms, and access to the multiple alignment and sequence-based tree representation of each family, as well as lists of all proteins in a given organism that belong to a given family or subfamily (Thomas et al. 2003; http://panther.celera.com).
In this paper, we describe a method for relating protein sequence relationships to function relationships. We also describe the PANTHER/X ontology that we have developed for summarizing and navigating molecular function and biological process. We then apply PANTHER to three areas of active research:
The family and subfamily classes are used to derive statistics on the present sizes and sequence diversity of protein families and subfamilies, in terms of the number of members in the nonredundant protein database in GenBank.
The associations of proteins with function ontology terms are used to visualize the relative number of human and mouse genes with a given molecular function or participating in a given biological process.
Position-specific scores in the family HMMs are used to rank known or putative missense SNPs according to their likelihood of affecting protein function.
RESULTS
Size and Sequence Similarity Distribution of Protein Families and Subfamilies in GenBank
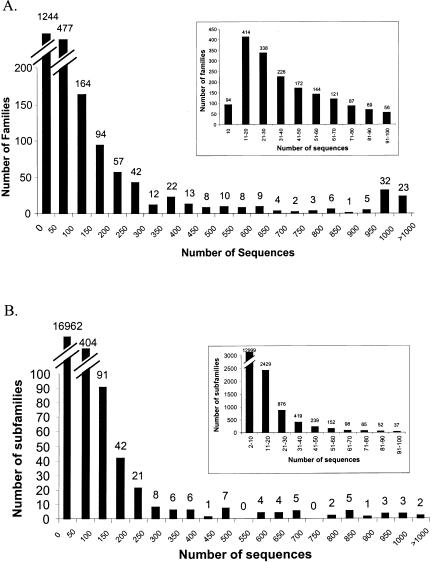
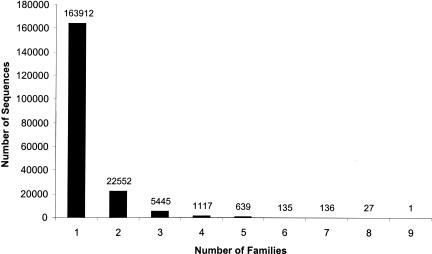
How many different sequences are presently in the PANTHER families and subfamilies? Figure 1 shows the histogram of sizes of PANTHER Version 3.0 families and subfamilies in the nonredundant protein database (NRDB) from GenBank. Before calculating these numbers, we first attempted to remove (“filter”) engineered sequences and fragments (see Methods). In total, 30.4% of the sequences in NRDB were used as PANTHER training sequences. The histogram accurately reflects the size distribution of protein families and subfamilies, except for the smallest and largest groups. In PANTHER Version 3, we have required a minimum of 10 members to define a family, and have limited large families to 1000 members (for efficiency of tree construction and curation). Note also that families can overlap by up to 90% in terms of their training sequences, thus members of some larger superfamilies (or sequences containing domains found in many different multidomain arrangements) are represented in more than one PANTHER family. Figure 2 shows the distribution of the number of families per sequence. Most sequences (85%) appear in only one family, and no sequence appears in more than nine families. The sequence that appears in nine families is, not surprisingly, an immunoglobulin variable region (Ig-V), simply because there are several thousand Ig-V regions in the nonredundant database (many differing from each other at only a single position), which are artificially divided into different families by the 1000-sequence limit. Other sequences that appear in the largest number of PANTHER families are myosins, and Notch-related proteins.
Figure 1.
Number of sequences in PANTHER families and subfamilies. (A) the distribution of the sizes of PANTHER/LIB families. Note that families are limited to no less than 10 sequences, and no more than 1000 sequences. (B) distribution of the sizes of PANTHER/LIB subfamilies. Singleton subfamilies are not included in the figure. The insets show a more detailed view of the distributions for sizes smaller than 100 sequences.
Figure 2.
Overlap of PANTHER families. Some sequences appear in more than one family, and this figure shows the distribution of the number of families in which a given sequence appears. Most sequences (163,912, 85%) appear in only one family, and no sequence appears in more than nine families.
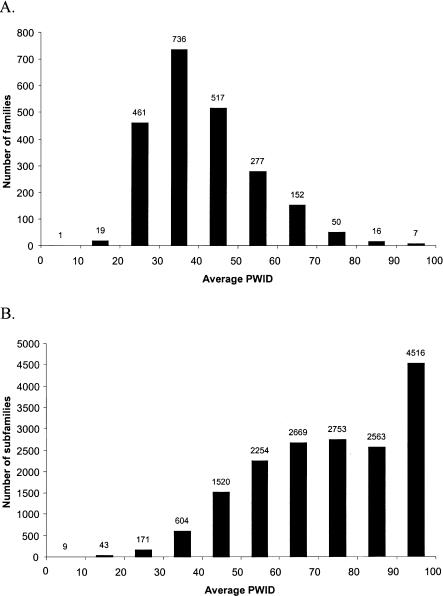
How similar are the sequences in a PANTHER family or subfamily? Figure 3 shows the histogram of the average percentage identity of sequence pairs in the same PANTHER family or subfamily. The average pairwise identity clearly peaks at 30%–40% for families, with 1714 (77%) of the families falling between 20% and 50% average pairwise identity. Some protein families have clearly diverged more than others in sequence. For subfamilies, which are defined more by functional (rather than just sequence) conservation, the distribution is broader. The number of functionally defined subfamilies is approximately constant across the range of pairwise identities from 50% to 90%. This suggests that different protein families have very different constraints on the average number of sequence changes required to alter their biological function. For this reason, PANTHER subfamilies are defined on a case-by-case basis by expert curators, rather than by using a computational algorithm. The peak at an average pair-wise identity of >95% probably primarily reflects the fact that to date, sequencing projects have focused on a few key model organisms, and sampled others very nonrandomly.
Figure 3.
Pairwise identity within PANTHER families and subfamilies. (A) Average pair-wise identity within PANTHER families. (B) Average pairwise identity within PANTHER subfamilies. Singleton subfamilies are not included. Pairwise identity is calculated over only the region of the sequences that aligns to the family HMM.
Molecular Function and Biological Process Classifications of Human and Mouse Genes
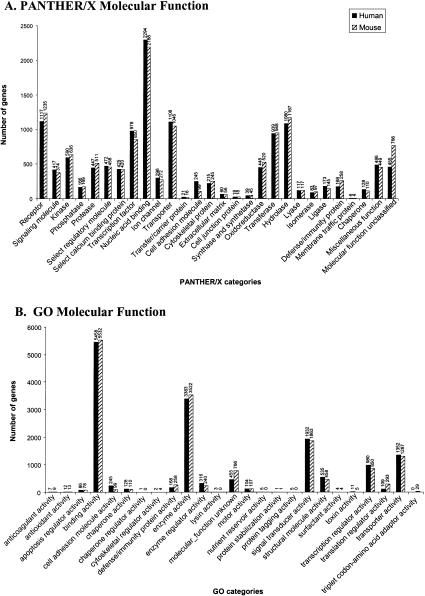
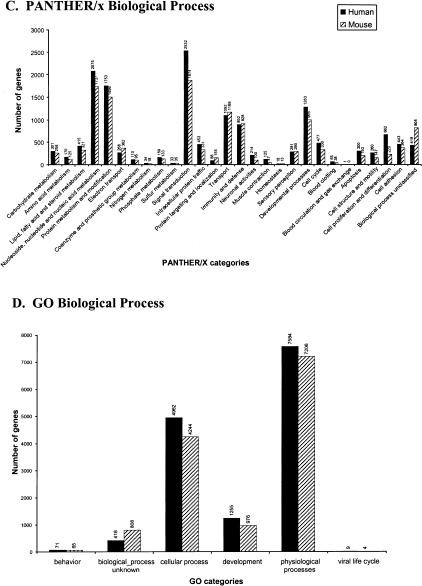
Among the applications of PANTHER/X is visualizing, in biological terms, inventories across databases or genomes (Venter et al. 2001). Nearly every whole-genome sequencing effort has presented a pie or bar chart of functions, or similar representation of predicted genes across the genome (or chromosome, in some cases). However, no standard set of categories is used across more than one or a few different publications. Figure 4,Figure 4 shows the categorization of LocusLink (Pruitt et al. 2000; Pruitt and Maglott 2001) human genes and mouse genes using two different ontologies, GO (Fig. 4B,4D) and PANTHER/X (Fig. 4A,4C). The functions represented are the same—the only difference is the structure of the ontology (see Methods and Mi et al. 2003). In brief, for GO, each GO association is represented as its highest-level (most general) term, derived by tracing up the edges of the DAG. For PANTHER/X, the GO association was first mapped to the closest matching term in PANTHER/X and then traced to its highest-level term. For molecular function, GO contains 28 level 1 terms. The distribution of gene products in different GO categories is very uneven: Nine categories contain no LocusLink associations at all, and taken together the emptiest 15 categories (54% of the categories) contain only a total of 14 LocusLink assignments (0.17% of the associations). On the other hand, three categories (enzyme activity, binding activity, and signal transducer activity) contain ∼70% of the LocusLink gene associations. Using PANTHER/X, there is a more even distribution of association across categories (Fig. 4A,4C) without sacrificing biological meaning, facilitating visual analysis. There is also significantly more detail, especially for biological process terms (note that if GO level 2 terms had been used instead of level 1 terms, there would have been 468 molecular function and 261 biological process terms, making a bar chart difficult to reproduce here). Although most categories contain approximately the same number of human genes as mouse genes, it is apparent in Figure 4,Figure 4 that there are several categories with significant differences between the genomes. This can be either because of real differences in gene number in the mouse and human genomes, or because of inconsistencies of associations in LocusLink. Three PANTHER biological process categories each contain more than two times as many human genes as mouse genes: neuronal activities, muscle contraction, and cell proliferation and differentiation.
Figure 4.
Comparing classifications of human and mouse LocusLink genes using GO terms and their mapped PANTHER/X terms. Top-level molecular function categories for (A) PANTHER/X and (B) GO. Top-level biological process terms for (C) PANTHER/X and (D) GO. The set of gene classifications is identical for PANTHER/X and GO; the difference is in organization (relationships between ontology terms).
Several GO terms cannot be mapped to PANTHER/X, even in an abbreviated form. This has relatively little impact on the number of proteins that can be meaningfully classified. In total, <0.5% of classified LocusLink genes have a GO term but no mapped PANTHER/X term. For human biological process classifications, ∼25% of the unmapped terms have actually been made obsolete in GO, and the other unmapped terms are generally detailed terms that have not yet been mapped to PANTHER/X. For unmapped human molecular function classifications, ∼33% represent terms that are now obsolete. No less than 51% of the unmapped terms are “binding proteins” defined under binding activity (GO:0005488), such as zinc binding (GO:0008270), protein binding (GO:0005515), or transcription factor binding (GO: 0008134). When designing PANTHER/X, we decided that these categories did not carry the same degree of functional meaning as receptor or transcription factor, for example, and were not as useful for dividing sets of proteins. To take the most extreme example, nearly all proteins can be categorized under protein binding. In other cases, PANTHER/X terms can be found in a different section of GO. For example, cytoskeletal proteins and extracellular matrix proteins are found in the GO cellular component ontology, but were included in the PANTHER/X molecular function ontology because they also have functional implications.
Predicting the Effect of Missense SNPs on Protein Function Using HMMs
At the level of protein sequences, each step in the process of evolution can be viewed (usefully if simplistically) as a random mutation followed by an in vivo functional assay. If we observe enough different proteins that have the same function to some degree of approximation, we can assume that we have sampled most of the “neutral” mutations (those that do not impair function). In real alignments, of course, we do not typically have a broad enough sampling of different sequences to assume that all possible functional variants have been observed. Fortunately, statistical solutions to this problem have already been developed for HMM modeling of protein families, and we suggest that they can also be fruitfully applied to the problem of “missense SNP scoring” in proteins. Indeed, the method proposed by Ng and Henikoff (2001) uses statistical methods that are very similar to those implemented in the SAM HMM modeling package (Hughey and Krogh 1996; Karplus et al. 1998), including the use of Dirichlet mixture priors (Sjolander et al. 1996).
The PANTHER/LIB HMMs can be viewed as a statistical method for scoring the “functional likelihood” of different amino acid substitutions on a wide variety of proteins. Because it uses evolutionarily related sequences to estimate the probability of a given amino acid at a particular position in a protein, the method can be referred to as generating “position-specific evolutionary conservation” (PSEC) scores. For the preliminary analysis presented here, we use the PANTHER Version 3.0 family-level HMMs (not subfamily-level). To demonstrate the utility of this view of the HMM probabilities, we analyzed the missense allele pairs obtained from two different databases. The first is the Human Gene Mutation Database (HGMD; Krawczak and Cooper 1997; Cooper et al. 1998), a curated database of mutations in human genes, most of which are linked to a disease. The second is dbSNP (Sherry et al. 2001), a database of human gene variations, most of which were collected randomly. We can then score the likelihood of a single amino acid at a particular position (amino acid PSEC, aaPSEC), or the likelihood of the transition of one amino acid to another (substitution PSEC, subPSEC). Formally, we define the scores as follows:
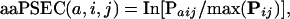 |
(1) |
where Paij signifies the probability of amino acid type a at position i in HMM j, the maximum is taken over the probabilities of all amino acids at position i of HMM j, and
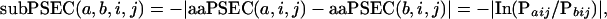 |
(2) |
for a substitution of amino acids a and b.
When aaPSEC = 0, this is the evolutionarily most common allele (inferred to be definitely functional), whereas more negative values of aaPSEC indicate that the allele is less likely to be observed across evolution (inferred to be less likely to conserve function). The substitution PEC score is simply the difference between the aaPSEC scores for the two alleles. We take the absolute value in order to make the scores symmetric, and then multiply by –1 to adhere to the substitution matrix convention that more negative scores correspond to more severe substitutions. When subPSEC = 0, the substitution is interpreted as functionally neutral, whereas more negative values of subPSEC predict more deleterious substitutions.
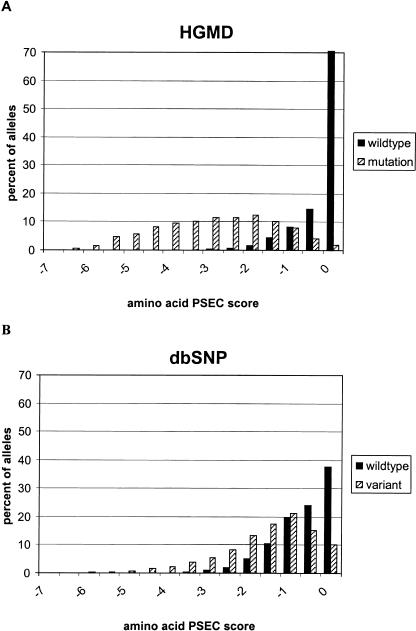
First, we compare aaPSEC scores for wild-type and mutant alleles in HGMD. If we assume that all HGMD mutations are causative for a disease, then the wild-type allele is assumed to be functional, whereas the mutant is impaired. In other words, we can use wild-type aaPSEC scores to represent a set of functional variants, and mutant aaPSEC scores to represent nonfunctional variants (Fig. 5A). The PANTHER Version 3.0 library assigned PSEC scores to 76% of the pairs (other positions could not be aligned to a PANTHER HMM). As expected, the distribution of HGMD mutant alleles extends to very negative aaPSEC scores, whereas the wild-type allele distribution is peaked at 0. Only 0.1% (a total of 12) of the wild-type alleles have aaPSEC < –3. These exceptions may help prove the rule, with some wild-type alleles actually encoding functionally impaired proteins. An interesting example is the Y897S mutation in Tie2, for which there are data at both the phenotypic level (an association with inherited venous malformations) and the molecular level (tyrosine kinase activity). The evolutionary conservation pattern at position 897 predicts that the disease-associated mutant with serine at this position should be “functional” while the wild type is not, exactly the reverse of what we would naively expect. Significantly, Y897S was shown to be a gain-of-function mutation, resulting in an eightfold increase in ligand-dependent autophosphorylation (Calvert et al. 1999), and is an interesting example of how a wild-type protein that is functionally impaired at the molecular level can be more “functional” at the phenotypic level. In contrast to the small number of wild-type alleles with aaPSEC < –3, >40% of the mutant alleles fall below this cutoff, suggesting this may be a useful cutoff value. The simple model of considering aaPSEC < –3 indicates that at least 0.76 × 0.41 = 31% of the mutant alleles in HGMD have impaired function and are therefore likely to be causative for the disease they are linked to. The number is likely to be significantly higher than this. If we consider progressively higher score intervals, the ratio of mutant alleles to wild-type alleles decreases rapidly, but even for the interval –1.0> aaPSEC > –1.5, there are more mutant alleles than wild-type alleles.
Figure 5.
Distribution of amino acid scores (aaPEC) for different missense SNP alleles in HGMD and dbSNP. (A) The distribution from HGMD shows that >40% of the disease-associated mutant alleles (hatched bars) are rare (aaPEC < –3) in alignments of related sequences, whereas >70% of the wild-type alleles (blackbars) are the most common allele across evolutionarily related sequences (aaPEC = 0). (B) The distribution from dbSNP (presumably randomly sampled SNPs) is very different from A, containing four times fewer evolutionarily rare alleles (aaPEC < –3) and more than one-third fewer evolutionarily most common alleles (aaPEC = 0).
Figure 5B shows a similar analysis of the set of missense variations listed in dbSNP. The wild-type here is defined by the allele represented in the primary RefSeq sequence. The distributions for different dbSNP alleles are not as well segregated as they are for HGMD, implying that the dbSNP substitutions are much less likely to occur at evolutionarily conserved sites in proteins. In addition, PANTHER HMMs (which represent the most highly conserved regions in proteins) cannot provide scores for as large a fraction of dbSNP variations as for HGMD. Of the 16,076 missense variations in dbSNP, 9920 occur in proteins with significant scores to a PANTHER HMM (NLL-NULL score < –100), of which 6508 align to a position in the top-scoring PANTHER HMM. This means we can analyze 40% of the dbSNP missense SNPs, as compared with 76% for HGMD. Of alleles we can score, 9.2% (598/6508) fall above our cutoff of aaPSEC < –3, as compared with 41% for the HGMD set. This indicates that our score correlates well with functional effect. However, there are still several low-scoring alleles in the dbSNP set, which we would predict to have impaired function. This will require detailed investigation outside the scope of this paper. Our list of potentially deleterious missense SNPs in dbSNP includes several well-characterized alleles known to have functional effect, such as R145C in apoE2* and apoE4-Philadelphia (aaPSEC = subPSEC = –5.06) and R158C in apoE2 (aaPSEC = sub-PSEC = –3.25). In addition, ∼5% of these low-scoring missense SNPs occur in olfactory receptor genes, in which mutations deleterious at the molecular level are not likely to affect survival in humans.
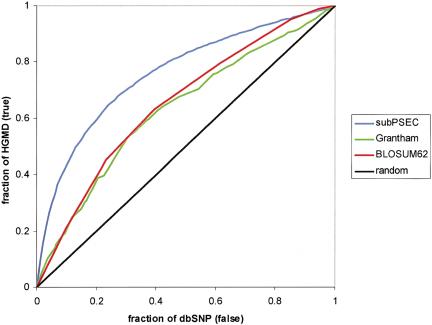
We now assess the ability of the substitution PEC scores, subPSEC, to separate neutral from deleterious missense SNPs. Figure 6 shows a relative operating characteristic (ROC) plot (Swets 1988) to compare position-specific substitution scores with two of the most commonly used amino acid substitution scales. The first is BLOSUM62 (Henikoff and Henikoff 1993), the most highly used substitution matrix for comparing protein sequences, and the second is the physico-chemical distance score proposed by Grantham (1974). In the ROC plot for assessing diagnostic accuracy, the “signal” of correct predictions (true positives) is shown as a function of the “noise” of incorrect predictions (false positives). We use the HGMD mutations as an approximate set of functionally deleterious missense SNPs, and the set of db-SNP variations as an approximate set of neutral missense SNPs. In actuality, of course, not all HGMD alleles are deleterious, nor are all dbSNP alleles neutral. Nevertheless, we expect HGMD to be significantly enriched in deleterious alleles relative to dbSNP; therefore, different scoring schemes can be compared with each other based on how well they segregate the alleles in these different sets.
Figure 6.
Predicting whether a missense SNP will have an effect on protein function: comparison between position-specific scores (subPEC) and “average” substitution scores. Position-specific scores from PANTHER HMMs (blue line) make a larger number of correct predictions (true positives shown on Y-axis) for a given number of errors (false positives shown on X-axis) than scores from the two most commonly referenced substitution scores: the Grantham scale (green line) and the BLOSUM62 substitution matrix (red line). The black line shows the curve for a random prediction, as a reference. HGMD mutations are used to approximate a set of functionally impaired proteins, and dbSNP variations are used to approximate a set of functional proteins (see text for more details).
In the ROC plot, a perfect prediction method would give a vertical line (infinite slope) with a noise of 0, and a completely random prediction would give a line with a slope of 1. The position-specific scores have a much higher slope than for BLOSUM62 scores, particularly at low error values. For example, using position-specific scores, in order to predict 10% of the HGMD (presumably deleterious) alleles, one would also incorrectly predict ∼1% of the dbSNP (presumably neutral) to be deleterious as well. To predict 10% of the deleterious alleles with BLOSUM62 scores, one would expect a number of errors roughly equal to 5% of the neutral alleles. This means that the false-positive prediction rate in this range is five times greater for BLOSUM62 than for position-specific scores. Our results are consistent with Ng and Henikoff (2001), showing that the “average” substitution probabilities in a substitution matrix are not as well suited as position-specific scores for scoring the functional likelihood of missense SNPs.
The position-specific scores are particularly effective at “rescuing” false-negative predictions by BLOSUM62 or Grantham scores. For example, the F294Y mutation in the GALT gene product has a high BLOSUM62 score, but has a very low position-specific score. In other words, the substitution appears conservative in an average sense, but at this particular position in the galactose-1-phosphate uridylyltransferase family, phenylalanine is absolutely conserved and any mutation should score poorly. Not surprisingly, then, F294Y is associated with galactosemia. Another example is the apparently conservative D203E mutation in the CHST6 gene product, which leads to type 1 macular corneal dystrophy.
A significantly smaller but fully experimentally validated set of SNPs is available from the Whitehead Institute (Cargill et al. 1999). We considered all 115 missense SNPs scored by Ng and Henikoff (2002). We found that 100/115 aligned to a position in an HMM from PANTHER Version 3.0, and could be given sub-PSEC scores. Of the 10 lowest-scoring missense SNPs, seven are also predicted by Ng and Henikoff to be deleterious, thus there is significant agreement on the strongest predictions. However, whereas Ng and Henikoff predict that 19/100 of the missense SNPs in this set are deleterious, only 5/100 have subPSEC < –3 (L57P in the INTGB3 gene product, R163W in F3, F291S in ANX3, L88R in DRD5, and I173N in CYP21), indicating that our cutoff is considerably more conservative. We therefore expect a lower false-positive prediction rate, but a higher false-negative prediction rate, than Ng and Henikoff.
DISCUSSION
We have described PANTHER, a comprehensive database for classifying protein sequences (see next page). PANTHER Version 3.0 includes >2200 protein families, which are further subdivided into >30,000 subfamilies. A subfamily is defined as groups of proteins that can be annotated as having a similar name, and identical biological function, as judged by biologist curators. Each family is represented as a tree, a multiple sequence alignment, and an HMM for searching. Subfamilies are curator-defined subtrees of the family tree, and also represented as HMMs. Both families and subfamilies have been named by biologist curators and associated with ontology terms describing function. It is hoped that the broader scientific community can help to ensure that the names and ontology associations are correct and up to date.
We have characterized the size and sequence similarity distributions for PANTHER/LIB families and subfamilies. Consistent with previous results from several studies, as family size increases, the number of families of that size decreases rapidly. Also consistent with previous studies, the distribution of sequence similarity within families is peaked sharply around 30%–40% identity, owing primarily to the practical limits of aligning related protein sequences. We report for the first time the corresponding distributions for protein subfamilies, where subfamilies are defined as comprising proteins that have the same function (to the best of our biological knowledge at present). We find that the sequence similarity distribution for subfamilies is much broader than for families, indicating that the relationship between sequence and functional plasticity varies widely for different protein families.
We have also illustrated the utility of our abbreviated ontology (PANTHER/X) for high-level analysis of large lists of proteins. Compared with a slice through a given depth of the Gene Ontology (GO), PANTHER/X divides mammalian genes into functional bins containing a relatively consistent (and tractable) number of sequences, allowing us to identify biological processes for which the number of associated human and mouse genes differs significantly (e.g. neuronal activities). It is important to emphasize that PANTHER/X was designed primarily for mammalian (or vertebrate, at least) proteins, and will need to be augmented to provide a more comprehensive classification of proteins from a broader range of organisms.
Finally, we have used the position-specific amino acid probabilities in the PANTHER/LIB HMMs to score single nucleotide polymorphisms in human proteins that lead to an amino acid substitution (missense SNPs). We have scored mutant alleles from the Human Gene Mutation Database (HGMD), using both dbSNP (a database of mostly randomly sampled variation) as well as wild-type HGMD alleles as controls. Our results indicate that HMM scores, derived from observing amino acid substitutions in a specific position in related protein sequences, can be usefully applied to the problem of predicting whether a given allele will be functionally neutral or deleterious. Our results demonstrate on a database-wide scale that position-specific scores are more effective at this task than substitution matrices such as BLOSUM62 and the Grantham scale. If we choose a very conservative prediction cutoff based on wild-type allele scores in HGMD, roughly 4% of the missense SNPs in dbSNP (we were able to make predictions for 40% of the missense SNPs, and ∼9% of those were below our cutoff) are likely to affect protein function. These predictions will require further analysis.
METHODS
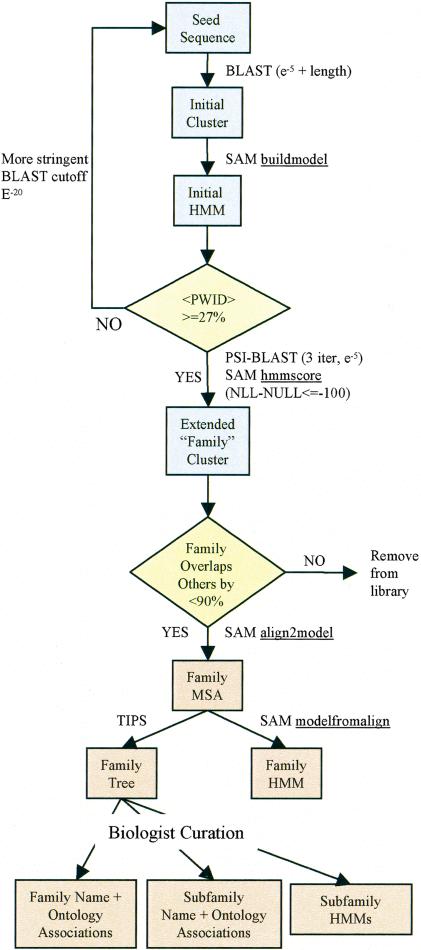
The overall process for building the PANTHER classification is shown in Figure 7. The basic steps are:
Figure 7.
Schematic illustration of the process for building PANTHER families.
Family clustering.
Multiple sequence alignment (MSA), family HMM, and family tree building.
Family/subfamily definition and naming.
Subfamily HMM building.
Molecular function and biological process association.
Of these, steps 1, 2, and 4 are computational, and steps 3 and 5 are human-curated (with the extensive aid of software tools).
Family Clustering
In PANTHER, families are defined as clusters of related proteins for which a good multiple sequence alignment can be made. The clusters are built around “seed” sequences, in two steps. In the first step, we use BLASTP to find sequences related to the seed in both sequence and overall length. An HMM is then constructed from this “initial cluster,” which is used to find additional members of the family to define an “extended cluster.” The two-step process allows us to avoid the problems of training an initial HMM from a diverse set of sequences, yet still capture the diversity of the larger set in the final HMM.
Seed Selection
Seed selection involves choosing the proteins that will serve as “seeds” around which we will build initial HMMs. For PANTHER Version 3.0, we focused on annotating mammalian genomes, and thus we biased our seed set accordingly. We defined our starting set as all human, mouse, and rat proteins in the GenBank Nonredundant (NR) Protein Database Release 122 (February 15, 2001). From this set, we removed (“filtered”) very short sequences (<30 amino acids), sequences annotated as partial (having an NR definition line containing the words “partial” or “fragment”) or mutants (definition line containing the strings “mutant,” “mutation,” “engineer,” or “synthetic”). Engineered mutants often contain changes to key functional residues, and including them can weaken the residue conservation profiles. We then sorted the sequences from longest to shortest, and used BLASTP alignments to split them into clusters defined by a percent identity cutoff (25%) and length-based cutoff (the length of the aligned region must be at least 70% of the length of the shorter sequence). From each cluster, the representative seed was defined as the sequence having the median length.
Initial Cluster and Initial HMM Building
The goal of this step is to generate a cluster of sequences that are globally homologous to the seed, in order to generate the initial HMM to reflect the seed's domain arrangement.
The seed is used to query the NR database (filtered to remove fragments and mutants, as above) using BLASTP. A sequence “hit” is accepted into the initial cluster if (1) it has an E-value < 10–5 and (2) the length of the BLASTP alignment is at least 70% of both the query and hit sequences. This is important because each cluster must contain related proteins that are all of roughly equal length, so that they are likely to share the same domain structure. All related sequences passing these thresholds are brought into the initial cluster (up to 500, sorted by E-value, for computational efficiency), and any sequences that are exact subsequences of another sequence in the cluster are removed (these are likely to be fragments).
The initial cluster is used as input into the buildmodel procedure of the UCSC SAM 2.1 package using the Dirichlet mixture prior parameter file -prior_library uprior9.plib. This creates a temporary HMM that is used to provide (1) an alignment that can be used to estimate the weights of the sequences in the initial HMM (using the SAM align2model procedure with the -sw2 option), (2) the length of the region conserved among the sequences in the family (using the “surgery” option in buildmodel). Sequences are weighted relatively using the Henikoff weighting scheme (Henikoff and Henikoff 1994), and given an absolute weight using the formula 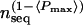 , where nseq is the number of sequences in an alignment and Pmax is the average probability for the most common amino acid at each position (Karplus et al. 1997). If >3.0, the absolute weight (i.e., number of independent counts) for all sequences is scaled to equal 3.0; otherwise, the HMM parameters will contain negligible contributions from the priors. The intial (weighted) HMM is built by using the resulting sequence weight file and the initial cluster file as input into buildmodel along with the following parameters: -nsurgery 0, -nmodels 1, and -modellength 0. These particular parameters are used so that the model length is constrained to remain the same as in the temporary HMM (this is done to reduce the computation time). The sequences in the initial cluster are then aligned to the initial HMM to produce an initial MSA.
, where nseq is the number of sequences in an alignment and Pmax is the average probability for the most common amino acid at each position (Karplus et al. 1997). If >3.0, the absolute weight (i.e., number of independent counts) for all sequences is scaled to equal 3.0; otherwise, the HMM parameters will contain negligible contributions from the priors. The intial (weighted) HMM is built by using the resulting sequence weight file and the initial cluster file as input into buildmodel along with the following parameters: -nsurgery 0, -nmodels 1, and -modellength 0. These particular parameters are used so that the model length is constrained to remain the same as in the temporary HMM (this is done to reduce the computation time). The sequences in the initial cluster are then aligned to the initial HMM to produce an initial MSA.
QA on Initial MSA
It is essential that the MSA be of high quality; otherwise, the resulting tree structure is unlikely to accurately reflect functional relationships. We have observed empirically that potentially poor alignments can be reliably identified by calculating the average pairwise identity over the regions of the sequences that align to the HMM. If an MSA has an average pairwise identity of <27%, the family-building process is restarted around the seed using a more stringent BLAST E-value cutoff (10–20). We find that ∼5% of the PANTHER Version 3.0 families fail this first QA step and must be rebuilt.
Extended Cluster Building
The goal of this step is to extend the clusters to include as many related sequences as possible. This will (1) make the resulting HMMs much more powerful because there will be more “observed” sequences to provide residue substitution statistics, and (2) bring more sequences into the family trees, providing as much information as possible about relationships that biologist curators can use to infer function.
We use the initial family HMM to search for new cluster members. Because it would be computationally prohibitive to score the resulting HMMs against the entire NR protein set, we need to define a smaller “search set” of proteins that are potentially related to the seed. We take the seed and run PSI-BLAST for three iterations (using an E-value cutoff of 10–5), and define the search set as the set of all proteins that appear in any of the PSI-BLAST iterations (not just the final iteration, because for some seed sequences PSI-BLAST can “wander” to very different protein families). For this step, we filter out mutants from NR, but we allow fragments as they can provide additional observations to refine the HMM parameters.
We then score the initial HMM against the search set using SAM hmmscore (with the local alignment parameter -sw2). There is no length restriction to hits here—any protein is brought into the cluster if it shares even a local (partial) match to the HMM as long as the resulting alignment is of high quality. Empirically, we find that for most families, a related protein that has a SAM (NLL-NULL) score better than –100 (units are natural logarithms or “nats”) has a high-quality alignment, and sequences scoring better than this cutoff are added to the initial cluster to define the extended cluster.
Removing Overlapping Clusters
The family clustering procedure described above naturally produces overlapping clusters for many protein superfamilies. Our goal for clustering was to span protein space well, not necessarily to partition it such that each sequence can appear in only one family. Because of the domain arrangement of proteins, as well as the broad evolutionary distances spanned by some families, the rigorous partitioning approach does not provide as much context as the spanning approach. However, we do want to remove any clusters that are essentially completely contained in other clusters, biasing our set toward larger clusters. To do this, we sort the clusters from largest to smallest, and then go down this list to choose which clusters are accepted into the library. The largest cluster is automatically accepted. The next largest cluster is accepted if <90% of its sequences are contained in the set spanned by all accepted clusters; this step is iterated until all clusters have been either accepted or rejected. Because we allow up to 90% overlap in sequence clusters, there are several examples of overlapping PANTHER families (Fig. 2).
Family MSA Building and HMM Re-estimation
The goal of this stage is to obtain a multiple sequence alignment for the extended cluster, and to re-estimate the parameters of the family HMM given all of the new sequences brought into the cluster during the extension step.
The final multiple sequence alignment for the family (the sequences in the extended cluster) is then created. Sequences are aligned (using SAM align2model) to the HMM from the initial cluster to produce a multiple sequence alignment. Recall that the extension process can bring in proteins that only match locally (over a single region, such as a domain) if the match is close enough to pass the score threshold. Therefore, it is critical that this alignment step be a local–local, or Smith-Waterman, type of alignment. Sequences are then reweighted as above, and these weights are used to re-estimate the family HMM parameters from the final multiple sequence alignment (using modelfromalign). Note that, unlike for the initial temporary HMM, the model length is constrained to remain the same as in the initial model. Because the extended alignment is local, poor or truncated statistical models can often result if the model length is allowed to vary during this step.
Sequence-Based Family Tree Building
Once all family clusters are obtained and the highly overlapping clusters removed, each remaining family MSA is used to build a tree representation of the sequence relationships between family members. The TIPS (Tree Inferred from Profile Scores) algorithm is described elsewhere (K. Diemer, B. Lazareva-Ulitsky, T. Hatton, and P.D. Thomas, in prep.). In overview, the method follows an agglomerative clustering process. For each cluster at any step in the process, a statistical profile is built that describes those sequences. The two most similar clusters are joined at each step. The similarity S between any two clusters K and M is defined by the equation:
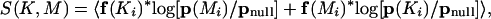 |
where the average is taken over all columns i that belong to the overlap of two alignments K and M, f(Ki) is the frequency vector of amino acids in the i-th match position in the alignment in cluster K, p(Ki) is the Dirichlet mixture profile vector built around sequences in cluster K in that position, and pnull is the background distribution (average probabilities of observing different amino acid types). In the above formula, we use a short-hand notation for the vector-derived quantities, where
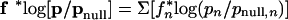 |
summing over the 20 amino acid types n. It should be noted here that both profiles p and frequencies f are calculated from weighted sequences using Henikoff-style sequence weighting.
In words, this translates to defining the similarity between clusters K and M as the average score of the sequences in K versus the profile for M, added to the average score of the sequences in M versus the profile for K (note that the profile score is effectively the HMM score, except that only aligned positions, and not insertions and deletions, are considered). The two clusters that have the maximum value of this function are joined. If the sequences in group K all score well against the profile for M, and vice versa, then the groups have similar residue conservation patterns and should be joined. Branch lengths for the join are estimated using symmetrized total relative entropy (Sjolander 1998). Note also that the similarity function is scaled according to the length of the match between a sequence and a profile, and therefore does not penalize partial (local) alignments.
Biologist Curation
After the family trees are built, they are reviewed and annotated by a team of expert curators. Unlike any other approaches toward curation that we are aware of, curation is performed in the context of a tree; that is, a family of sequences is annotated in the context of the set of related proteins. This allows curators to make inferences that could not be made if they were looking at a single sequence at a time, as well as perform consistency checks on the incoming data as well as the annotations they make themselves. Also, most families are reviewed by curators who have expert knowledge of the relevant family, molecular function, or biological process.
One of the curator's tasks is to decide how to divide the tree into subtrees, or subfamilies. This is done using software called a “tree-attribute viewer,” that shows a table of annotations (attributes) for sequences in a tree (Thomas et al. 2003), allowing for rapid curation. Each subtree should be the largest possible subtree for which all of the sequences in the subtree share the following properties: (1) the same name (or a consistent name that can be applied to all sequences in the subtree); (2) the same molecular function(s); and (3) the same biological process(es). Note that not all sequences must be individually annotated in GenBank in exactly the same way for the curator to decide that they all, in fact, are likely to share the same attributes. In fact, the lack of standards for nomenclature, the wide range of annotation quality, and the years of transitive sequence annotation have made biologist interpretation an imperative. The curator's ability to infer the functions of proteins that are either incorrectly or inadequately annotated is precisely what we wish to exploit. The tree representation is a powerful means of grouping sequences together—each subtree is a possible subfamily. If an unannotated sequence is placed deep within a branch of sequences known to have a particular function, it is very likely that this unannotated sequence shares that function as well.
Naming the Families and Subfamilies
After deciding which subtrees should be designated as distinct subfamilies, the biologist curators give each subfamily a biologically meaningful name. In some cases, because all sequences within a subfamily have the same definition, naming the subfamily is trivial. Often, different synonyms may have been used for each of the sequences in a subfamily. In that case, curators will use their expert knowledge to pick the most informative name. If a SWISS-PROT (Bairoch and Apweiler 2000) sequence is present in a subfamily, that name is often chosen because of its high quality.
Often there are subfamilies in which none of the individual sequences has a clear function. However, that subfamily is present in a family because there is significant sequence similarity with other subfamilies. The convention used for naming these subfamilies is to determine the closest subfamily whose function is clear (X), and to name the uncertain subfamily X-RELATED. Information about the organisms from which the sequences derive is also useful in naming subfamilies. It is not uncommon for a tree to contain orthologs from a wide variety of organisms. In this case, the protein names are often inconsistent (often because of organism-specific naming conventions), but it is clear from the MSA and tree that all sequences are orthologs. In many cases, a single name is selected (the most biologically informative, sometimes biased toward nomenclature for human gene products). This rule is not applied universally because sometimes there can be well-known names in different species that the curator is uncomfortable overwriting.
Biologically meaningful names are also given to each of the families. Occasionally, a family will consist of a single subfamily: that is, given the present state of biological knowledge, all sequences have the same name and functions. More often, there are several different functions across subfamilies of an evolutionarily conserved protein family. If the protein family has a well-established name, then the PANTHER family is given that name (e.g., ANTP/PBX FAMILY OF HOMEOBOX PROTEINS). Often there is no well-established name. In this case, the curator either gives the protein a more general name that applies to all proteins in a family (e.g., NUCLEAR HORMONE RECEPTOR) or finds the largest subfamily name (Y) and names the family Y-RELATED.
Creating the PANTHER/X Abbreviated Ontology
The PANTHER/X ontology comprises two types of classifications: molecular function and biological process. The molecular function schema classifies a protein based on its biochemical properties, such as receptor, cell adhesion molecule, or kinase. The biological process schema, on the other hand, classifies a protein based on the cellular role or process in which it is involved, for example, carbohydrate metabolism (cellular role), signal transduction (cellular role), TCA cycle (pathway), neuronal activities (process), or developmental processes (process). Oncogenesis is, in fact, a pathological process, but because it is such an important field, it is included in the PANTHER/X biological process schema.
There are no more than three levels of categories in either PANTHER/X schema. Level 1 categories are broad and general functional terms, such as receptor, protease, or transcription factor in the molecular function schema, and carbohydrate metabolism, signal transduction, or developmental processes in the biological process schema. Level 2 and 3 categories are subcategories of level 1 categories, and are more specific functional terms, such as G-protein-coupled receptor, serine-type protease, or zinc finger transcription factor in the molecular function schema, and glycolysis, MAPKKK cascade, or neurogenesis in the biological process schema. Under parent categories having more than one child, we have introduced an “other” category, such as other receptor or other carbohydrate metabolism process, to avoid generating an excessive number of categories with few subfamilies classified in them.
One important point is that, properly speaking, the ontology is a DAG (directed acyclic graph) rather than a true hierarchy. In practice, this means that a given category can have more than one parent. For simplicity, we have attempted to minimize the number of instances in which the schema deviates from a hierarchy, but there are still several cases in which a child category has multiple parents. Unlike the full GO schema, a child must appear at the same level under each parent so that depth has a consistent correlation with specificity. For example, nuclear hormone receptor (level 2) is classified under the parents receptor (level 1) and transcription factor (level 1).
Associating Families and Subfamilies With Ontology Terms
This step is performed by a biologist curator. Curators use many different pieces of information while performing the classification, such as textbooks, PubMed abstracts, SWISS-PROT keywords and definitions, OMIM records (http://www.ncbi.nlm.nih.gov/omim/), GenBank records, and their own expert knowledge of the field. Because they are curating in the context of the family tree, they may also infer function based on what is known about adjacent subfamilies. Curators may only place subfamilies into existing PANTHER/X categories; they may not create a new category unless it is cooperatively decided that there is a compelling reason to do so.
Proteins having related sequences also generally have a common biochemical (molecular) function. The same is often not the case for proteins participating in the same biological process—that is, most pathways are comprised of a series of different biochemical reactions. In general, then, molecular function tends to change less dramatically within a family than does the biological process. Therefore, inferences about molecular function can more often be made than can inferences about biological process. Again, knowledge of the biological context is important. For example, an expert may be hesitant to infer the biological process of a serine/threonine kinase, but not that of citrate synthase. The number of pathways a biochemical reaction is used in affects one's ability to infer biological process.
After the subfamily-level classification was completed, categories were associated with the family-level models. Because many families contain subfamilies with diverse functions, only the categories that were common to all subfamilies were associated with the families. It is therefore possible for a family to have no function association at all, even if all of the subfamilies are associated with functions.
Quality Control of Ontology Associations
After the initial classification effort, all the ontology associations underwent a two-step quality control process: (1) validation and (2) consistency check. During the validation step, biologist curators reviewed all subfamily assignments in each category. That is, rather than making classifications family by family as in the initial assignment process, classifications were checked category by category, generally by experts with knowledge of the relevant area. In cases that were not obviously correct, textbooks, PubMed, as well as other available tools were used to resolve discrepancies. If a subfamily was incorrectly classified, or was not classified in a category it belonged in, reviewers were encouraged to provide reclassifications. These classifications were reviewed by our internal team. After the validation step was completed, a consistency check was performed. Subfamilies that shared common sequences but had not been consistently classified across different families were reviewed. Depending on the context of the subfamilies, the reviewer would decide whether to make them consistent. For example, if 4 sequences were shared by two subfamilies with 5 sequences each, these two subfamilies should have basically the same classification. However, if 4 sequences were shared by two subfamilies of very different size, say with 5 and 200 sequences, the functional classification of these two subfamilies could be different (the smaller subfamily might be much more specific as it spans fewer sequences).
Using HMMs to Classify Sequences
Query sequences can be scored against the PANTHER library of HMMs. The search takes advantage of the hierarchical structure of the library. Instead of scoring every sequence against all ∼35,000 family and subfamily HMMs, a sequence is first scored only against the 2236 family HMMs. Only if the family HMM score is marginal or significant (we use an NLL-NULL score cutoff of –20) is the sequence scored against the subfamily HMMs for that family. The PANTHER database at Celera stores all HMM scores (family or subfamily) more significant than –20. For the purposes of classification, however, the highest scoring HMM (either family or subfamily) is used. One of the key advantages of PANTHER is that a protein can be recognized as being a close relative of training sequences (subfamily member), or a more distant one (family member), and that these two cases can mean very different things for the purposes of function prediction. For example, a novel serine/threonine kinase receptor family member can only be inferred to have only the general function of a protein kinase, whereas a member of the BMPR1 subfamily of S/T kinases can be inferred to be involved in the specific biological process of skeletal development.
Comparing GO and PANTHER/X Associations for LocusLink
LocusLink GO associations were taken from the file LL_tmpl.gz downloaded from NCBI at ftp://ftp.ncbi.nih.gov/refseq/LocusLink/ (May 20, 2003). The sources of the GO associations were GOA and Proteome for human, and MGD for mouse.
In the LL_tmpl file, 6925 LocusLink entries are associated with at least one GO term each. For the high-level overview presented in Figure 4, A and 4C, each GO term was converted to its most general (top-level progenitor) category (or categories) in GO (May 5, 2003, Molecular function Revision 2.679, Biological process Revision 2.762) by tracing it up the DAG structure. For example, long-chain acyl-CoA dehydrogenase (GO:0004466) traces to enzyme activity (GO:0003824), and both angiotensin II receptor activity (GO:0004945) and hormone activity (GO:0005179) trace to a common progenitor, signal transducer activity (GO:0004871) in the molecular function ontology. Likewise, fatty acid metabolism regulation (GO:0006632) traces to cell growth and/or maintenance (GO:0008151) in the biological process ontology. If two GO associations for the same LocusLink entry were converted to the same top-level progenitor, the top-level category is counted only once. To derive the equivalent PANTHER/X associations, all LocusLink GO terms were first mapped to PANTHER/X terms. The mapping file is available as Supplemental Material (http://panther.celera.com/publications/gr7724_03=suppl). The PANTHER/X terms were then traced up the DAG to their top-level progenitor categories. Note that because both GO and PANTHER/X are DAGs, a given LocusLink association can have more than one top-level progenitor in either ontology.
Predicting the Effect of Missense SNPs on Protein Function
The PANTHER/LIB HMMs were used as a statistical method for scoring the “functional likelihood” of different amino acid substitutions on a wide variety of proteins. The set of missense SNPs associated with Mendelian diseases was taken from the Human Gene Mutation Database (HGMD; Krawczak and Cooper 1997; Cooper et al. 1998; release date March 11, 2003). The set of missense SNPs representing “normal” variation were taken from NCBI's dbSNP, which provides a mapping to RefSeq protein sequences (Sherry et al. 2001; release date May 20, 2003). A smaller but fully validated set of missense SNPs sampled from healthy individuals was taken from resequencing data generated by Cargill et al. (1999).
For missense SNPs in these sets, the protein sequence containing the missense SNP was scored against the PANTHER family HMMs using the UCSC SAM package (Baum-Welch scoring, local–local alignment sw2). The HMM with the most significant score was selected for the analysis, if the NLL-NULL score was less than –100. For missense SNPs associated with multiple proteins, the analysis was performed on the protein with the most significant HMM score. Proteins were then aligned to the most significant HMM using the UCSC SAM package (local–local alignment). Proteins that scored greater than –100 against all PANTHER HMMs were excluded from the analysis, as the alignments are less reliable. The position of the missense SNP in the protein determined the corresponding position in the aligned HMM model. If the missense SNP position aligned to an insert state, then it was excluded from our analysis. If it aligned to an HMM “match state,” then that position is represented by a vector of 20 probabilities, one for each amino acid. The appropriate amino acid probabilities were then inserted into equations 1 and 2 (see Results section) to generate position-specific evolutionary conservation scores aaPSEC and subPSEC. All aaPSEC and subPSEC scores are available as Supplemental Material.
Acknowledgments
We thank Richard Mural, Michael Ashburner, and Mark Adams for helpful comments on the manuscript. We thank Kimmen Sjolander for early discussions, particularly on the tree-attribute viewer concept and HMM building; Betty Lazareva for helping to develop scoring functions for the tree-building algorithm; Thomas Hatton for assembling the attribute table data and for assistance in curation; and Jody Vandergriff for assistance with figures. We thank Olivier Doremieux, Nan Guo, Steven Rabkin, and Shinji Sato for critical software engineering. We give special thanks to all the other biologists who helped to curate the PANTHER version 3.0 library: Elizabeth Alcamo, Michelle Arbeitman, Vivien Bonazzi, Zuoming Deng, Kent Duncan, Vikas Duvvuri, Marcos E. Garcia-Ojeda, Doug Guarnieri, Joy Hatzidakis, Caroline A. Heckman, Guy Hermans, Karen Ho, Karen Ketchum, Chinnappa Kodira, Lingyun Li, Catherine Liu, Ning Liu, Devan- and S. Manoli, Mark Melville, Natalia Milshina, Susan Prohaska, Samara Reck-Peterson, Aylin Rodan, Iain Russell, Lisa Ryner, Chris Smith, Gangadharan Subramanian, Alex Szidon, Jon Tupy, Michael Vagell, Ursula Vitt, Richard Wagner, Jian Wang, James H. Whalen, Paul Woo, Jennifer Wortman, and Jianbo Yue.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
[Supplemental material is available online at http://panther.celera.com/publications/gr7724_03=suppl.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.772403.
References
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M., Ball, C.A., Blake, J.A., Botstein, D., Butler, H., Cherry, J.M., Davis, A.P., Dolinski, K., Dwight, S.S., Eppig, J.T., et al. The Gene Ontology Consortium. 2000. Gene ontology: Tool for the unification of biology. Nat. Genet. 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwood, T.K., Beck, M.E., Bleasby, A.J., and Parry-Smith, D.J. 1994. PRINTS—A database of protein motif fingerprints. Nucleic Acids Res. 22: 3590–3596. [PMC free article] [PubMed] [Google Scholar]
- Bairoch, A. 1991. PROSITE: A dictionary of sites and patterns in proteins. Nucleic Acids Res. 19 Suppl: 2241–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch, A. and Apweiler, R. 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28: 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S.R., Griffiths-Jones, S., Howe, K.L., Marshall, M., and Sonnhammer, E.L. 2002. The Pfam protein families database. Nucleic Acids Res. 30: 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert, J.T., Riney, T.J., Kontos, C.D., Cha, E.H., Prieto, V.G., Shea, C.R., Berg, J.N., Nevin, N.C., Simpson, S.A., Pasyk, K.A., et al. 1999. Allelic and locus heterogeneity in inherited venous malformations. Hum Mol. Genet. 8: 1279–1289. [DOI] [PubMed] [Google Scholar]
- Cargill, M., Altshuler, D., Ireland, J., Sklar, P., Ardlie, K., Patil, N., Shaw, N., Lane, C.R., Lim, E.P., Kalyanaraman, N., et al. 1999. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 22: 231–238. [DOI] [PubMed] [Google Scholar]
- Chiu, I.M., Yaniv, A., Dahlberg, J.E., Gazit, A., Skuntz, S.F., Tronick, S.R., and Aaronson, S.A. 1985. Nucleotide sequence evidence for relationship of AIDS retrovirus to lentiviruses. Nature 317: 366–368. [DOI] [PubMed] [Google Scholar]
- Cooper, D.N., Ball, E.V., and Krawczak, M. 1998. The Human Gene Mutation Database. Nucleic Acids Res. 26: 285–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayhoff, M.O., Barker, W.C., and McLaughlin, P.J. 1974. Inferences from protein and nucleic acid sequences: Early molecular evolution, divergence of kingdoms and rates of change. Orig. Life 5: 311–330. [PubMed] [Google Scholar]
- Eddy, S.R. 1996. Hidden Markov models. Curr. Opin. Struct. Biol. 6: 361–365. [DOI] [PubMed] [Google Scholar]
- Grantham R. 1974. Amino acid difference formula to help explain protein evolution. Science 185: 862–864. [DOI] [PubMed] [Google Scholar]
- Gribskov, M., McLachlan, A.D., and Eisenberg, D. 1987. Profile analysis: Detection of distantly related proteins. Proc. Natl. Acad. Sci. 84: 4355–4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft, D.H., Selengut, J.D., and White, O. 2003. The TIGRFAMs database of protein families. Nucleic Acids Res. 31: 371–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannenhalli, S.S. and Russell, R.B. 2000. Analysis and prediction of functional sub-types from protein sequence alignments. J. Mol. Biol. 13: 61–76. [DOI] [PubMed] [Google Scholar]
- Henikoff, S. and Henikoff, J.G. 1991. Automated assembly of protein blocks for database searching. Nucleic Acids Res. 19: 6565–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1993. Performance evaluation of amino acid substitution matrices. Proteins 17: 49–61. [DOI] [PubMed] [Google Scholar]
- ____. 1994. Position-based sequence weights. J. Mol. Biol. 243: 574–578. [DOI] [PubMed] [Google Scholar]
- Hughey, R. and Krogh, A. 1996. Hidden Markov models for sequence analysis: Extension and analysis of the basic method. Comput. Appl. Biosci. 12: 327–345. [DOI] [PubMed] [Google Scholar]
- Jongeneel, C.V., Bouvier, J., and Bairoch, A. 1989. A unique signature identifies a family of zinc-dependent metallopeptidases. FEBS Lett. 242: 211–214. [DOI] [PubMed] [Google Scholar]
- Karp, P.D. and Riley, M. 1993. Representations of metabolic knowledge. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1: 207–215. [PubMed] [Google Scholar]
- Karplus, K., Sjolander, K., Barrett, C., Cline, M., Haussler, D., Hughey, R., Holm, L., and Sander, C. 1997. Predicting protein structure using hidden Markov models. Proteins Suppl 1: 134–139. [DOI] [PubMed] [Google Scholar]
- Karplus, K., Barrett, C., and Hughey, R. 1998. Hidden Markov models for detecting remote protein homologies. Bioinformatics 14: 846–856. [DOI] [PubMed] [Google Scholar]
- Krawczak, M. and Cooper, D.N. 1997. The Human Gene Mutation Database. Trends Genet. 13: 121–122. [DOI] [PubMed] [Google Scholar]
- Krogh, A., Brown, M., Mian, I.S., Sjolander, K., and Haussler, D. 1994. Hidden Markov models in computational biology. Applications to protein modeling. J. Mol. Biol. 235: 1501–1531. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Letunic, I., Goodstadt, L., Dickens, N.J., Doerks, T., Schultz, J., Mott, R., Ciccarelli, F., Copley, R.R., Ponting, C.P., and Bork, P. 2002. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 30: 242–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewes, H.W., Albermann, K., Heumann, K., Liebl, S., and Pfeiffer, F. 1997. MIPS: A database for protein sequences, homology data and yeast genome information. Nucleic Acids Res. 25: 28–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi, H., Vandergriff, J., Campbell, M., Narechania, A., Lewis, S., Thomas, P.D., and Ashburner, M. 2003. Assessment of genome-wide protein function classification for Drosophila melanogaster. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. [DOI] [PubMed] [Google Scholar]
- Ng, P.C. and Henikoff, S. 2001. Predicting deleterious amino acid substitutions. Genome Res. 11: 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2002. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 12: 436–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt, K.D. and Maglott, D.R. 2001. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 29: 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt, K.D., Katz, K.S., Sicotte, H., and Maglott, D.R. 2000. Introducing RefSeq and LocusLink: Curated human genome resources at the NCBI. Trends Genet. 16: 44–47. [DOI] [PubMed] [Google Scholar]
- Rollins, B.J., Morton, C.C., Ledbetter, D.H., Eddy Jr., R.L., and Shows, T.B. 1991. Assignment of the human small inducible cytokine A2 gene, SCYA2 (encoding JE or MCP-1), to 17q11.2–12: Evolutionary relatedness of cytokines clustered at the same locus. Genomics 10: 489–492. [DOI] [PubMed] [Google Scholar]
- Schultz, J., Milpetz, F., Bork, P., and Ponting, C.P. 1998. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. 95: 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry, S.T., Ward, M.H., Kholodov, M., Baker, J., Phan, L., Smigielski, E.M., and Sirotkin, K. 2001. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 29: 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjolander, K. 1997. “Theoretic method for evolutionary inference in proteins.” Ph.D thesis, University of California at Santa Cruz, Santa Cruz, CA.
- ____. 1998. Phylogenetic inference in protein superfamilies: Analysis of SH2 domains. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6: 165–174. [PubMed] [Google Scholar]
- Sjolander, K., Karplus, K., Brown, M., Hughey, R., Krogh, A., Mian, I.S., and Haussler, D. 1996. Dirichlet mixtures: A method for improved detection of weak but significant protein sequence homology. Comput. Appl. Biosci. 12: 327–345. [DOI] [PubMed] [Google Scholar]
- Sonnhammer, E.L., Eddy, S.R., and Durbin, R. 1997. Pfam: A comprehensive database of protein domain families based on seed alignments. Proteins 28: 405–420. [DOI] [PubMed] [Google Scholar]
- Swets, J.A. 1988. Measuring the accuracy of diagnostic systems. Science 240: 1285–1293. [DOI] [PubMed] [Google Scholar]
- Thomas, P.D., Kejariwal, A., Campbell, M.J., Mi, H., Diemer, K., Guo, N., Ladunga, I., Ulitsky-Lazareva, B., Muruganujan, A., Rabkin, S., et al. 2003. PANTHER: A browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 31: 334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304–1351. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- ftp://ftp.ncbi.nih.gov/refseq/LocusLink/; NCBI LocusLink.
- http://panther.celera.com; PANTHER Protein Classification.
- http://www.geneontology.org; Gene Ontology Consortium.
- http://www.ncbi.nlm.nih.gov/omim/; OMIM, Online Mendelian Inheritance in Man.