Abstract
We report the construction of the mouse full-length cDNA encyclopedia,the most extensive view of a complex transcriptome,on the basis of preparing and sequencing 246 libraries. Before cloning,cDNAs were enriched in full-length by Cap-Trapper,and in most cases,aggressively subtracted/normalized. We have produced 1,442,236 successful 3′-end sequences clustered into 171,144 groups, from which 60,770 clones were fully sequenced cDNAs annotated in the FANTOM-2 annotation. We have also produced 547,149 5′ end reads,which clustered into 124,258 groups. Altogether, these cDNAs were further grouped in 70,000 transcriptional units (TU),which represent the best coverage of a transcriptome so far. By monitoring the extent of normalization/subtraction, we define the tentative equivalent coverage (TEC),which was estimated to be equivalent to >12,000,000 ESTs derived from standard libraries. High coverage explains discrepancies between the very large numbers of clusters (and TUs) of this project,which also include non-protein-coding RNAs,and the lower gene number estimation of genome annotations. Altogether,5′-end clusters identify regions that are potential promoters for 8637 known genes and 5′-end clusters suggest the presence of almost 63,000 transcriptional starting points. An estimate of the frequency of polyadenylation signals suggests that at least half of the singletons in the EST set represent real mRNAs. Clones accounting for about half of the predicted TUs await further sequencing. The continued high-discovery rate suggests that the task of transcriptome discovery is not yet complete.
One of the primary goals of genome sequencing projects is to identify the genome sequences that are transcribed into functional mRNAs, so that full-length cDNAs can be isolated to allow further downstream biology, and functional and structural genomics. The limitations of a priori genome annotation dictate that the transcriptome needs to be identified experimentally via cDNA cloning and sequencing. Although expressed sequence tags (ESTs) (Adams et al. 1991, 1995; Hillier et al. 1996; Marra et al. 1999; Kargul et al. 2001) and ORESTES (Camargo et al. 2001) have been extremely valuable for new gene discovery, these approaches have not allowed high-throughput recovering of full-length cDNA clones nor definition of protein sequence derived from actual cDNA clones. To overcome such problems, we undertook from the year 1995, a strategic project aimed at the comprehensive collection of at least one full-length cDNA derived from each mouse gene, a strategy that is recently becoming useful in similar projects to collect full-length gene collections (Stapleton et al. 2002; Strausberg et al. 2002).
Because of the limited processivity of reverse transcriptase and other limitations, standard cDNA libraries generally contain a majority of truncated transcripts. The introduction of full-length cDNA libraries by Cap-Trapper or other technologies that take advantage of the peculiarity of the cap-structure, (Carninci et al. 1996, 1997, 1998; Suzuki et al. 1997; Carninci and Hayashizaki 1999; Mizuno et al. 1999) did not solve this problem completely.
To enable more effective cloning of longer transcripts and representative sampling of the full spectrum of mRNA lengths, we first developed methods on the basis of thermo-activation of the reverse transcriptase (Carninci et al. 1998, 2002b) to enable synthesis of long first-strand cDNAs exceeding 23 Kb. We have also shown that our cloning vectors, λ-FLC-I, λ-FLC-II, and λ-FLC-III (Carninci et al. 2001), which allow routine bulk excision of cloned cDNA into plasmids for sequencing, also permit improved cDNA library diversity. This is due to the peculiar preferential cloning of a larger insert size (2.5∼3.0 Kb), which is markedly larger than cDNA libraries prepared with other available vectors and resembles the size of starting mRNA. We have shown that the gene discovery of libraries prepared with λ-FLC vectors was improved by >60% compared with conventional vectors and that, before the mouse draft genome sequence was available, the rate of 5′ sequence novelty was about 3.5-fold improved (Carninci et al. 2001).
To maximize the gene discovery, we have also developed methods for normalization and subtraction of full-length cDNAs (Carninci et al. 2000), and in this work, we have fully extended the capability of normalization/subtraction to the highest extent of coverage rate of any transcriptome described so far, supported by an extensive tissue sampling.
We have also prepared this project by developing a sequencing line (RISA sequencing analyzer system), which is ad hoc designed for the routing, quality control, and large-scale sequencing of full-length cDNA clones (Shibata et al. 2000). Complementary technologies included modifications of cDNA library preparation to facilitate sequencing, such as the removal of the original G/C stretches used to clone the 5′ ends (Shibata et al. 2001a) and the removal of poly(A) stretches (Shibata et al. 2001b). In later stages, we have improved the quality of the starting substrate, by developing a method for the extraction of cytoplasmic RNA from fresh and frozen tissues (Carninci et al. 2002a), so that the starting material for library construction is essentially devoid of unspliced introns.
To enable rapid prioritization for full-length sequencing (Osato et al. 2002), we developed a strategy to promptly cluster sequences from newly sequenced cDNAs into preclustered groups (Konno et al. 2001), which are the backbone of our database and allow prompt evaluation of newly constructed cDNA libraries necessary for monitoring the library quality and gene discovery rate.
In this work, we summarize the gene discovery process (Phase I) on the basis of sequencing and characterization of cDNA clones derived from 246 cDNA libraries, including 380 sublibraries. We analyze the factors that are necessary to target a complex transcriptome and evaluate the current transcriptome coverage. We used the concept of clusters to identify provisional transcriptional units (TU). A TU is computationally defined to be a group of transcripts that contain a common core of genetic information having the same orientation, which does not necessarily correspond to protein-coding regions (Okazaki et al. 2002). From this collection, 60,770 fully sequenced cDNA clones have been function annotated (Kawai et al. 2001; Okazaki et al. 2002) and used for genetics studies, expression profiling (Miki et al. 2001), protein–protein interaction studies (Suzuki et al. 2003), and alternative splicing (Kochiwa et al. 2002). The analysis of polyadenylation signals also suggests that a majority of the 3′ ends are likely to be true ends. The 5′ and 3′ ESTs, together with the fully sequenced cDNA (Okazaki et al. 2002), constitute the most comprehensive description of any transcriptome, allowing us to identify the borders of the transcribed regions, and are, therefore, essential to describe the transcriptional units and transcriptional starting points and promoters. The majority of the libraries were derived from the strain C57Bl6/J, whose genome sequence has just became available (Waterston et al. 2002).
RESULTS
Gene Discovery by Sequencing
We based the gene discovery on large-scale sequencing of libraries enriched for full-length subtracted/normalized cDNA. We clustered cDNA sequences from 3′-end reads (Konno et al. 2001), because 3′ cDNA priming is relatively reliable due to reduced internal priming of cDNA (Mizuno et al. 1999) with the trehalose thermoactivated reverse transcriptase (Carninci et al. 1998). At the outset of this project, there was neither genome sequence draft nor extensive information on full-length cDNA, so cDNA grouping-based 5′-end clustering was not undertaken, because we anticipated that, beside alternative promoters and multiple transcriptional starts, non-full-length clone from low-quality libraries would artificially increase the apparent number of clusters leading to redundant full-length sequencing. It became evident from the first round of annotation of full length sequences (Kawai et al. 2001) that 3′-end clustering of deeply sequenced libraries still overestimates the number of Tus, due to the high frequency of alternative polyadenylation and accumulation of internal priming. In later stages of this project, we sequenced the 5′ ends of representative clones of 3′-end clusters before full-insert sequencing (Okazaki et al, 2002). Subsequently, 5′ sequencing was carried out routinely, because of greater frequency of known full-insert cDNA sequences and the mouse genome sequence.
The quality of each library was first evaluated by sequencing both ends of 2∼10 of 384-well plates for CDS integrity, novelty (appearance of new clusters), and problems (Table 1, complete table at www.genome.org). Thereafter, massive 3′-end sequencing of satisfactory libraries was continued, and the output was evaluated continuously until the internal redundancy was 2 (i.e., on average each sequence was represented twice) or until <10% of the new sequences produced new clusters/singletons. CDS integrity was scored by aligning 5′ ESTs to mouse known sequences annotated as mRNA and complete CDS, assuming that the average quality of the other cDNA was the same (Sugahara et al. 2001). The CDS score (Table 1, complete table at www.genome.org) is then important for choosing clones for full-insert sequencing. To statistically support the full-length rate evaluation of strongly subtracted libraries, we have also been using the EST score, obtained by aligning the RIKEN libraries sequences to clusters of 5′ ESTs (Sugahara et al. 2001). Although this correlated well with the CDS score, the EST score became unnecessary at a later stage, when a large quantity of complete mRNA sequences became available. Similarly, the 3′ CDS score considered positive the cDNA clones that showed alignment extending downstream to the annotated stop codon. The final analysis (Table 2) of 5′ and 3′ ends against a larger data set versus complete mouse CDS suggests that 89.1% and 96.5% of the protein-coding cDNAs are, respectfully, 5′ and 3′ intact in relationship to CDS, suggesting that ∼85.7 % of clones have both ends complete.
Table 1.
cDNA Libraries and Their Sequencing
| Library ID | Develop. stage | Strain | Organ/tissue (E) embryo, L) Lactation, (Po) Postnatal (Pr) Pregnancy | Age (E) embryo, (L) Lactation, (Po) Postnatal (Pr) Pregnancy | Other conditions | Vector: λ if not specified | Insert size | Size sel. | RNA notes | Normalization and sub. status | Normarization drivers (RoT) | Norm. RoT (CoT) | Sub. drivers (RoT) | Sub. RoT (CoT) | Library ID | Total RoT (CoT) | Yield = % that was not sub. | Sublib ID | 3′ seq. clones | 3′ clusters + singl. | 3′ red. | 3′ cluster without hit to known genes | 3′ unique clusters + singl. | Pres. of poly A signals | 3′ complete CDS | 5′ seq. clones | 5′ clusters + singl. | 5′ red. | 5′ cluster without hit to known genes | 5′ unique clusters + singl. | 5′ complete CDS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | stage1 | C57BL/6J | blastocyst | TripIEx | N.A. | total | Std | 01 | Std | 01-000 | 156 | 154 | 1.01 | 10 | 21 | 100 | (82.00) | 954 | 869 | 1.10 | 49 | 290 | 75.05 | ||||||||
| Zap | 1.27 K | Std | Std | 06-000 | — | — | — | — | — | — | — | 256 | 179 | 1.43 | 7 | 27 | 95.93 | ||||||||||||||
| 06 | stage28 | C57BL/6J | kidney | 06 | |||||||||||||||||||||||||||
| Zap | 1.40 K | Nor | mRNA (5) | 5 | N.A. | 06-100 | 4491 | 1422 | 3.16 | 43 | 97 | 3867 | 99.31 | 1292 | 1013 | 1.28 | 23 | 171 | 92.72 | ||||||||||||
| Zap | 1.09 K | Std | Std | 07-000 | — | — | — | — | — | — | — | 78 | 71 | 1.10 | 3 | 13 | (91.67) | ||||||||||||||
| 07 | stage28 | C57BL/6J | brain | 07 | |||||||||||||||||||||||||||
| Zap | 1.02 K | Nor | mRNA (5) | 5 | N.A. | 07-100 | 461 | 294 | 1.57 | 3 | 22 | 374 | 99.00 | 67 | 66 | 1.02 | 2 | 19 | (84.85) | ||||||||||||
| 08 | stage28 | C57BL/6J | lung | Zap | 0.80 K | Nor | 08 | N.A. | 08-100 | — | — | — | — | — | — | — | 77 | 52 | 1.48 | 12 | 19 | (97.78) | |||||||||
| 09 | stage28 | C57BL/6J | spleen | Zap | 0.80 K | Nor | 09 | N.A. | 09-100 | 366 | 128 | 2.86 | 3 | 9 | 344 | 99.64 | 384 | 108 | 3.56 | 5 | 25 | 96.70 | |||||||||
| Zap | 0.99 K | Std | Std | 10-000 | — | — | — | — | — | — | — | 115 | 67 | 1.72 | 4 | 8 | 94.34 | ||||||||||||||
| 10 | stage28 | C57BL/6J | heart | Zap | 0.76 K | Nor | 10 | N.A. | 10-100 | 271 | 160 | 1.69 | 2 | 3 | 245 | 97.60 | 260 | 158 | 1.65 | 3 | 31 | 93.10 | |||||||||
| FLCI | 1.80 K | Std | Std | 10-200 | 666 | 447 | 1.49 | 24 | 26 | 551 | 98.53 | 553 | 395 | 1.40 | 26 | 75 | 91.82 | ||||||||||||||
| Zap | 0.75 K | Std | Std | 11-000 | 243 | 179 | 1.36 | 5 | 5 | 209 | 99.32 | 303 | 230 | 1.32 | 13 | 88 | 79.58 | ||||||||||||||
| 11 | stage26 | C57BL/6J | E 18 whole body | Zap | 0.97 K | Nor | 11 | N.A. | 11-100 | 10887 | 4057 | 2.68 | 189 | 277 | 9458 | 99.20 | 1565 | 1504 | 1.04 | 46 | 811 | 62.01 | |||||||||
| Zap | 0.99 K | Nor | N.A. | 11-900 | 6665 | 1978 | 3.37 | 78 | 70 | 5799 | 99.29 | 185 | 182 | 1.02 | 15 | 102 | 57.75 | ||||||||||||||
| Zap | 2.36 K | Std | Std | 12-000 | 2343 | 1204 | 1.95 | 104 | 52 | 1972 | 99.80 | 588 | 547 | 1.07 | 32 | 132 | 89.30 | ||||||||||||||
| 12 | stage28 | C57BL/6J | lung | 12 | |||||||||||||||||||||||||||
| Zap | 1.31 K | Nor | N.A. | 12-100 | 161 | 148 | 1.09 | 4 | 2 | 132 | 99.03 | 298 | 269 | 1.11 | 9 | 68 | 92.05 | ||||||||||||||
| 13 | stage28 | C57BL/6J | liver | Zap | 2.42 K | Std | 13 | Std | 13-000 | 1665 | 807 | 2.06 | 34 | 47 | 1461 | 99.77 | 343 | 329 | 1.04 | 7 | 52 | 92.35 | |||||||||
| FLCI | 3.73 K | Yes | Std | Std | 14-210 | 40 | 32 | 1.25 | 4 | 1 | 31 | (100.00) | 28 | 24 | 1.17 | 7 | 7 | (100.00) | |||||||||||||
| 14 | stage28 | C57BL/6J | brain | FLCI | 2.53 K | Yes | Std | 14 | Std | 14-220 | 40 | 36 | 1.11 | 2 | 2 | 34 | (100.00) | 22 | 20 | 1.10 | 2 | 6 | (100.00) | ||||||||
| FLCI | 3.61 K | Yes | Nor | N.A. | 14-320 | 35 | 18 | 1.94 | 2 | 1 | 33 | (100.00) | 29 | 15 | 1.93 | 1 | 3 | (100.00) |
Summary of libraries, clones, and sequencing resources. Libraries are listed by ID order. (Sub-libraries ID) Different cloning events form the same mRNA/first-strand cNDA reaction. See Methods for details on subtraction drivers. (Unique) Number of cDNAs that derive from a given library only. (Poly(A) signal) The presence of the top 6 signal as in Table 5. (Subtraction and normalization) Subtraction is with RNA (RoT) unless specified (CoT), which corresponds to DNA. (N.A.) Not available; (Std) standard; (Sub.) subtraction; (Nor) normalization; (Seq.) sequenced; (Singl.) singletons; (red.) redundancy; (pres.) presence; (Size sel.) size selection; (ssDNA) single strand DNA; (Total RNA) no poly(A) selection
Table 2.
Summary of Sequencing and Full-Length Rate
| Successful sequences | 1,989,385 |
| Transcriptional Units | 70,214 |
| 3′ end sequences | 1,442,236 |
| 3′ end clusters + singletons | 171,144 |
| 5′ end sequences | 547,149 |
| 5′ end clusters + singletons | 164,915 |
| Full length rate at 5′ ends (clones analyzed) | 89.15% (247,085/277,131) |
| Full length rate at 3′ ends (clones analyzed) | 96.52% (591,174/612,464) |
For quality control, we did not consider the completeness of the UTR regions, because variability of the starting site (Suzuki et al. 2001a,b), 5∼50 bp trimming of 5′ ends in the reference cDNA clones generated by conventional methods (Gubler and Hoffman 1983) and variation of polyadenylation sites (Beaudoing et al. 2000; Iseli et al. 2002), which also suggest that public databases probably do not contain all of the existing full-length UTR variations.
Multiple Strategies Are Necessary for Gene Discovery
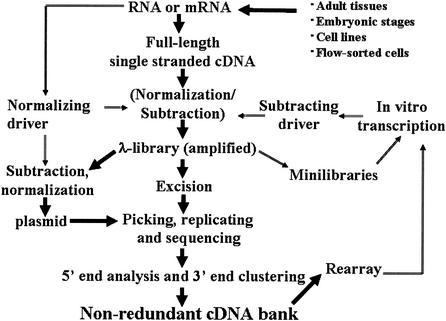
Gene discovery was monitored by 3′-end clustering (Konno et al. 2001). Except for the library 01 (Sasaki et al. 1998; Table 1, complete table at www.genome.org), the remaining 245 libraries were enriched for full-length cDNAs by Cap-Trapper (Carninci et al. 1996, 1997; Carninci and Hayashizaki 1999; Tables 1 and 3). Such libraries, represented in chronological order (Table 1, complete table at www.genome.org), include 390 sublibraries that represent different cDNA cloning events from the same starting mRNA and usually differ for cDNA preparation protocols, such as normalization, subtraction, cloning vector, or size selection. Mixed libraries were prepared by mixing first-strand cDNAs carrying 6-base sequence tags between the oligo-dT primer and the cloning site, and were not counted as separated sublibraries. A general schema of the library preparation and sequencing pipeline is shown in Figure 1. Monitoring the overall gene discovery efficiency between sublibraries has been instrumental in improving the strategy throughout the project. Figure 2 shows the result of monitoring the gene discovery throughout the project. An expanded analysis displaying gene discovery for each library (Fig. 2) was used to monitor and manage the gene discovery, which relied on frequent update of clustering.
Table 3.
Importance of Protocols and Vectors for the Gene Discovery
| A: Protocols | 3′ Sequences | 3′ Clusters | 3′ Singletons | 3′ Group specific clusters | 3′ coding | 5′ Sequences | 5′ Clusters | 5′ Singletons | 5′ Group specific clusters | 5′ coding |
|---|---|---|---|---|---|---|---|---|---|---|
| Normization and subtraction | 772,209 | 61,732 | 47,509 | 21,947 | 96.14 | 140,707 | 30,911 | 61,555 | 7528 | 79.92 |
| Small-scale | 372,132 | 42,316 | 29,322 | 8238 | 96.51 | 228,086 | 32,249 | 31,851 | 7878 | 91.41 |
| Standard | 259,267 | 33,119 | 19,421 | 3943 | 96.65 | 78,407 | 18,883 | 15,779 | 1668 | 90.57 |
| Normalization | 168,383 | 21,178 | 12,057 | 1389 | 97.87 | 130,004 | 22,825 | 14,036 | 3252 | 89.97 |
| Subtraction | 157,238 | 27,792 | 11,799 | 3625 | 95.63 | 79,027 | 16,526 | 12,011 | 3444 | 91.17 |
| Cytoplasmic RNA | 144,752 | 21,348 | 7167 | 947 | 97.8 | 96,967 | 21,278 | 10,675 | 2347 | 90.15 |
| Library subtraction | 65,223 | 13,092 | 4710 | 577 | 98.23 | 94,781 | 17,012 | 10,037 | 2311 | 92.02 |
| Size-selection, normalization and subtraction | 59,750 | 16,837 | 2983 | 352 | 98 | 8698 | 3639 | 4184 | 146 | 77.13 |
| Size-selection | 22,342 | 7870 | 1241 | 102 | 96.76 | 5414 | 2068 | 1854 | 110 | 84.42 |
| (details unavailable) | 19,760 | 6556 | 920 | 222 | 95.68 | 2718 | 1472 | 652 | 67 | 86.01 |
| Capfinder/SMART | 156 | 133 | 21 | 0 | (82) | 954 | 589 | 280 | 12 | 75.05 |
| Size-selection-normalization | 35 | 17 | 1 | 0 | (100) | 29 | 13 | 2 | 1 | (100) |
| B: Vectors | ||||||||||
| λ FLCI | 1,178,336 | 70,945 | 84,780 | 46,073 | 96.59 | 481,725 | 48,407 | 103,631 | 34,504 | 89.09 |
| λ ZAP | 190,064 | 20,775 | 6458 | 2328 | 96.24 | 19,500 | 7559 | 5927 | 467 | 83.07 |
| λ FLCII | 48,656 | 12,287 | 3425 | 997 | 96.45 | 13,571 | 6174 | 3081 | 354 | 84.95 |
| λ FLCIII | 24,931 | 7128 | 1747 | 303 | 96.74 | 10,763 | 4395 | 1421 | 147 | 91.82 |
| λ TripIEx | 156 | 133 | 21 | 0 | (82) | 954 | 589 | 280 | 12 | 75.05 |
| pBS | 93 | 71 | 6 | 0 | 93.55 | 85 | 62 | 10 | 1 | 85.45 |
Figure 1.
Overall strategy for preparation of cDNA libraries and routing of rearrayed clones to prepare drivers for subtracting new cDNA libraries. From small samples of tissue (i.e., <15 μg of total RNA as template), the resulting cDNA could neither be normalized nor subtracted. When at least 15 μg of starting total RNA was available, cDNA was subtracted with a driver derived from a minilibrary set and nonredundant rearrayed library. For larger tissues, cDNA was prepared from mRNA. In this case, cDNA was also normalized by using an aliquot of the starting mRNA together with the subtraction step. Any newly prepared libraries went through this routine, making libraries prepared at a later time more strongly subtracted than those prepared earlier.
Figure 2.
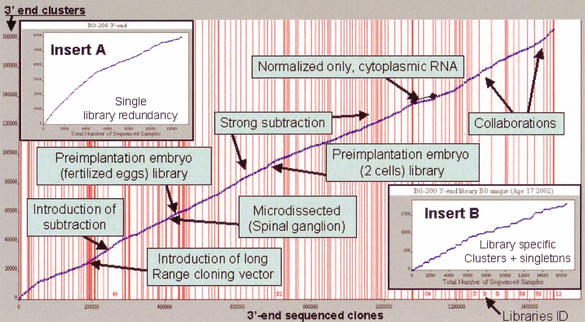
Overall sequencing growth during the course of the project: x axis shows the number of sequences and y the number of 3′ clusters. Vertical lines indicate switches between sequenced libraries. Strong subtraction is intended with RoT larger than 2000 and up to 500. Library number is displayed only when space allows. We highlighted the most productive factors that influenced gene discovery. (Top, left, inset) The internal redundancy of one library and (bottom, left, inset) the number of library specific clusters plus singletons (the gene discovery rate per library). The overall curve is derived by summing many curves as in B. (A, B) The B0 library, 2-cell stage.
Sources of mRNA
We assumed that discrete mRNA expression of specific genes is often restricted to particular tissues or developmental stage, or to a subset of cells within a large tissue. Therefore, we collected mRNA from mouse samples from a very wide variety of tissue and developmental stages, sacrificing more than 35,000 mice (embryos and adults).
In the absence of a suitable cytoplasmic mRNA extraction protocol at the beginning of the project, we used whole RNA or mRNA derived from whole RNA (including nuclear RNA). An analysis of fully sequenced cDNA clones (Okazaki et al 2002) of known protein-coding genes suggested that 7.3∼9.7% of the cDNAs retain unspliced introns when, respectively, they are manually curated or computationally analyzed. To overcome this problem, in a later stage we used cytoplasmic RNA, which is devoid of nuclear RNA and contaminating of incompletely spliced mRNA, when cytoplasmic RNA from a large variety of tissues became available (Carninci et al. 2002b). From cytoplasmic RNA, we have sequenced 96,967 5′ ends and 144,752 3′ ends, producing 21,348 3′-end clusters and 10,675 3′-end singletons.
Microdissection
We prepared full-length libraries from small tissues, including 71 microdissected tissues (Tables 1 and 3). To this end, we modified the Cap-Trapper for small quantities of total RNA, without PCR, which might skew representation, especially at expanses of long cDNAs and would affect comprehensiveness of gene discovery. In fact, library 01 (Blastocyst) (Sasaki et al. 1998; Table 1, complete table at www.genome.org), constructed with a PCR-based cap-switch kit, produced only 924 clusters after 3187 sequencing passes (redundancy 3.45). In contrast, cap-trapped cDNA libraries produced from embryo mRNA without PCR amplification allowed much better gene discovery, for example, library I1 (blastocyst) with 4513 clusters after 8128 sequencing passes (redundancy 1.8), library 74 (fertilized egg) with 3285 clusters/7602 passes (redundancy 2.31), and library B0 (embryo, 2 cell stage) with 5810 clusters/16,483 passes (redundancy 2.84). These cap-trapped libraries showed even lower redundancy after sequencing ∼3000 clones, highlighting the advantages of not using PCR. Globally, small-scale libraries allowed the isolation of >39,000 of the 3′-end singletons (Table 3).
Vectors
Introduction of the λ-FLC vectors (Carninci et al. 2001) resulted in improved gene discovery (Fig. 2), due to the larger average cloning size (2.5–3Kb) if compared with conventional vectors (λ-Zap or plasmid), which resulted in average insert size of 1.0∼1.5 Kb (Table 1, complete table at www.genome.org). Table 3 shows that the majority of the clusters plus singletons of the cDNA could be cloned in one of the three λ-FLC vectors, except for 6458 3′ singleton and 2328 3′ clusters (total, 4.7% of the total groups) that seem to be represented only in the λ-Zap vector. Uniqueness rate of clusters plus singletons has been considerably higher for the λ-FLC libraries (11%) compared with λ-Zap libraries (4.6%), both because of strong subtraction and larger insert cloning. Clones in λ-FLC-II and λ-FLC-III, which can be excised into functional vectors without insert cleavage (Carninci et al. 2001), constitute, respectively, 15,712 and 8,549 groups, and are promptly amenable for transferring in expression vectors.
Normalization and Subtraction
To further select by sequencing rarely expressed mRNAs, we normalized/subtracted the cDNAs before cloning, after developing a method that does not cause cDNA degradation during these procedures (Carninci et al. 2000). Generally, normalization was used when the quantity of starting mRNA (>5 μg) was sufficient to spare an aliquot for the preparation of a normalizing driver (Carninci et al. 2000), and, therefore, was not possible for small tissues. Subtraction was used either alone or in combination (single step) with normalization (Carninci et al. 2000) whenever we had more than 200–300 ng of cap-trapped cDNA. Subtraction was omitted when preparing normalized cDNA libraries from cytoplasmic mRNA, introduced late in the project, to prepare a set of cDNA clones that have low probability of retained unspliced introns, which were then prioritized for full-length insert sequencing. Late in the project, a subtraction method based on amplified libraries became available (T. Hirozane-Kishikawa and P. Carninci, unpubl.), which is essentially adapted from another technology (Bonaldo et al. 1996), but using as starting substrate, plasmid extracted from amplified λ libraries rather than plasmid-amplified libraries to minimize size bias (Fig. 1).
Subtraction drivers were routinely prepared by promptly rearraying representative clones of all new clusters (Fig. 1) used to prepare RNA run-off drivers by bulk in vitro transcription. We also used nine minilibraries (see Methods) that were prepared by cloning the abundant fraction of cDNA, which is the side product of a normalization procedure (Carninci et al. 2000) of early normalized libraries, followed by amplification of 1000–2000 clones to eliminate rarely expressed genes incidentally present in the mini-library. Therefore, mini-library clones consist mainly of highly expressed mRNAs.
Normalization almost doubled the gene discovery compared with non-normalized cDNA libraries (Carninci et al. 2000), and this is further improved for normalized/subtracted libraries (e.g., compare standard versus normalized/subtracted sublibraries from libraries 24, 30, and 31; Table 1, complete table at www.genome.org). Furthermore, the reiterative, combined subtraction/normalization strategy supported by prompt rearray of clones allowed preparation of libraries by which the gene discovery rate has been consistently high (Fig. 2). Satisfactory libraries showed redundancy ≤2 after sequencing 15,000–20,000 cDNA clones.
The specificity of the normalization/subtraction was proven by showing that the frequency of B1 and B2 repeats in 3′ ends does not change with subtraction, ruling out nonspecific removal of related but different sequences (Carninci et al. 2000). In a further analysis, among 208,668 sequences (June 2000) from normalized/subtracted libraries, 982 sequences (0.41%) corresponded to 26 actin family members. Of these, 117 sequences only (11.9%) had a correspondent in the subtracting driver, whereas the majority were derived from the genes absent in the driver. In comparison, of 39,971 sequences sampled from standard cDNA libraries, 204 clones (0.51%) represented actin family members, of which half (102) had a correspondent in the driver used at that time, suggesting that subtraction caused at least a 4.2-fold enrichment of sequences absent in the driver. Failure to subtract the remainder may be due to incomplete hybridization or removal of the hybrid, exceptional over-representation of the tester, and failure to replicate or efficiently transcribe RNA drivers for certain clones. This analysis also ensures that different members of gene families were spared from nonspecific hybridization.
Generally, these data also demonstrate that the normalization/subtraction procedures are relatively free of flaws, which have been supposed to impair gene discovery (Wang et al. 2000), probably because the poly(A) tail of cap-trapped libraries have controlled length, and, therefore, cDNAs are not removed nonspecifically by 3′-end interactions.
Clusters Distribution
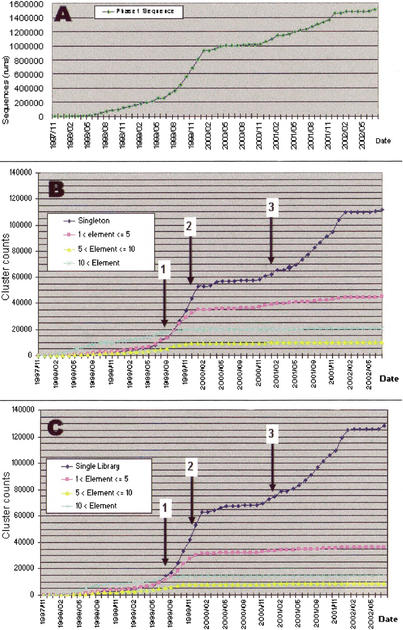
Monitoring the appearance of new singletons and clusters shows a clear trend (Fig. 3). Highly expressed mRNAs appeared early in the project and from multiple libraries and reached a plateau of about 20,000 clusters with more than 10 sequences, of which 14,316 appeared in at least 10 different libraries. Similarly, the intermediately expressed clusters were found early, although this group is less represented than the highly expressed ones. These classes did not further increase, even following deep sequencing of later libraries. Instead, the class of intermediate rare clusters, with 2–5 appearances in the whole project and from 2 to 5 libraries, has been steadily growing. Altogether, there are more than 58,000 clusters that appeared from different libraries, which can be considered bona fide 3′ ends with high confidence. However, 3′ ends are not limited to this group because of tissue restriction of gene expression. Finally, a large quantity of singletons or clusters derived exclusively from a single library appeared concomitantly to strong subtraction, special tissue sampling, and choice (Fig. 3). Alternatively, as there were few switches between these classes of expression, even when nonsubtracted libraries (small-scale and cytoplasmic mRNA libraries) were used, we can conclude that a consistent portion of mRNA expression is restricted to specific tissue or development stages.
Figure 3.
In the course of the gene discovery project (top), we monitored the appearance of singletons vs. clusters of various classes of size (middle) and number of libraries in which they appeared (bottom). Arrows 1, 2, and 3 show the date when increasingly larger subtracting drivers, respectively, consisted of 13,500, 37,500, and 126,000 rearrayed clusters, and were introduced for cDNA library subtraction.
Coverage
Altogether, we have successfully sequenced 1,989,385 ESTs (5′ + 3′) (Tables 1 and 2), and have produced 720,959,679 bp of ESTs + finished full-length cDNA sequence data. As suggested in Table 3, a transcriptome should be addressed by multiple approaches, including normalization, subtraction, and aggressive sampling of microdissected tissues. For instance, >400,000 3′-end ESTs were produced from small-scale libraries (including 71 microdissected plus small-scale cDNA libraries), which resulted in isolation of >29,300 3′ singletons (Table 3), as well as 29,332 3′-end singletons and 8,238 clusters that appeared only in small-scale libraries. This complemented the data obtained by sequencing 772,209 3′ ends from strongly subtracted/normalized standard-scale cDNA libraries, which allowed isolation of almost 22,000 specific 3′-end clusters and ∼47,500 3′ singletons.
Throughout the project, we have tracked the extent of hybridization for most of the cDNA libraries by checking the CPM (counts per minute) of radiolabeled cDNA before and after the normalization/subtraction. For instance, in library B2-300 (corpora quadrigemina), up to 95% of the mass of cDNA has been subtracted and the remaining 5% of cDNA was cloned and sequenced. For this library, we sequenced 4609 3′ ends, identifying 428 unique clusters, which would be obtained by roughly sequencing >92,000 clones of a non-normalized/subtracted library, allowing a much deeper collection of rare transcripts than would be allowed by a standard library. Similar calculation for all of the normalized/subtracted libraries prompted us to define the tentative equivalent coverage (TEC), which was obtained by dividing the number of clones sequenced from each library by the fraction of cDNA that escaped removal by subtraction/normalization. By keeping unchanged the sequences of the remaining nonsubtracted libraries, the total TEC exceeds 12,000,000 3′ sequences. Such estimations are supported by improved gene discovery. For instance, the standard cerebellum library 15 shows ∼2.5% of unique clusters versus ∼10% of the subtracted library A7 (Table 1, complete table at www.genome.org). See Table 1, complete table at www.genome.org, to compare the low rate of unique clusters in early nonsubtracted libraries versus the 10-fold or better rate of uniqueness of recent strongly subtracted libraries.
Although such estimations should be verified experimentally, this approach seems to provide a coverage that is not inferior to SAGE, for which there are currently more than 6,000,000 tags in current databases (Boon et al. 2002).
Our sequences cluster with >79% of the embryo-specific cDNA clone set from NIA (Kargul et al. 2001). We have not investigated further whether the nonoverlapping representative clones of the NIA represent alternative isoforms of our cDNA clones or are obtained because of deeper sequencing of preimplantation embryo libraries.
The use of fully sequenced FANTOM clones, on the basis of this collection, sequence of clones for which we had pairs of reads from 5′ and 3′ ends together with sequences in public databases, allowed us to define a set of more than 37,000 TU that mapped to the genome (Okazaki et al. 2002). If we include singletons, the number of candidates to be independent TU increased to ∼70,000, which certainly represent the most extensive coverage of a mammalian transcriptome so far. However, we have not attempted to reconvert the current clusters number to TU. We are confident that the number of TU will increase further, but this will require experimental validation on the basis of continued full-length cDNA sequencing.
Transcriptional Starting Site and Promoter Identification
We have shown previously that Cap-Trapper is at least as valuable as the Capfinder/Smart (Sasaki et al. 1998) and the Oligo-Capping technique (Maruyama and Sugano 1994; Suzuki et al. 1997) in identifying transcriptional starting points (TSP) (Sugahara et al. 2001), which are very valuable for identifying promoter regions (Suzuki et al. 2001a,b). Validation of the Cap-Trapper was performed with clusters derived from the same gene across several libraries, which also confirmed that certain genes have fixed TSP, whereas others have multiple TSP. For instance, cyclophilin (gi50620; clustered with the RIKEN 0610007C08) appeared to have a 44-nucleotide 5′UTR short isoform that is Blastocyst specific (20 of 96 clones), 41 blastocyst clones were within +6 nucleotides, compared with the sequence of clone 0610007C08, and six were up to 29 nucleotides longer. Longer and shorter variants were never observed in 29 clones from 16 different tissues (data not shown). Also, 19 of the clones that appear from normal tissue showed a splicing variant from nucleotides 122–179 that never appeared in blastocyst (data not shown). Other genes showed a much more conserved TSP across libraries, such as the heat-shock protein 70 mRNA, which has essentially the same 5′ end (longer than U27129, respectively, 45∼52 nucleotides for 19 clones, 33 and 32 nucleotides for 2 more clones) including clones from the blastocyst (library 01).
About 88.8% 5′ ends matching known genes are longer than the CDS, allowing the identification of new upstream TSP (Okazaki et al. 2002). Here, we mapped the potential TSP of 8637 known genes (Table 4), including tissue-specific ones. As 92.7% of clusters of more than two sequences extend the CDS, we can conclude that the majority of the remaining 67,186 of 5′ end clusters (excluding singletons) also represent TSPs, In about 18% of cases, a new TSP was found to be >100 nucleotides upstream on the genome. A total of 609 clusters (7.2%) appeared only in one tissue type (as in Table 6, below) and 1781 (21%) in a single developmental stage. The value of this resource was illustrated in our recent experimental validation of the transcription start sites of the tartrate-resistant acid phosphatase locus, in which we identified four separate transcription start sites used in different tissues (Walsh et al. 2003).
Table 4.
Estimation of Transcriptional Starting Points Identified by 5′ EST Mapping
| 5′ ESTs extending the CDSs | Clusters matching same genes | Singletons that extend CDSs | |
|---|---|---|---|
| TSP <100 nt longer on genome | 17,073 | 2350 | 42 |
| New TSP >100 bp longer on genome | 1669 | 516 | 29 |
| Single tissue type (1) | 4488 | 609 | 1269 |
| Multiple tissue type (1) | 224,934 | 7902 | n.a. |
| Single developmental stage (1)(2) | 16,418 | 1781 | 1269 |
| Multiple developmental stage (1)(2) | 213,509 | 6747 | n.a. |
| Single library ID | 2,078 | 269 | 1269 |
| Multiple Library ID | 227,849 | 8259 | n.a. |
| Total, >10 bp (longer) | 121,444 | 4934/8637 | 635 |
| Total, ±10 bp | 66,192 | 5115/8637 | 224 |
| Total, <10 bp (shorter) | 59,449 | 7030/8637 | 367 |
Only matches that extend existing CDS were considered
Reference mRNAs with complete CDS without hit are not shown
Clusters were prepared without using genome alignment and later aligned to the genome sequence
Without taking into account library IDs 21, 23 and unrecognized mixed libraries
Divided as preimplantation embryo, embryo after day 6, and adult
Table 6.
Expression in Various Types of Tissues (Non-mutually Exclusive)
| Tissue type | 3′ singletons | 3′ clusters | 3′ specific clusters | 5′ singletons | 5′ clusters | 5′ specific clusters |
|---|---|---|---|---|---|---|
| Adult | 60,161 | 66,212 | 26,829 | 77,873 | 44,393 | 20,834 |
| Embryo, 6 days or later | 23,478 | 41,915 | 5106 | 25,949 | 23,012 | 2922 |
| Embryo, before 6 days | 4999 | 11,431 | 1516 | 3015 | 3524 | 332 |
| Cell lines | 5523 | 16,045 | 569 | 4777 | 12,787 | 929 |
| Nervous tissues | 21,290 | 38,794 | 7906 | 28,157 | 21,706 | 5847 |
| Peripheral nervous tissues | 5407 | 20,836 | 2180 | 5510 | 7186 | 1009 |
| Immune tissues | 16,144 | 27,257 | 2257 | 16,631 | 22,624 | 1284 |
| Stimulated/induced | 11,203 | 17,184 | 187 | 9761 | 17,737 | 42 |
| Reproduction | 8278 | 23,068 | 3444 | 9335 | 6754 | 1338 |
| Respiratory | 5038 | 19,768 | 1125 | 7728 | 10,524 | 1223 |
| Digestive organs | 2692 | 13,779 | 1054 | 3595 | 6376 | 755 |
| Circulation | 2561 | 13,188 | 739 | 3221 | 4674 | 458 |
| Secretory organs | 1740 | 9599 | 617 | 2412 | 6300 | 715 |
| Renal, urinary | 1616 | 9438 | 339 | 1999 | 4873 | 390 |
| Epithelial tissue | 1027 | 9162 | 287 | 1531 | 1643 | 170 |
| Cancer | 1016 | 7943 | 248 | 1918 | 4869 | 340 |
| Endocrine | 765 | 6262 | 116 | 1202 | 1180 | 79 |
| Renal, urinary (II) | 75 | 1347 | 38 | 182 | 944 | 68 |
| Other | 5148 | 21,864 | 1455 | 7041 | 5960 | 796 |
Cap-Selection Does Not Enrich for 3′ Truncated Clones
Because 3′ truncation of cap-selected libraries and oligo-dT primed libraries has been a major concern (Bashiardes and Lovett 2001; Nam et al. 2002), we evaluated the extent of this problem in our set by aligning 3′ ends with the mouse RefSeq data set with annotated polyadenylation (685 sequences taken in September 2000). There were 44,614 RIKEN sequences that aligned to the RefSeq data set, belonging to 1160 clusters. Of them, 995 sequences (2.2%) and 103 clusters (8.8 %) started inside the annotated CDS (3′ truncated); 12.7% of the clones and 46% of the clusters started more than 50 nucleotides internally from the annotated polyadenylation site and were potential 3′ UTR primed (PUP). These transcripts represent either alternative polyadenylation or 3′ UTR nonspecific priming due to the relatively high A/T content (∼40%). To the same RefSeq data set we compared unclustered publicly available sequences derived from cDNA libraries that were not cap selected (Soares libraries, indicated as Unigene libraries Mm 30, 66, 70, 72, 86, and 90). These were no better than cap-trapped libraries, showing 3.4% 3′ truncation rate and 18.4% of PUP. Similarly, Oligo-Capped libraries (Mm 132, 135, and 135), showed 6.1% and 29.8% of 3′ truncation and PUP and NCI-CGAP libraries (Mm 332, 333, 337, 340, and 341), showed 4.2% and 23.5% of 3′ truncation and PUP. Beside providing reassurance that Cap-Trapper does not enrich for 3′ truncation, these data show the limitations of clustering only on the basis of one-end sequencing.
Classes of Polyadenylation Sites
The 3′ ends of full-length cDNAs are also essential to permit annotation of functional signals within the 3′UTRs, which contribute to stability regulation, polyadenylation, and localization within the cell and translation efficiency. This project identified >171,000 3′ ends, of which 75,056 were clusters of two or more elements. In a preliminary analysis, we visually inspected 120 randomly selected 3′-end clusters corresponding to known mouse genes with an annotated poly(A) site. Among these, 99 clusters (82.5%) contained polyadenylation signals, of which 89 clusters (74.2%) corresponded to the sequence annotated previously and 17 (14.1%) represented alternative polyadenylation signals. Internal priming in A-rich regions accounted for eight clusters (6.7%), and six clusters (5%) were either internally primed on non-A-rich sequences or represented new unconventional polyadenylation sites. Among the 120 clusters, 70.74% also showed canonical polyadenylation sequences (AAUAAA and AUUAAA). This proportion is identical to the observed frequency in human annotated mRNAs (Gautheret et al. 1998). By EST analysis, alternative polyadenylation seems to occur in at least 29%–40% of genes (Gautheret et al. 1998; Beaudoing et al. 2000), and many more such variable polyadenylation events probably escape the analysis, due to long-range heterogeneity of the 3′ ends (Iseli et al. 2002).
We can classify 3′ ends on the basis of polyadenylation signals. A total of 78.3% of the clones carried a signal, and in 76.3% of the cases, the signal was among the nine most represented (Konno et al. 2001). We further classify the appearance of poly(A) signals on the basis of the expression level by tracking the subtraction data. cDNA clones from libraries that were intermediately and strongly subtracted (200>RoT>50 and RoT≥200) showed a significant decrease of the frequency of the strong polyadenylation sites (AAUAAA), but a slight increase of the weakest AUUAAA signal, as well some of the other weaker polyadenylation signals (Table 5). Singletons, which represent generally rare mRNAs that appeared only once in the course of the project, confirmed this trend, showing the presence of recognizable polyadenylation signals in only ∼49% of cases. Interestingly, the ratio of the strongest AAUAAA signal versus rarer, weaker signals was notably altered. AAUAAA represents only 56% of the signals, with a concomitant slight increase of the AUUAAA, whereas the remaining weaker polyadenylation signals showed two- to more than fourfold increased frequency compared with clones in multiple clusters (Table 5). The strength of the polyadenylation signal is correlated to the elongation of the poly(A) tail, which in turn is correlated to translation efficiency (Wahle 1995) One might assume that strong polyadenylation/translation is not required for rarely expressed mRNAs, many of which appear to be noncoding RNAs (Okazaki et al. 2002). At least half of the singletons showed an unequivocal polyadenylation signal (Table 5), thus suggesting that at least 48,000 of the cDNAs in this class are genuine processed mRNAs.
Table 5.
Use of Polyadenylation Signals Varies Depending on the Class of Expression
| Polyadenylation signals | AATAAA % | ATTAAA % | AATTAA % | AAATAA % | AGTAAA % | AATATA % | CATAAA % | TAATAA % | AATAAT % | Others % | Total with signals | No signals | Total sequences |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total 3′ Est collection | 71.377 | 16.872 | 0.948 | 0.997 | 2.830 | 2.076 | 1.818 | 0.419 | 0.191 | 2.473 | 1,128,930 | 313,306 | 1,442,236 |
| 3′ clusters of 2 or more | 72.000 | 16.801 | 0.891 | 0.912 | 2.763 | 1.977 | 1.731 | 0.380 | 0.170 | 2.377 | 1,083,955 | 261,843 | 1,345,798 |
| 3′ singletons | 56.371 | 18.604 | 2.324 | 3.046 | 4.451 | 4.465 | 3.920 | 1.339 | 0.691 | 4.789 | 44,975 | 51,463 | 96,438 |
| Standard libraries | 74.749 | 15.130 | 0.642 | 0.785 | 2.763 | 1.754 | 1.587 | 0.376 | 0.165 | 2.051 | 207,626 | 55,442 | 263,068 |
| Mild subtr/norm (5 ≤ RoT ≤ 50) | 73.830 | 15.718 | 0.462 | 0.779 | 3.021 | 1.700 | 1.866 | 0.370 | 0.165 | 2.089 | 167,664 | 36,294 | 203,958 |
| Strong subt/norm (200 > RoT > 50) | 69.613 | 18.003 | 1.000 | 0.995 | 2.697 | 2.218 | 1.721 | 0.407 | 0.199 | 3.148 | 202,835 | 41,692 | 244,527 |
| Very strong subt/norm (RoT ≥ 200) | 68.960 | 17.890 | 1.370 | 1.250 | 2.758 | 2.422 | 2.071 | 0.456 | 0.221 | 2.599 | 446,662 | 158,715 | 605,377 |
| Other protocols | 74.466 | 15.809 | 0.391 | 0.681 | 3.381 | 1.512 | 1.568 | 0.520 | 0.120 | 1.552 | 54,778 | 12,527 | 67,305 |
| Library conditions unavailable | 74.551 | 15.446 | 0.464 | 0.689 | 3.061 | 1.612 | 1.019 | 0.357 | 0.152 | 2.650 | 49,365 | 8636 | 58,001 |
Tissue cDNA Encyclopedia
Subtraction, and to a lesser extent, normalization strategies detract from the usefulness of library of origin information as a source of insight into tissue-specific expression of particular transcripts. Nevertheless, there is still invaluable information to be obtained by tracking such information. To this end, we kept track of the original tissues even in mixed cDNA libraries, by using tagging cDNAs even in mixed libraries. Several libraries included in the main groups of tissue (Table 6) were prepared before a large number of clones were rearrayed for subtraction (Table 1, complete table at www.genome.org). Therefore, grouping libraries from tissues of similar origin may provide a de facto gene-expression profile.
Embryonic tissues produced fewer clusters than adult ones, perhaps because adult clusters allowed sampling by microdissection of very specialized tissues (such as inner ear), whereas in embryo, many whole tissues were used. The nervous tissue highlights the importance of microdissecting within a large tissue. In fact, 38,794 3′-end clusters (of which 7906 were unique to the central nervous tissue) and 21,290 3′-end singletons were produced from nervous tissue. Sampling of the peripheral nervous system (such as retina, inner ear, sympathetic ganglion), further produced 2180 3′-end unique clusters and 5407 3′-end singletons. The cDNA obtained by preimplantation and early postimplantation from fertilized egg to 4.5 dpc, including 4999 3′-end singletons and 1515 specific 3′-end clusters required the largest effort in terms of sampling, and yet deserve more future attention. The reproduction represents a heterogeneous category of libraries from tissues such as ovary and testis at various stages of development and contains genes involved in germ-cell maintenance, but a very large complexity is produced by testis. During spermatogenesis, the methylation status is rearranged, and we could eliminate the possibility that such variability is caused by deregulated transcription, although in the annotation we did not observe particular over-representation of non-coding RNA specifically derived from testis libraries. It is also evident from Table 6 that there is room for gene discovery in certain tissues and categories, such as cancer.
Further work lies ahead to verify the global expression of such mRNAs, similar to what has been done for an initial set, the RIKEN 19K, analyzed for their expression in 50 tissues, expanded recently to a 60K set for 21 tissues, which gave additional clues to gene function (http://genome.gsc.riken.go.jp/READ/) (Miki et al. 2001; Bono et al. 2003). Correlating expression analysis by sequencing subtracted libraries, microarray, and other methods such as SAGE will present further computational challenges. To this end, we have started grouping the tissues by adapting the nomenclature and tissue classification of The Mouse Anatomical Dictionary (http://www.informatics.jax.org/menus/expression_menu.shtml) (Ringwald et al. 2001; Table 7, complete table at www.genome.org), and have further correlated the microarray signals to EST counts in gene discovery (http://genome.gsc.riken.go.jp/matrics/ef/fantom2/READ/). Expression by EST counting aims complement-expression analysis by microarray; absence from libraries cannot discern lack of expression from subtraction, but ESTs are valuable for identifying rare mRNAs expression. Finally, a tissue and developmental stage-based grouping better displays tissues that should be the target of remaining gene discovery (Table 7, complete table at www.genome.org).
Table 7.
Tissue Anatomical Dictionary With Sequencing Coverage
| stage | Tissue type I | Definition I | Definition II | Definition III | Definition IV | Definition V | Definition VI | Definition VII | ID | Strain | Organ, tissues, condition, age (E, embryo, L, lactation, Po, postnatal, Pr, pregnancy) | Clones (5′ + 3′) | Clusters (5′ + 3′) | Redundancy |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ES cells | C3 | ES cells | 11,918 | 6139 | 1.94 | |||||||||
| ES cells | 24 | ES cells | 17,811 | 5569 | 3.20 | |||||||||
| stage1 | one-cell stage | body | 74 | C57BL/6J | in vitro fertilized eggs | 18,123 | 6296 | 2.88 | ||||||
| stage1 | one-cell | body | BI | C57BL/6J | 1 cell | 1499 | 936 | 1.60 | ||||||
| stage2 | two-cell stage | body | BO | C57BL/6J | 2 cells | 12,531 | 6972 | 1.80 | ||||||
| stage2 | 3-cell stage | body | BJ | C57BL/6J | 3 cells | 9 | 7 | 1.29 | ||||||
| stage3 | 4-8 cell stage | body | BK | C57BL/6J | 4 cells | 309 | 197 | 1.57 | ||||||
| stage3 | 4-8 cell stage | body | E8 | C57BL/6J | embryo 8 cells | 10,371 | 5593 | 1.85 | ||||||
| stage4 | morula | body | BL | C57BL/6J | morula | 854 | 526 | 1.62 | ||||||
| stage4 | blastocyst | body | I1 | C57BL/6J | blastocyst | 10,671 | 5822 | 1.83 | ||||||
| stage5 | blastocyst | body | O1 | C57BL/6J | blastocyst | 1110 | 1023 | 1.09 | ||||||
| stage7 | embryo | body | BN | C57BL/6J | E5 whole body | 1297 | 822 | 1.58 | ||||||
| stage8 | embryo | body | 56 | C57BL/6J | E6 whole body | 565 | 513 | 1.10 | ||||||
| stage10 | embryo | body | C4 | C57BL/6J | E7 whole body | 11,016 | 6117 | 1.80 | ||||||
| stage12 | embryo | body | 57 | C57BL/6J | E8 whole body | 21,971 | 9466 | 2.32 | ||||||
| stage14 | embryo | body | D0 | C57BL/6J | E9 whole body | 14,853 | 8885 | 1.67 | ||||||
| stage15 | embryo | body | B1 | C57BL/6J | E 9.5 partenogenotic embryo | 9904 | 5903 | 1.68 | ||||||
| stage16 | embryo | body | 26 | C57BL/6J | E10 whole body | 15,322 | 6536 | 2.34 | ||||||
| stage16 | embryo | body | 34 | C57BL/6J | E10 whole body | 1801 | 1338 | 1.35 | ||||||
| stage16 + stage17 | embryo | body | 28 | C57BL/6J | E10 + E11 whole body | 17,265 | 7330 | 2.36 | ||||||
| stage18 | embryo | body | 27 | C57BL/6J | E11 whole body | 12,053 | 4397 | 2.74 | ||||||
| stage18 | embryo | body | upper body | 62 | C57BL/6J | E11 upper body | 475 | 431 | 1.10 | |||||
| stage18 | embryo | head | 62 | C57BL/6J | E11 head | 3507 | 2886 | 1.22 |
Nonredundant Set of cDNAs for Full-Length Sequencing
Clusters identify candidates for full-length cDNA sequencing (Phase II), which are generally selected from best scoring and cytoplasmic RNA libraries. Library 74-204 (fertilized egg) is an exception and was sequenced even though only a suboptimal fraction of cDNAs (52.9%) were predicted to be full length, because it was not feasible to recollect 5000 embryos (Table 1, complete table at www.genome.org). Selection of clones for full-insert sequencing took place in real time and from libraries that showed best full-length score. About 67% of the 60,770 resequenced clones appeared to be full length, and the discrepancy with Table 2 can be attributed to better sequencing and finishing success rate for short cDNA clones and failures of recognizing clusters of truncated clones as such. In fact, due to clustering limitations previous to genome sequence availability, truncated cDNAs often became candidates for full-insert sequencing. Additionally, full-length sequencing operations show higher sequencing and finishing success rates for short cDNA clones.
Function Assignment
The contribution of these sequences to known protein sets is described elsewhere, as well as the completely curated assignment of cDNA function at the FANTOM1 and FANTOM2 annotations (Kawai et al. 2001; Okazaki et al. 2002) and accompanying studies, and is therefore beyond the scope of this work. Before curated annotation, all ESTs are preliminarily analyzed by BLAST to assign temporary function when hitting a known gene, which currently happens for 65.5% of 5′ and 54.7% of 3′ EST clusters. In subtracted libraries, on average, 65% of the 5′ end reads do also hit protein databases at E = 10-19, allowing additional temporary functional assignment for clones that are otherwise not curated. Relatively higher efficiency of 5′-end annotation may be due to truncated clones matching CDS regions, short 5′ UTRs that do not prevent identification of CDS regions, and different coverage of the 5′ and 3′ data sets. Many of the clusters that are not annotated may simply be noncoding RNAs. Similarity search-based functional information is available by searching our Web site (http://genome.gsc.riken.go.jp/) for homology information. Retrieved sequences are displayed, together with features such as relevant homology to public databases and multiple alignments, with other clones in the mouse full-length cDNA encyclopedia. All of the sequences have been deposited with DDBJ, and are from there propagated to other databases, and then incorporated into the Unigene and various genome browsers.
DISCUSSION
Obtaining a complete view of a transcriptome is arguably more complex than sequencing a genome because of the huge dynamic range of mRNA expression, which requires tailored cDNA library construction and the nature of the mRNA itself, including alternative transcript, promoter, termination sites, and the presence of noncoding RNAs. Furthermore, clone identification, quality control, tracking, storage, and prioritization cannot be automated in the same way as shotgun sequencing of a genome. Full-length sequencing of individual cDNAs is also more painstaking than shotgun sequencing of genomic BACS. To gain complete representation of a transcriptome, one needs to sample the full diversity of tissues and the full diversity of inducible states. The extent to which this task is still ongoing is reflected in the high frequency of new transcripts identified when we recently sampled cells of the innate immune system responding to microbial stimulus (Wells et al. 2003; see Tables 1 and 7, complete tables at www.genome.org). The rewards of transcriptome sequencing are proportionally great, as the full-length cDNA sequence gives an experimental validation of predictions that can only be made in silico from genome annotation or EST assembly. In this project, we have provided the model that can be applied to humans and to other mammals, and as other mammalian transcriptomes are completed, we will gain proportionally great insight into mammalian functional genomics.
One issue that occupies the minds of scientists and non-scientists alike is the number of genes (Ewing and Green 2000; Hogenesch et al. 2002; Crollius et al. 2000). Because clustering tends to overpredict the number of mRNAs, we had combined initial clustering with genome-draft mapping and reintroduced the concept of TU (Okazaki et al. 2002). We estimated that there are ∼70,000 of them in the current data set. The TU counting simply differs with previous conservative gene counting, which focuses on protein-coding genes (Dunham et al. 1999; Hattori et al. 2000), The set of fully sequenced cDNAs annotated in FANTOM2 was clustered into ∼37,000 TU along with sequences from the public domain. Of these, ∼18,000 encoded proteins, which is significantly less than the 30,000 estimated, were based upon mouse (and human) genome annotation. This assessment supports the view that there are many additional and genuine TU still to be fully sequenced in the RIKEN cDNA set. In the most recent libraries, the rate of discovery of new singletons is still close to the linearity, suggesting that despite the depth of sampling, even this transcriptome cannot be considered finished, and other tissues/differentiation states remain to be explored. This conclusion is also in keeping with recent direct analysis suggesting that the transcriptional output of the genome in humans greatly exceeds gene predictions (Kapranov et al. 2002). Representation of a comprehensive collection of ESTs (Fig. 3) resemble an in silico representation of classical studies on the nature and complexity of the transcriptome by cDNA/RNA reassociation experiments (Galau et al. 1977). Although our figure does not discern the most abundant mRNAs, the distribution of gene discovery resembles the renaturation profile and derived distribution of highly expressed mRNAs, intermediately expressed and rarely expressed (singletons), applied on the whole organism. Beside differences such as the subtraction, this analysis vindicates in silico pioneering observations on the basis of reassociation technologies of the existence of such expression classes.
The appearance of a large scale of clusters only in small-scale, microdissected libraries suggests that along with the universally expressed mRNAs, there is a considerable set of genes for which expression seems to be restricted to a subset of cells. We sampled many tissues extensively to obtain the largest variety of mRNAs and categorize genes involved in various biological phenomena. Further, the mouse afforded the great opportunity of sampling any developmental stage and preparing high-quality RNA from freshly dissected tissues. Beside libraries for all relevant adult tissues and biological phenomena such as proliferation, apoptosis, cell–cell communication, and lineage differentiation, pre-implantation and post-implantation embryonic development is covered extensively with 81 libraries that provide full-length cDNA clones and information for almost 80% of the clones of an existing set or embryonic libraries (Kargul et al. 2001). Clearly, for these tissues, mouse full-length cDNAs will be essential to identify equivalent human mRNAs that are not readily accessible in human, especially for early post-implantation specimens.
Brain has been hypothesized to express at least half of the genes and, therefore, we sampled 52 neuronal tissues. Some peripheral nervous tissues or sensory system libraries, such as the sympathetic ganglion, retina, and inner ear showed surprising diversity from the central nervous system. We also characterized a relatively simple type of neural tissue, the cerebellum, and its development from birth to neonatal stage to define the variability of mRNA expression in a simplified neuronal tissue. Therefore, we prepared and sequenced seven libraries from neonate (P0 to 1 month), stages, when production of new cell types and extensive apoptosis occur, plus one adult library. A characterization of the cerebellar development was done previously (Diaz et al. 2002). According to what has been described in this analysis, we have found genes that appear to be expressed differently. For instance, the Cam kinase II β, which was reported to increase during development of the cerebellum, appeared only once in adult cerebellum and peaks at postnatal day 10 with seven sequences. Math-1, reported to be expressed after birth and to decrease after 1 wk. Accordingly, two Math-1 cDNA clones were sampled only from the postnatal 0 day libraries. Similarly, most of the other genes reported to have specific expression patterns (Diaz et al. 2002) appeared in cerebellum libraries but, due to normalization/subtraction, their count was not sufficient for statistical analysis. On the contrary, normalization/subtraction strategies have produced ∼5700 clusters that are unique to various stages of cerebellum development and await further characterization. It is very likely that other tissues, if deeply sampled similarly, could show such complexity.
Among other phenomena, we monitored five cDNA libraries covering the development and regression of the mammary gland, because the gland development resembles breast cancer tissue for the ability of a mass of proliferating cell to invade a stromal tissue, protecting it from premature apoptosis and final apoptotic regression (Wiseman and Werb 2002). We also investigated the development of the reproductive system with 10 libraries, whose source tissues include regions of primordial germ cells and those of later development. Other biological cycles include nine thymus libraries, representing developmental stages from E14 through the P3 neonatal stage, when clonal deletion of self-reacting lymphocytes is highest, to adult and the associated regression of thymic tissues. In the continuation of our work, methods to resubtract amplified cDNA libraries prepared by normalizing and subtracting cDNA before cloning will be a complementary strategy to select for very rarely expressed mRNAs in order to sequence as close as possible to saturation (T. Hirozane-Kishikawa and P. Carninci, in prep.). Such technologies, based on amplified λ cDNA library subtraction, should allow targeting a class of transcripts that all together represent 7 < 1% of the mass of mRNA in a tissue. Considering different strains may also play a role, for instance, C57Bl6/J has a mutated CDK-3 (Ye et al. 2001), which may affect downstream pathways. Further we are selecting new, rare/very long cDNAs by size selection (>6.5 Kb), stabilization of long cDNAs and improved sequencing operation for long cDNAs. To produce functional proteins and simplify annotation, we are using, as much as possible, cytoplasmic RNA devoid of unspliced introns, polysomal, and membrane-bound polysomal RNA; this latter effort will facilitate identifying secreted and membrane proteins. In the generation of new full-length libraries, we are also looking at vectors that allow prompt transfer of clones into expression vectors (Carninci et al. 2001).
Despite prioritization of sequencing on the basis of clustering, the FANTOM2 full-length sequence set contained numerous clusters and redundancy, which was used to enable a comprehensive analysis of the frequency of alternative splicing (Zavolan et al. 2003). A real insight into the function of the transcriptome will not be complete until alternative splice forms are identified comprehensively. The frequency of functional alternative splicing is so high, that additional sequencing of even one additional member of each multimember cluster is likely to generate many additional variant full-length sequences. We are developing strategies to identify TU clusters in which such variation is more prevalent on the basis of EST and genomic information to prioritize additional sequencing.
Our approach can be readily applied to organisms other than mouse, including numerous model organisms, animals of alimentary interest, and plants, and to some extent, to clone the missing human full-length cDNAs. However, we anticipate that getting a complete set of human full-length cDNAs will be very challenging, due to the difficulty of sampling and restricted availability of high-quality samples. Perhaps nonhuman primates may provide mRNA for similar approaches or, alternatively, remaining human cDNAs could be collected with RT–PCR on the basis of the presence of mouse homolog mRNA sequences.
The mouse sequence information we have generated is freely available at http://genome.gsc.riken.go.jp/. These data are updated periodically, and the sequences are deposited in public databases through the DNA Database of Japan (DDBJ). Representative clones are, at the moment, available, but the complete cDNA collection (∼2 million clones) does not allow prompt distribution, although mechanisms are being considered to make it happen. Request for clones should be sent to http://genome.gsc.riken.go.jp and the e-mail address therein specified. At the moment, the first set of representative 60,770 clones fully sequenced (the FANTOM 2 set) is available.
METHODS
Preparation of cDNA Libraries
Mice (mostly C57Bl6/J) were sacrificed according to institutional guidelines. We used Cap-Trapper technology (Carninci et al. 1996, 1997; Carninci and Hayashizaki 1999) and trehalose-thermoactivated reverse transcriptase (Carninci et al. 1998; Carninci and Hayashizaki 1999) to prepare libraries enriched for full-length inserts (full-length libraries). Published protocols were adapted so that we could prepare cDNA libraries from small samples (1.5–50 μg of RNA) from microdissected tissues. Most cDNA populations were normalized (Carninci et al. 2000). Abundant cDNAs were also subtracted by using biotinylated mRNA or in vitro-transcribed RNA driver prepared from linearized plasmid vectors as a driver with XhoI, SstI, or BamHI, depending on the cloning orientation and vector usage (Carninci et al. 2000), or by PCR from rearrayed clone sets using primer adapters on the vector carrying T7 and T3 RNA polymerase promoters. Single-strand cDNA, full-length cDNA was normalized and subtracted by hybridization with biotinylated aliquots of mRNA. Hybridized, abundant cDNA were removed by using streptavidin-coated magnetic beads and, after synthesis of the second-strand cDNA and restriction digestion, cDNA inserts were cloned as described (Carninci and Hayashizaki 1999). We prepared nine minilibraries to use as sources of subtraction drivers. The highly expressed cDNAs were stripped from beads and cloned in parallel to normalized libraries. Approximately 1000 clones (2000 clones for brain) were amplified on SOB-ampicillin plates; plasmids from these clones were used to prepare RNA drivers. In addition, nonredundant or low-redundancy drivers were prepared from rearrayed bacteria as described. RNA was biotinylated by use of the Mirus kit (Panvera).
Subtraction Drivers
Other than what was published (Carninci et al. 2000), the driver used to prepare strongly subtracted libraries was not constituted by run-off transcripts from rearrayed clones linearized by using the BamHI site that is the 3′-end cloning site. Instead, we used PCR to rearray pooled clones with primers containing T7 RNA polymerase promoters, and then synthesized run-off transcripts from PCR products. This was done to overcome potential problems due to the presence of BamHI in the inserts, which would cause the drivers to be limited to the 5′ ends of clones, and, therefore, the enrichment for truncated cDNAs corresponded to the collected genes. The subtraction drivers, as shown in Table 1, complete table at www.genome.org, are as follows: mini set0: liver, lung, and brain minilibraries; mini set1: liver, lung, brain, and placenta minilibraries; or mini set2: liver, lung, brain, placenta, testis, pancreas, small intestine, stomach, and tongue minilibraries. Rearrayed clones were Nm1, 4000 clones; Nm2, 1600 clones; Nm3, 13,440 clones (rearrayed plates 2XB00001–2XB00035); Lm1, 8832 clones (low-redundancy rearray); Nm4, or 1920 clones (rearrayed plates PXB00002–PXB00006). Mixed cDNA libraries were prepared in the majority of cases by using first-strand primers that carried as a tag an arbitrary sequence of 6 bp, which differs at least two nucleotides from all of the other tags. This was recognized during the vector-masking procedures and, therefore, tissue (mRNA sample) origin of the cDNA was possible. Mixed cDNA are advantageous for stronger subtraction and tracking. Cloning vectors (λFL-C-1, λFL-C-II, and λFL-C-III), whose cloning capacity exceeds 15 Kb, have been described (Carninci et al. 2001). Phage libraries underwent bulk excision to yield plasmid libraries. Libraries were mainly electroporated into DH10b or DH5α. Early libraries (Table 1, complete table at www.genome.org) were cloned in pBluescript (Stratagene), either after bulk excision from a λ vector (Short et al. 1988) or by direct cloning. Library 01 was prepared by using the Capfinder kit (Clontech) and cloned in Lambda Triplex(Sasaki et al. 1998). A complete updated set of protocols is described in Bowtell and Sambrook (2002) and in the book Web site.
Sequencing Reagents and Procedures
Bacteria were picked by using Q-bot and Q-pix(Genetics) and transferred to 384-microwell plates. The bacteria were grown either at 30°C or 37°C; growth at 30°C increased the stability of clones with long inserts. Replicates of the primary plates were sent to the plasmid extraction team. The clones from each 384-well plate were grown in four 96-well plates with deep wells. After overnight incubation, plasmids were extracted with a filter-based plasmid preparation system, either manually (Itoh et al. 1997) or automatically (Itoh et al. 1999). To minimize the ID errors of early studies based on slab-gel, we used the RISA 384 capillary sequencer (Shibata et al. 2000). Additionally, we added known samples at fixed positions in the 384-well plates. When the control samples were not recognized, which indicates potential ID errors, we did not include the clones in the collection. A detailed list of other artifacts and quality control is available in Supplementary Table 1). Resequencing of random clones from the FANTOM-1 (Kawai et al. 2001) showed that the ID error was <3% (data not shown). The RISA sequencer used an ad hoc basecaller. For each peak, the basecaller calculates and assigns a value (0–10) to the ratio between the amplitudes of the primary (signal) and secondary (noise) peaks; in addition, the basecaller assigns a value (0–4) to the amplitude of the peak. The sum of these two values (0–14) represents the confidence value of each peak. With regard to evaluating the entire sequence of a sample, the sum 0–7 corresponds to score 0 (unreliable) and 8–14 to 1.
Sequence Analysis
If not otherwise described, the clustering was made following Konno et al. (2001), and other computer analysis, if not otherwise specified, following Okazaki et al. (2002), and references therein. For clustering, the linkers and vector sequences were removed from sequences, then the first 200-bp regions were selected and used as TAG sequences. Different from what was published (Konno et al. 2001), we updated the system using BLAST 2.2.2 or later versions to create the groups (clusters + singletons) to make the nonredundant TAG library. BLAST values 1e-20 or lower, with options (-F F S- 1) values were preclustered together. Next, the BLAST-aligned sequences were checked for identity >90%, overlap >160 nucleotides, and overhang <10 nucleotides. To the nonredundant representative member of a cluster, new members were routinely (usually weekly) added, and sequences that did not follow such conditions became new groups.
To evaluate normalization/subtraction, actin clones that appeared in the Unigene database on April 25, 2000 were selected and compared with BLAST with 1e20 or lower value versus the standard cDNA libraries 66–204, 67–204, 68–204, 74–204, 74–207, B0–200, C8–200, and C9–200 (29,685 sequences) and the recent satisfactory normalized-subtracted libraries A0–A9, B0–B9, and E0–E6 (237,099 sequences). For mapping our clusters on the 292 genomic sequences, we used BLAST values lower than 1e-55. Transcriptional starting point analysis was done by using EST clusters subsequently aligned onto the genome.
Acknowledgments
We thank Saiko Takaku-Akahira, Chitose Sakai, Fumiko Takahashi, Akihiro Tagawa, Tomoko Toya, Tomoaki Yamamura, Takako Akimura, Akiko Tomaru, and Rie Numazaki, for technical contributions; Hirozumi Nishibe for helping in the preparation of the manuscript. This project has been supported by a Research Grant for the RIKEN Genome Exploration Research Project from the MEXT of the Japanese Government to Y.H. and ACT-JST (Research and Development for Applying Advanced Computational Science and Technology) of Japan Science and Technology Corporation (JST) to Y.H. This work is supported by the RIKEN presidential fund for Intra RIKEN collaboration to T.K.H., and a presidential fund for innovative tool development to P.C.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1119703.
Footnotes
[Supplemental material available online at www.genome.org.]
References
- Adams, M.D., Kelley, J.M., Gocayne, J.D., Dubnick, M., Polymeropoulos, M.H., Xiao, H., Merril, C.R., Wu, A., Olde, B., Moreno, R.F., et al. 1991. Complementary DNA sequencing: Expressed sequence tags and human genome project. Science 252: 1651-1656. [DOI] [PubMed] [Google Scholar]
- Adams, M.D., Kerlavage, A.R., Fleischmann, R.D., Fuldner, R.A., Bult, C.J., Lee, N.H., Kirkness, E.F., Weinstock, K.G., Gocayne, J.D., White, O., et al. 1995. Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature 377: 3-174. [PubMed] [Google Scholar]
- Aparicio, S.A. 2000. How to count... human genes. Nat. Genet. 25: 129-130. [DOI] [PubMed] [Google Scholar]
- Bashiardes, S. and Lovett, M. 2001. cDNA detection and analysis. Curr. Opin. Chem. Biol. 5: 15-20. [DOI] [PubMed] [Google Scholar]
- Beaudoing, E., Freier, S., Wyatt, J.R., Claverie, J.M., and Gautheret, D. 2000. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 10: 1001-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaldo, M.F., Lennon, G., and Soares, M.B. 1996. Normalization and subtraction: Two approaches to facilitate gene discovery. Genome Res. 6: 791-806. [DOI] [PubMed] [Google Scholar]
- Bono, H., Nikaido, I., Kasukawa, T., Hayashizaki, Y., RIKEN GER Group and GSL Members, and Okazaki, Y. 2003. Comprehensive analysis of the mouse metabolome based on the transcriptome. Genome Res. 13: (this issue). [DOI] [PMC free article] [PubMed]
- Boon, K., Osorio, E.C., Greenhut, S.F., Schaefer, C.F., Shoemaker, J., Polyak, K., Morin, P.J., Buetow, K.H., Strausberg, R.L., De Souza, S.J., et al. 2002. An anatomy of normal and malignant gene expression. Proc. Natl. Acad. Sci. 99: 11287-11292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowtell, D. and Sambrook, J. 2002. DNA microarrays: A molecular cloning manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Camargo, A.A., Samaia, H.P., Dias-Neto, E., Simao, D.F., Migotto, I.A., Briones, M.R., Costa, F.F., Nagai, M.A., Verjovski-Almeida, S., Zago, M.A., et al. 2001. The contribution of 700,000 ORF sequence tags to the definition of the human transcriptome. Proc. Natl. Acad. Sci. 98: 12103-12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci, P. and Hayashizaki, Y. 1999. High-efficiency full-length cDNA cloning. Methods Enzymol. 303: 19-44. [DOI] [PubMed] [Google Scholar]
- Carninci, P., Kvam, C., Kitamura, A., Ohsumi, T., Okazaki, Y., Itoh, M., Kamiya, M., Shibata, K., Sasaki, N., Izawa, M., et al. 1996. High-efficiency full-length cDNA cloning by biotinylated CAP trapper. Genomics 37: 327-336. [DOI] [PubMed] [Google Scholar]
- Carninci, P., Westover, A., Nishiyama, Y., Ohsumi, T., Itoh, M., Nagaoka, S., Sasaki, N., Okazaki, Y., Muramatsu, M., Schneider, C., et al. 1997. High efficiency selection of full-length cDNA by improved biotinylated cap trapper. DNA Res. 4: 61-66. [DOI] [PubMed] [Google Scholar]
- Carninci, P., Nishiyama, Y., Westover, A., Itoh, M., Nagaoka, S., Sasaki, N., Okazaki, Y., Muramatsu, M., and Hayashizaki, Y. 1998. Thermostabilization and thermoactivation of thermolabile enzymes by trehalose and its application for the synthesis of full length cDNA. Proc. Natl. Acad. Sci. 95: 520-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci, P., Shibata, Y., Hayatsu, N., Sugahara, Y., Shibata, K., Itoh, M., Konno, H., Okazaki, Y., Muramatsu, M., and Hayashizaki, Y. 2000. Normalization and subtraction of cap-trapper-selected cDNAs to prepare full-length cDNA libraries for rapid discovery of new genes. Genome Res. 10: 1617-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci, P., Shibata, Y., Hayatsu, N., Itoh, M., Shiraki, T., Hirozane, T., Watahiki, A., Shibata, K., Konno, H., Muramatsu, M., et al. 2001. Balanced-size and long-size cloning of full-length, cap-trapped cDNAs into vectors of the novel λ-FLC family allows enhanced gene discovery rate and functional analysis. Genomics 77: 79-90. [DOI] [PubMed] [Google Scholar]
- Carninci, P., Nakamura, M., Sato, K., Hayashizaki, Y., and Brownstein, M.J. 2002a. Cytoplasmic RNA extraction from fresh and frozen mammalian tissues. Biotechniques 33: 306-309. [DOI] [PubMed] [Google Scholar]
- Carninci, P., Shiraki, T., Mizuno, Y., Muramatsu, M., and Hayashizaki, Y. 2002b. Extra-long first-strand cDNA synthesis. Biotechniques 32: 984-985. [DOI] [PubMed] [Google Scholar]
- Diaz, E., Ge, Y., Yang, Y.H., Loh, K.C., Serafini, T.A., Okazaki, Y., Hayashizaki, Y., Speed, T.P., Ngai, J., and Scheiffele, P. 2002. Molecular analysis of gene expression in the developing pontocerebellar projection system. Neuron. 36: 417-434. [DOI] [PubMed] [Google Scholar]
- Dunham, I., Shimizu, N., Roe, B.A., Chissoe, S., Hunt, A.R., Collins, J.E., Bruskiewich, R., Beare, D.M., Clamp, M., Smink, L.J., et al. 1999. The DNA sequence of human chromosome 22. Nature 402: 489-495. [DOI] [PubMed] [Google Scholar]
- Ewing, B. and Green, P. 2000. Analysis of expressed sequence tags indicates 35,000 human genes. Nat. Genet. 25: 232-234. [DOI] [PubMed] [Google Scholar]
- Galau, G.A., Klein, W.H., Britten, R.J., and Davidson, E.H. 1977. Significance of rare mRNA sequences in liver. Arch. Biochem. Biophys. 179: 584-599. [DOI] [PubMed] [Google Scholar]
- Gautheret, D., Poirot, O., Lopez, F., Audic, S., and Claverie, J.M. 1998. Alternate polyadenylation in human mRNAs: A large-scale analysis by EST clustering. Genome Res. 8: 524-530. [DOI] [PubMed] [Google Scholar]
- Gubler, U. and Hoffman, B.J. 1983. A simple and very efficient method for generating cDNA libraries. Gene 25: 263-269. [DOI] [PubMed] [Google Scholar]
- Hattori, M., Fujiyama, A., Taylor, T.D., Watanabe, H., Yada, T., Park, H.S., Toyoda, A., Ishii, K., Totoki, Y., Choi, D.K., et al. 2000. The DNA sequence of human chromosome 21. Nature 405: 311-319. [DOI] [PubMed] [Google Scholar]
- Hillier, L.D., Lennon, G., Becker, M., Bonaldo, M.F., Chiapelli, B., Chissoe, S., Dietrich, N., DuBuque, T., Favello, A., Gish, W., et al. 1996. Generation and analysis of 280,000 human expressed sequence tags. Genome Res. 6: 807-828. [DOI] [PubMed] [Google Scholar]
- Hogenesch, J.B., Ching, K.A., Batalov, S., Su, A.I., Walker, J.R., Zhou, Y., Kay, S.A., Schultz, P.G., and Cooke, M.P. 2001. A comparison of the Celera and Ensembl predicted gene sets reveals little overlap in novel genes. Cell 106: 413-415. [DOI] [PubMed] [Google Scholar]
- Iseli, C., Stevenson, B.J., de Souza, S.J., Samaia, H.B., Camargo, A.A., Buetow, K.H., Strausberg, R.L., Simpson, A.J., Bucher, P., and Jongeneel, C.V. 2002. Long-range heterogeneity at the 3′ ends of human mRNAs. Genome Res. 12: 1068-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, M., Carninci, P., Nagaoka, S., Sasaki, N., Okazaki, Y., Ohsumi, T., Muramatsu, M., and Hayashizaki, Y. 1997. Simple and rapid preparation of plasmid template by a filtration method using microtiter filter plates. Nucleic Acids Res. 25: 1315-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, M., Kitsunai, T., Akiyama, J., Shibata, K., Izawa, M., Kawai, J., Tomaru, Y., Carninci, P., Shibata, Y., Ozawa, Y., et al. 1999. Automated filtration-based high-throughput plasmid preparation system. Genome Res. 9: 463-470. [PMC free article] [PubMed] [Google Scholar]
- Kapranov, P., Cawley, S.E., Drenkow, J., Bekiranov, S., Strausberg, R.L., Fodor, S.P., and Gingeras, T.R. 2002. Large-scale transcriptional activity in chromosomes 21 and 22. Science 296: 916-919. [DOI] [PubMed] [Google Scholar]
- Kargul, G.J., Dudekula, D.B., Qian, Y., Lim, M.K., Jaradat, S.A., Tanaka, T.S., Carter, M.G., and Ko, M.S. 2001. Verification and initial annotation of the NIA mouse 15K cDNA clone set. Nat Genet. 28: 17-18. [DOI] [PubMed] [Google Scholar]
- Kawai, J., Shinagawa, A., Shibata, K., Yoshino, M., Itoh, M., Ishii, Y., Arakawa, T., Hara, A., Fukunishi, Y., Konno, H., et al. 2001. Functional annotation of a full-length mouse cDNA collection. Nature 409: 685-690. [DOI] [PubMed] [Google Scholar]
- Kochiwa, H., Suzuki, R., Washio, T., Saito, R., Bono, H., Carninci, P., Okazaki, Y., Miki, R., Hayashizaki, Y., and Tomita, M. 2002. Inferring alternative splicing patterns in mouse from a full-length cDNA library and microarray data. Genome Res. 12: 1286-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno, H., Fukunishi, Y., Shibata, K., Itoh, M., Carninci, P., Sugahara, Y., and Hayashizaki, Y. 2001. Computer-based methods for the mouse full-length cDNA encyclopedia: Real-time sequence clustering for construction of a nonredundant cDNA library. Genome Res. 11: 281-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra, M., Hillier, L., Kucaba, T., Allen, M., Barstead, R., Beck, C., Blistain, A., Bonaldo, M., Bowers, Y., Bowles, L., et al. 1999. An encyclopedia of mouse genes. Nat. Genet. 21: 191-194. [DOI] [PubMed] [Google Scholar]
- Maruyama, K., and Sugano, S. 1994. Oligo-capping: A simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene 138: 171-174. [DOI] [PubMed] [Google Scholar]
- Miki, R., Kadota, K., Bono, H., Mizuno, Y., Tomaru, Y., Carninci, P., Itoh, M., Shibata, K., Kawai, J., Konno, H., et al. 2001. Delineating developmental and metabolic pathways in vivo by expression profiling using the RIKEN set of 18,816 full-length enriched mouse cDNA arrays. Proc. Natl. Acad. Sci. 98: 2199-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, Y., Carninci, P., Okazaki, Y., Tateno, M., Kawai, J., Amanuma, H., Muramatsu, M., and Hayashizaki, Y. 1999. Increased specificity of reverse transcription priming by trehalose and oligo-blockers allows high-efficiency window separation of mRNA display. Nucleic Acids Res. 27: 1345-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam, D.K., Lee, S., Zhou, G., Cao, X., Wang, C., Clark, T., Chen, J., Rowley, J.D., and Wang, S.M. 2002. Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription. Proc. Natl. Acad. Sci. 99: 6152-6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., Nikaido, I., Osato, N., Saito, R., Suzuki, H., et al. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563-573. [DOI] [PubMed] [Google Scholar]
- Osato, N., Itoh, M., Konno, H., Kondo, S., Shibata, K., Carninci, P., Shiraki, T., Shinagawa, A., Arakawa, T., Kikuchi, S., et al. 2002. A computer-based method of selecting clones for a full-length cDNA project: Simultaneous collection of negligibly redundant and variant cDNAs. Genome Res. 12: 1127-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringwald, M., Eppig, J.T., Begley, D.A., Corradi, J.P., McCright, I.J., Hayamizu, T.F., Hill, D.P., Kadin, J.A., and Richardson, J.E. 2001. The mouse gene expression database (GXD). Nucleic Acids Res. 29: 98-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crollius, R.H., Jaillon, O., Bernot, A., Dasilva, C., Bouneau, L., Fischer, C., Fizames, C., Wincker, P., Brottier, P., Quetier, F., et al. 2000. Estimate of human gene number provided by genome-wide analysis using Tetraodon nigroviridis DNA sequence. Nat. Genet. 25: 235-238. [DOI] [PubMed] [Google Scholar]
- Sasaki, N., Nagaoka, S., Itoh, M., Izawa, I.M., Konno, H., Carninci, P., Yoshiki, A., Kusakabe, M., Moriuchi, T., Muramatsu, M., et al. 1998. Characterization of gene expression in mouse blastocyst using single-pass sequencing of 3995 clones. Genomics 49: 167-179. [DOI] [PubMed] [Google Scholar]
- Shibata, K., Itoh, M., Aizawa, K., Nagaoka, S., Sasaki, N., Carninci, P., Konno, H., Akiyama, J., Nishi, K., Kitsunai, T., et al. 2000. RIKEN integrated sequence analysis (RISA) system—384-format sequencing pipeline with 384 multicapillary sequencer. Genome Res. 10: 1757-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata, Y., Carninci, P., Sato, K., Hayatsu, N., Shiraki, T., Ishii, Y., Arakawa, T., Hara, A., Ohsato, N., Izawa, M., et al. 2001a. Removal of polyA tails from full-length cDNA libraries for high-efficiency sequencing. Biotechniques 31: 1042, 1044, 1048-1049. [DOI] [PubMed] [Google Scholar]
- Shibata, Y., Carninci, P., Watahiki, A., Shiraki, T., Konno, H., Muramatsu, M., and Hayashizaki, Y. 2001b. Cloning full-length, cap-trapper-selected cDNAs by using the single-strand linker ligation method. Biotechniques 30: 1250-1254. [DOI] [PubMed] [Google Scholar]
- Short, J.M., Fernandez, J.M., Sorge, J.A., and Huse, W.D. 1988. Lambda ZAP: A bacteriophage λ expression vector with in vivo excision properties. Nucleic Acids Res. 16: 7583-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton, M., Carlson, J., Brokstein, P., Yu, C., Champe, M., George, R., Guarin, H., Kronmiller, B., Pacleb, J., Park, S., et al. 2002. A Drosophila full-length cDNA resource. Genome Biol. 3: RESEARCH0080-0. [DOI] [PMC free article] [PubMed]
- Strausberg, R.L., Feingold, E.A., Grouse, L.H., Derge, J.G., Klausner, R.D., Collins, F.S., Wagner, L., Shenmen, C.M., Schuler, G.D., Altschul, S.F., et al. 2002. Proc. Natl. Acad. Sci. 99: 16899-16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugahara, Y., Carninci, P., Itoh, M., Shibata, K., Konno, H., Endo, T., Muramatsu, M., and Hayashizaki, Y. 2001. Comparative evaluation of 5′-end-sequence quality of clones in CAP trapper and other full-length-cDNA libraries. Gene 263: 93-102. [DOI] [PubMed] [Google Scholar]
- Suzuki, Y., Yoshitomo-Nakagawa, K., Maruyama, K., Suyama, A., and Sugano, S. 1997. Construction and characterization of a full length-enriched and a 5′-end-enriched cDNA library. Gene 200: 149-156. [DOI] [PubMed] [Google Scholar]
- Suzuki, Y., Taira, H., Tsunoda, T., Mizushima-Sugano, J., Sese, J., Hata, H., Ota, T., Isogai, T., Tanaka, T., Morishita, S., et al. 2001a. Diverse transcriptional initiation revealed by fine, large-scale mapping of mRNA start sites. EMBO Rep. 2: 388-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, Y., Tsunoda, T., Sese, J., Taira, H., Mizushima-Sugano, J., Hata, H., Ota, T., Isogai, T., Tanaka, T., Nakamura, Y., et al. 2001b. Identification and characterization of the potential promoter regions of 1031 kinds of human genes. Genome Res. 11: 677-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, H., Saito, R., Kanamori, M., Kai, C., Schönbach, C., Nagashima, T., Hosaka, J., and Hayashizaki, Y. 2003. The mammalian protein–protein interaction database and its viewing system that is linked to the main FANTOM2 viewer. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Wahle, E. 1995. 3′-end cleavage and polyadenylation of mRNA precursors. Biochim. Biophys. Acta 1261: 183-194. [DOI] [PubMed] [Google Scholar]
- Walsh, N.C., Cahill, M., Carninci, P., Kawai, J., Okazaki, Y., Hayashizaki, Y., Hume, D.A., and Cassady, A.I. 2003. Multiple tissue-specific promoters control expression of the tartrate-resistant acid phosphatase gene. Gene (in press). [DOI] [PubMed]
- Wang, S.M., Fears, S.C., Zhang, L., Chen, J.J., and Rowley, J.D. 2000. Screening poly(dA/dT)-cDNAs for gene identification. Proc. Natl. Acad. Sci. 97: 4162-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Wells, C.A., Ravasi, T., Sultana, R., Yagi, K., Carninci, P., Bono, H., Faulkner, G., Okazaki, Y., Quackenbush, J., Hume, D.A., et al. 2003. Continued discovery of transcriptional units expressed in cells of the mouse mononuclear phagocyte lineage. Genome Res. 13: (this issue). [DOI] [PMC free article] [PubMed]
- Wiseman, B.S. and Werb, Z. 2002. Stromal effects on mammary gland development and breast cancer. Science 296: 1046-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, X., Zhu, C., and Harper, J.W. 2001. A premature-termination mutation in the Mus musculus cyclin-dependent kinase 3 gene. Proc. Natl. Acad. Sci. 98: 1682-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavolan, M., Kondo, S., Schönbach, C., Adachi, J., Hume, D.A., RIKEN GER Group and GSL Members, Hayashizaki, Y., and Gaasterland, T. 2003. Impact of alternative initiation, splicing, and termination on the diversity of the mRNA transcripts encoded by the mouse transcriptome. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
WEB SITE REFERENCES
- http://genome.gsc.riken.go.jp/; Describes the overall activity of The RIKEN GER Group.
- http://www.informatics.jax.org/menus/expression_menu.shtml; Introduces the mouse tissue's classification.
- http://genome.gsc.riken.go.jp/READ/; Describes the microarray expression database of the RIKEN GER Group.