Abstract
Rhodobacter sphaeroides 2.4.1 is a facultative photoheterotrophic bacterium with tremendous metabolic diversity, which has significantly contributed to our understanding of the molecular genetics of photosynthesis, photoheterotrophy, nitrogen fixation, hydrogen metabolism, carbon dioxide fixation, taxis, and tetrapyrrole biosynthesis. To further understand this remarkable bacterium, and to accelerate an ongoing sequencing project, two whole-genome restriction maps (EcoRI and HindIII) of R. sphaeroides strain 2.4.1 were constructed using shotgun optical mapping. The approach directly mapped genomic DNA by the random mapping of single molecules. The two maps were used to facilitate sequence assembly by providing an optical scaffold for high-resolution alignment and verification of sequence contigs. Our results show that such maps facilitated the closure of sequence gaps by the early detection of nascent sequence contigs during the course of the whole-genome shotgun sequencing process.
Rhodobacter sphaeroides is an α-3, purple, nonsulfur photosynthetic eubacterium, which has significantly contributed to our understanding of the molecular genetics of photosynthesis and other important bioenergetic processes. This highly versatile organism can grow aerobically or anaerobically in the presence or absence of light, while using external electron acceptors such as hydrogen or reduced organic compounds. Furthermore, this bacterium has tremendous metabolic diversity, as it can oxidize a variety of compounds such as organic acids, sugars, polyols, and methanol, and toxic compounds such as formaldehyde. It can also reduce organic or inorganic compounds such as thymine, sulfate, and toxic metal oxides, etc. (Clayton and Sistrom 1978; Moore and Kaplan 1992; Mouncey et al. 1997; Barber and Donohue 1998). Besides their bioenergetic versatility and metabolic diversity, it is believed that this group of bacteria (α-3 subdivision of the Proteobacteria), and in particular an R. sphaeroides-like organism, may have been the ancestor of the primitive mitochondrion (Yang et al. 1985). Evidence supporting this view comes from the findings that this bacterium has two aminolevulinic acid synthases (Shamin type) and a benzodiazepine-like receptor (Neidle and Kaplan 1993; Yeliseev and Kaplan 1995), which share a high degree of sequence and functional homology with mammalian mitochondrial proteins.
R. sphaeroides presents a uniquely rich set of metabolic and phylogenetic questions that require detailed answers through a concerted effort to establish the fundamental genomic underpinnings for this organism. As such, the sequencing of the R. sphaeroides genome was funded by the U.S. Department of Energy (DOE), the National Institutes of Health (NIH), and other agencies, and was carried out by two sequencing centers, the Joint Genome Institute (JGI) and the University of Texas—Houston Medical School (UT). JGI adopted the whole-genome shotgun approach (Fleischmann et al. 1995) to sequence the entire genome, while UT focused on chromosome II (Mackenzie et al. 2001). Although a large number of microbial genomes have been completed using this sequencing strategy, much optimization remains to be done in terms of reducing the cost and effort required during the finishing stages of any sequencing project. In this regard, physical maps guide sequence assembly, characterize gaps, and validate finished sequence, especially when attempting to assemble genomic regions containing repeated DNA sequences, as cleavage patterns can frequently discern such sequence elements. Entire regions of a genome might be excluded from “completed” sequence data because of limited budgets and the absence of physical mapping data (Sensen 1999). Although physical maps had been constructed for R. sphaeroides, which established the existence of a complex genome containing two chromosomes (Suwanto and Kaplan 1989a,b, 1992; Choudhary et al., 1994, 1997), these maps are of modest resolution (the average fragment sizes of these maps are >150 kb). Indeed, nearly two thirds of chromosome II lacked any physical or genetic markers, thus these maps were minimally useful as a scaffold for sequence assembly or as a means for assembly validation. Therefore, optical maps (EcoRI and HindIII) were constructed to identify gaps, characterize assembled sequence contigs, and to ultimately validate finished sequence.
The optical mapping system is largely automated, and constructs ordered restriction maps from ensembles of genomic DNA molecules mounted on the specially derivatized glass surface (Meng et al. 1995; Anantharaman 1997, 1998, 1999; Aston et al. 1999a,b; Cai et al. 1998; Jing et al. 1999; Lai et al. 1999; Lim et al. 2001). DNA molecules are imaged using fluorescence microscopy after restriction enzyme digestion, but importantly, restriction fragments remain ordered. Because DNA molecules are stained with a fluorochrome, the mass of each restriction fragment is accurately determined using integrated fluorescence intensity measurements, which obviates the need for uniformly elongated molecules required for length-based mass measurements. Gap-free restriction maps of an entire microbial genome are generated using a map assembler in a process that is similar to shotgun sequence assembly (Anantharaman 1997, 1998, 1999).
Here, we present two genome-wide optical maps (EcoRI and HindIII) of R. sphaeroides strain 2.4.1 that were used as a scaffold for sequence assembly and to validate the finished sequence. Notably, the EcoRI optical map is a very high-resolution map with an average fragment size of ∼7 kb and represents the highest resolution attained thus far by optical mapping for an entire genome.
METHODS
DNA Preparation
R. sphaeroides strain 2.4.1 genomic DNA gel inserts (Schwartz and Cantor 1984) were prepared from a culture grown chemoheterotrophically at 32°C on a shaker using Sistrom's minimal medium A and stored in 0.5 M EDTA (pH 8.0) (Lueking et al. 1978). Prior to use, the DNA gel inserts were washed thoroughly overnight in TE (10 mM Tris, 1 mM EDTA; pH 8.0) to remove excess EDTA. The washed inserts were melted at 72°C for 7 min., and the melted agarose was digested at 42°C for 2 h in β-agarase solution (New England Biolabs; 100 μL of TE + 2 μL [1 Unit] β-agarase per 20 μL agarose). Suitable DNA dilutions were made from this sample with TE using centrifugation (microfuge; rotor radius 13 cm, 6000 rpm, 30 min), to ensure uniform dilution and to minimize the presence of supercoiled plasmid DNA on optical mapping surfaces. Lambda DASH II bacteriophage DNA (Stratagene) was added to the genomic DNA solution (10 pg/μL). Such DNA samples were mounted onto an optical mapping surface and examined by fluorescence microscopy to check molecular integrity and concentration.
Surface Preparation
Glass cover slips (22 × 22 mm, Fisher's Finest; Fisher Scientific) were cleaned and derivatized as before (Zhou et al., 2002). Surface properties were assayed by digesting λ DASH II bacteriophage DNA with 40 units of HindIII and EcoRI diluted in 100 μL of digestion buffer with 0.02% Triton X-100 (SIGMA) at 37°C to determine optimal digestion times, which ranged from 40–120 min.
DNA Mounting, Overlay, Digesting and Staining
DNA molecules were mounted on derivatized glass surfaces by capillary action using a microfluidic device (E. Dimalanta and D.C. Schwartz, unpubl.). Then, a thin layer of acrylamide (3.3% containing 0.02% Triton X-100 [SIGMA]) was applied to the surface, which upon polymerization was washed with 400 μL TE for 2 min, followed by washing with 200 μL digestion buffer for another 2 min. To set up the digestion, 200 μL of digestion buffer with enzyme (20 μL NEB [New England Biolabs] Buffer 2, 176 μL high purity water, 2 μL 2% Triton X-100 [SIGMA] and 2 μL NEB-HindIII or EcoRI; 2 units/μL) were added to the surface and incubated in a humidified chamber at 37°C for 40–120 min. After digestion, the surface was washed twice by adding 400 μL TE and aspirated off, 2–5 min each time. The surface was mounted onto a glass slide with 12 μL, 0.2 μM YOYO-1 solution {containing 5 parts YOYO-1; 1,1′-[1,3-propanediylbis[(dimethyliminio) -3,1-propanediyl]]bis[4–[(3-methyl-2(3H)- benzoxazolylidene)-methyl]]-, tetraiodide [Molecular Probes] and in 95 parts of β-mercaptoethanol in TE 20% v/v}. The sample was sealed with nail polish and incubated (4°C in the dark) for 20 min or overnight for the staining dye to diffuse before checking under the fluorescence microscope.
Image Acquisition and Processing
DNA samples were imaged by fluorescence microscopy as previously described using a 63× objective (Zeiss) and a high-resolution digital camera (Princeton Instruments; Lim et al. 2001). Images were collected using a fully automated image acquisition system developed by our laboratory (Autocollect). Comounted λ DASH II DNA molecules were used to estimate the digestion rate and to provide internal fluorescence standards for accurately sizing the DNA fragments (Anantharaman et al. 1997; Marra et al. 1997; Lin et al. 1999). The image files were processed to create maps using previously described software (Lim et al. 2001).
Optical Map Assembly
Individual molecule restriction maps were overlapped by aligning restriction sites based on fragment sizes using specially written software Gentig (Anantharaman 1997, 1998, 1999; Lai et al. 1999; Lin et al. 1999; Lim et al. 2001). Briefly, Gentig assembles single-molecule restriction maps into a genome-wide map contig using a greedy algorithm with limited backtracking for finding an almost optimal scoring set of map contigs in order to avoid the high computational complexity that would occur in attempting to find the optimal assembly. Bayesian inference was used to estimate the probability that two distinct single-molecule restriction maps could have been derived from the proposed placement while subject to various data errors such as sizing errors, missing restriction sites (missing cuts), and false cut errors. A Bayesian approach required fine tuning of these parameters and a known prior statistical distribution of error sources. These parameters, such as standard deviation, digestion rate, false cut, and false match probability, can be reestimated from the data using a limited number of iterations of Bayesian probability density maximization. After these parameters are correctly estimated from the data, the best offset and alignment between a pair of maps can be computed by an efficient dynamic programming algorithm.
DNA Sequence Assembly
DNA sequence trace files of the whole-genome were generated by the DOE microbial genome project and chromosome II-specific files were generated by UT. Assembly of the whole-genome data set by the DOE generated 195 contigs ranging in size from ∼0.5–100 kb. Scaffold sequence data (not to be confused with the optical scaffold data also described in this work) also supplied by the DOE, indicated sequencing subclones that had insert ends that assembled into two contigs. This information permitted the deduction of a tentative contig order and orientation around chromosome I and II, which was cross checked by optical mapping. PCR primers were then designed to the contig ends, and PCR products spanning the contig gaps were generated then sequenced and included in subsequent rounds of assembly.
The possession of chromosome II (∼0.9 Mb)-specific sequences permitted an initial assembly of this replicon independently from the whole-genome sequence data set. The in silico map of chromosome II was then checked against the optical map for accuracy. Assembly of chromosome I (∼3.0 Mb) was then undertaken as a part of a whole-genome assembly, which included the now complete chromosome II DNA sequence. This method was used to “encourage” the assembler into aligning trace files derived from chromosome II with the completed chromosome II sequence. In this way, all trace files not assembling with the chromosome II sequence must by default be derived from chromosome I or plasmids, which were then closed and finished in a targeted approach described above. All assemblies were made using the Phred, Phrap, and Consed package (Gordon et al. 1998). On average, the redundancy of the DNA sequence coverage of chromosomes I and II was about sevenfold and about ninefold, respectively. Regions that misassembled, as described here, consisted of areas of low sequence coverage (2–3×), that arose as a consequence of locally high %G+C composition with strong secondary structure. These regions, which were repeated at several places in the genome, proved highly recalcitrant to PCR and additional sequencing efforts.
Optical Maps Versus In Silico Maps
The EcoRI and HindIII optical maps of R. sphaeroides 2.4.1 genome chromosome I and chromosome II were aligned separately with the in silico EcoRI and HindIII maps of chromosome II and seven sequence contigs of chromosome I using Gentig. The possible misassembled parts of the chromosome I sequence contigs were removed in order to estimate the optical errors compared to the sequence in silico maps. The missing fragments and the false cuts or missing cuts were determined based on these alignments. The relative error associated with each consensus map fragment in the optical maps was calculated by the following formula: (| in silico map fragment size) – (optical map fragment size |)/(in silico map fragment size) × 100%, and was plotted against in silico map fragment masses to visualize the relationship between fragment size and the relative error.
RESULTS
Acquisition of Data and Construction of Optical Maps
Whole-genome shotgun optical mapping (Anantharaman 1997, 1998, 1999; Lai et al. 1999; Lin et al. 1999) was used to construct the restriction maps presented here, a mapping approach that bears many similarities to whole-genome shotgun sequencing (Fleischmann et al. 1995; Marra et al. 1997; Soderlund et al. 1997). The advantages afforded by this approach accrue mainly from the use of genomic DNA as the source of single molecules, which obviates the needs for libraries, PCR, or separations. As with shotgun sequencing, a large set of optical maps (vs. “sequence reads”) is used to multiply cover any given region to establish continuity and accuracy of the physical maps across a genome.
Genomic DNA molecules were randomly broken into large fragments (200–1,600 kb) during the course of sample preparation. Such fragments were elongated and bound to optical mapping surfaces for digestion by two restriction endonucleases (EcoRI and HindIII) in separate experiments and then imaged by fluorescence microscopy using Autocollect (see Methods). Maps derived from individual DNA molecules were then assembled into whole-genome consensus maps using the Gentig map assembler (Anantharaman 1997, 1998, 1999; Lai et al. 1999).
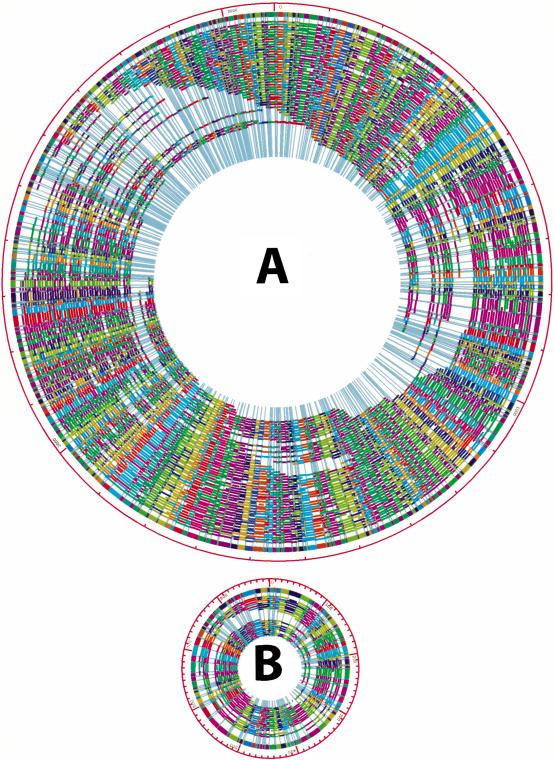
The EcoRI mapping began with the collection and processing of 576 molecules. In total, 256 of these molecules were assembled into two circular maps covering two distinct chromosomes (I and II); a statistical summary is presented in Table 1. Both maps were circularized without gaps and a typical restriction fragment reported within the consensus map was computed from a number of congruent molecules—37 (chromosome I) or 12 (chromosome II). Similar digestion rates were calculated for chromosome I (82.6%) and chromosome II (84.48%). The average fragment size of the EcoRI optical maps was 6.49 kb for chromosome I, and 6.93 kb for chromosome II, which represents the highest resolution optical maps thus far reported. Figure 1 shows the finished EcoRI maps (consensus maps) of the R. sphaeroides chromosomes and the underlying single-molecule maps used to construct the consensus map.
Table 1.
Optical Mapping Data of R. sphaeroides 2.4.1. Genome
| Data collection
|
Contigged molecules
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Enzyme | Chromosome | # Molecules | Mass (Mb) | # Molecules | Mass (Mb) | Contig rate (%) | Circular contig size (kb) | Average fragment size (kb) | Average fragment size SD (kb) | Circularization false probability |
| EcoRI | Chr I | 576 | 270.3 | 232 | 116.82 | 47.67 | 3107 | 6.49 | 1.18 | 0.02515 |
| Chr II | 1 | 24 | 12.05 | 890 | 6.93 | 1.02 | 0.00188 | |||
| HindIII | Chr I | 464 | 250.5 | 152 | 93.10 | 41.85 | 3190 | 17.78 | 2.21 | 0.02012 |
| Chr II | 6 | 22 | 11.77 | 938 | 24.10 | 3.53 | 0.00607 | |||
Figure 1.
Whole-genome EcoRI optical map contigs of chromosome I and II of R. sphaeroides 2.4.1. (A) Chromosome I circular map contig. The outermost multicolored ring shows the consensus map calculated by Gentig. The consensus map was built from the shown underlying maps, obtained from individual DNA molecules, and represented here as multicolored arcs. Congruent restriction fragments, shown in the consensus map are denoted by common color; the color coding scheme is random to enhance contrast. Missing cuts are displayed as fragments that span across one or more cut sites in the consensus map. (B) Chromosome II circular map contig; display scheme is the same as (A).
The precision of the EcoRI whole-genome maps (sizing error per single restriction fragment) was estimated from the standard deviation calculated from the sets of like restriction fragments used to calculate each position within the consensus maps (these maps report the average mass of each fragment set). Accordingly, the average standard deviation about the mean fragment size was 1.18 kb for chromosome I and 1.02 kb for chromosome II, based on the final circularized consensus maps.
The same methodologies and analyses were used to construct the HindIII optical map. Here, a total of 464 digested DNA molecules were imaged and processed to construct two separate maps (Table 1), with a total mass of 104.87 Mb, representing ∼26× coverage of the R. sphaeroides 2.4.1 genome. The average standard deviation, computed from all of the consensus map fragments, was 2.21 kb for chromosome I and 3.53 kb for chromosome II, based on the finished map contigs. Finally, both chromosomes were sized at 3.19 Mb and 938 kb, for chromosome I and II, respectively. These values were based on the summation of all restriction fragments comprising the HindIII consensus maps.
The correctness of a completed circularized map was statistically evaluated by the same Bayesian inference scheme, which underlies Gentig (see Methods). In essence, a series of hypothesized map alignments supplied by the map data set were statistically evaluated with prior probabilities that model common experimental error. As such, the false positive probabilities for the circularization of the maps reported here were 0.02515 and 0.00188 for the chromosome I and II EcoRI maps, while similar values were obtained for chromosome I (0.02012) and chromosome II (0.00607) HindIII maps (Table 1). Placing these values into an experimental context—from previous optical maps we have found that when the false-positive probabilities for the circularization were <0.05, reliable map closure was assured. Finally, the restriction patterns generated by both EcoRI and HindIII were apparently random, hence no specific restriction patterns or structural features were discerned.
Assessment of Optical Mapping Errors
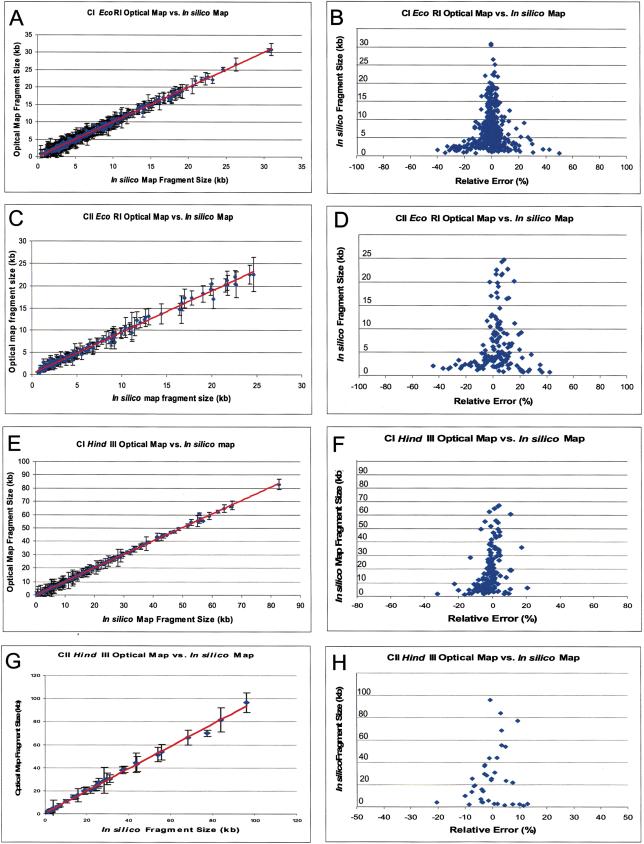
To assess the errors and accuracy of the EcoRI and HindIII maps, comparisons were made between optical mapping and sequence data (Fig. 2). The relative sizing error was shown by the plot of the optical map fragments against the corresponding in silico map fragments based on the alignment of optical maps with in silico maps constructed from the available sequence data of chromosomes I and II (Fig. 2A–H). The error bars in Figure 2A, C, E, and G indicate the standard deviation about the means of the restriction fragment sizes used to calculate consensus map fragments shown in Figure 2. The average absolute error was 1.18 kb for the chromosome I EcoRI map, 1.02 kb for the chromosome II EcoRI map, 2.10 kb for the chromosome I HindIII map, and 3.53 kb for the chromosome II HindIII map, relative to the in silico maps. Generally, a high degree of correspondence was found between optical map fragment sizes and the in silico map fragment sizes, and the trend line is almost identical to the diagonal line.
Figure 2.
Comparisons of the HindIII and EcoRI optical maps of R. sphaeroides chromosome I (CI) and chromosome II (CII) to sequence data. (A) (CI EcoRI), (C) (CII EcoRI), (E) (CI HindIII) and (G) (CII HindIII) are the plots of optical map fragment sizes versus the in silico map fragment sizes from finished sequence. The error bars represent the standard deviation of optical map fragment size on the means. (B) (CI EcoRI), (D) (CII EcoRI), (F) (CI HindIII), and (H) (CII HindIII) show plots of the absolute error of optical fragment size versus sequence.
Scatter plots were done to show details of how the relative fragment sizing error (|optical map fragment size—corresponding in silico map fragment size|/corresponding in silico map fragment size × 100%) varied with restriction fragment size (Fig. 2B,D,F,H). Overall, relative sizing errors were expectedly found to be inversely proportional to the restriction fragment size (Meng et al. 1995). From this analysis, the average relative sizing error for the EcoRI chromosome I optical map was 13.39% for fragments <3 kb, 6.46% for fragments 3–5 kb, and 3.38% for fragments >5 kb. While the EcoRI chromosome II optical map showed a range of average relative sizing errors of 16.34% for fragments <3 kb, 7.81% for fragments 3–5 kb, and 5.71% for fragments >5 kb. A similar assessment of the chromosome I HindIII map showed average relative sizing errors of 6.0% for fragments <5 kb and 1.69% for fragments >5 kb. Likewise, the HindIII map of chromosome II presented the following statistics: 7.74% for fragments <5 kb and 3.97% for fragments >5 kb.
Alignments of Optical Maps to In Silico Maps
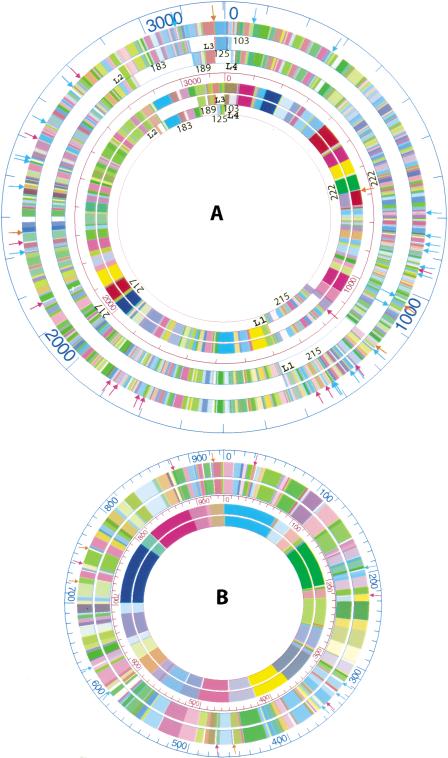
The above analyses deal solely with the optical fragment sizing errors, however, it is informative to assess such errors in the context of map alignments between optical maps and the in silico maps (from available sequence data). Such comparisons facilitate sequence assemblies and validate finished sequence. The alignment of the EcoRI optical map of chromosome I with the corresponding in silico maps of seven sequence contigs (Fig. 3A; contigs 125 and 103 were graphically combined) produced overlaps between the in silico restriction maps that were then carefully compared with the optical map. This comparison revealed errors as strings of in silico fragments at the distal ends of four sequence contigs (Fig. 3A; 103, 125, 183, 215, and 217), which sufficiently differed (>50%) from each other and the optical map to be noted (Table 2). Accordingly, the putatively errant fragments in the contig 103 were eliminated to enable its seamless joining with contig 125 at the 0 position (Fig. 3A). This is represented in Figure 3A as a single, continuous sequence contig.
Figure 3.
The alignments of the HindIII and EcoRI consensus optical maps with corresponding in silico maps. (A) The outer three rings show the alignment of the in silico EcoRI optical map (outermost ring) with the chromosome I in silico maps (7 total; numbered), while the inner three circles show the alignment of the HindIII optical map (the outer one) with the chromosome I in silico HindIII maps. The numbers indicate the sequence contig number. L1, L2, L3, and L4 indicate the four loci for adjacent sequence contigs, and detailed map alignments are shown in Tables 2 and 3. Orange arrows denote false cuts, red arrows denote missing cuts, and light blue arrows denote missing fragments (missing fragments less than 1 kb, or one of the two fragments for missing cuts and false cuts were not shown). (B) The outer two rings show the alignment of the EcoRI in silico map with the corresponding optical map (the outermost ring) of chromosome II, while the inner two rings show the alignment of the HindIII in silico map with the HindIII optical map (outer). Orange arrows denote false cuts, red arrows denote missing cuts, and light blue arrows denote missing fragments (missing fragments less than 1 kb, or one of the two fragments less than 1 kb for missing cuts and false cuts are not shown).
Table 2.
Problematic Alignments Between the Eco RI Chromosome I Optical and In Silico Maps
| Contig 215 Contig 217
|
Contig 217 Contig 183
|
Contig 189 Contig 125
|
Contig 103 Contig 222
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Optical map | Locus 1 | Optical map | Locus 2 | Optical map | Locus 3 | Locus 4 | ||||
| 1.08 | 1.08 | - | 16.18 | 15.86 | - | 3.89 | 3.53 | - | - | - |
| 11.23 | 10.89 | - | 10.07 | 9.71 | - | 10.16 | 10.37 | - | - | - |
| 1.94 | 0.48 | - | 16.83 | 16.78 | - | 3.8 | 3.89 | - | - | - |
| 3.7 | 2.17 | - | 3.45 | 3.63 | - | 30.64 | 27.45 | 3.08 | - | - |
| 2.69 | 1.3 | - | 2.67 | 2.42 | 15.75 | 1.73 | - | 1.71 | 1.14 | - |
| 2.31 | 0.77 | 1.09 | 8.83 | 5.32 | 8.84 | 24.59 | - | 25.17 | 2.01 | - |
| 2.66 | 2.4 | 2.61 | 15.14 | - | 14.98 | 3.06 | - | 2.83 | 1.05 | - |
| 1.56 | 0.85 | 0.57 | - | - | 0.8 | 5.39 | - | 2.32 | 6.3 | - |
| 3.38 | - | 1.92 | 2.3 | - | 1.58 | 2.83 | - | - | 8.38 | - |
| 3.04 | - | 2.01 | 23.21 | - | 22.1 | 9.25 | - | - | - | 1.67 |
| 1.29 | - | 0.79 | 9.39 | - | 8.82 | 5.26 | - | - | - | 5.91 |
| 2.87 | - | 0.94 | 2.41 | - | 2.6 | 6.33 | - | - | - | 1.97 |
| 12.08 | - | 12.61 | 5.1 | - | 5.01 | 6.5 | - | - | - | 5.99 |
| 3.41 | - | 2.43 | 1.21 | - | 1.36 | 10.99 | - | - | - | 10.69 |
| - | 1.41 | 5.17 | - | 5.03 | 14.85 | - | - | - | 14.39 | |
| 10.21 | - | 10.33 | 4.37 | - | 4.22 | 2.39 | - | - | - | 2.51 |
| 8.26 | - | 8.44 | 15.21 | - | 14.88 | 5.35 | - | - | - | 4.93 |
Significant differences are bolded.
To further confirm these errors we then analyzed the same set of in silico maps against the HindIII optical map of chromosome I. If two separate enzyme maps show discrepancies in the same region, then it is probable that the sequence data is problematic. This analysis showed errors in the in silico map at the same positions originally flagged by the EcoRI optical map at contigs 183, 215, and 217 (Table 3). Although sequence contig 103 presented no HindIII restriction sites, its size of 19.12 kb spanned the gap between sequence contig 125 and 222. Given this finding and the EcoRI data allowed us to confidently insert it at the beginning of sequence contig 222. Further comparisons in the problematic regions showed that the EcoRI map alignments revealed additional discrepancies, and these problematic regions are defined as loci 1–4 (L1 to L4) as shown in Tables 2 (EcoRI) and 3 (HindIII).
Table 3.
Problematic Alignments of the Chromosome I Hind III Optical and In Silico Maps
| Contig 215 Contig 217
|
Contig 217 Contig 183
|
Contig 189 Contig 125
|
Contig 103 Contig 222
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Optical map | Locus 1 | Optical map | Locus 2 | Optical map | Locus 3 | Locus 4 | ||||
| 23.07 | 22.77 | - | 35.58 | 36.17 | - | 10.52 | 10.62 | - | - | - |
| 25.8 | 25.7 | - | 21.7 | 21.51 | - | 9.87 | 8.92 | - | - | - |
| 19.13 | 18.94 | - | 22.55 | 22.6 | - | 27.36 | 11.42 | 15.31 | - | - |
| 2.92 | 1.47 | - | 9.81 | 9.44 | - | - | - | 0.54 | - | - |
| 3.01 | 0.41 | - | 34.42 | 35.04 | 12.9 | 7.89 | - | 7.58 | - | - |
| 3.74 | 2.18 | - | 2.61 | 2.87 | 2.11 | 10.34 | - | 10.14 | - | - |
| 2.19 | 2.43 | - | 3.15 | 3.18 | 2.88 | 52.81 | - | 1.53 | 19.12 | 30.33 |
| 3.43 | 1.13 | - | 3.14 | 0.5 | 3.17 | 7.8 | - | - | - | 7.36 |
| 2.01 | 0.64 | - | 8.51 | - | 8.33 | 56.29 | - | - | - | 56.29 |
| 2.97 | 0.4 | 1.49 | 64.21 | - | 66.45 | 9.22 | - | - | - | 9.22 |
| 3.77 | - | 3.29 | 27.41 | - | 27.57 | 4.34 | - | - | - | 4.34 |
| 25.83 | - | 26.15 | 13.24 | - | 12.25 | 28.49 | - | - | - | 28.49 |
| 53.03 | - | 54.34 | 32.14 | - | 31.94 | 59.2 | - | - | - | 59.2 |
Significant differences are bolded.
Figure 3A also shows the high-resolution EcoRI optical map alignments to the same chromosome I sequence data. For this error analysis, the putative sequence misassemblies were removed, and these filtered data revealed only a single false cut in the optical map. However, given a high-resolution map, we predicted a larger number of missing fragments. Here, we identified 40 missing cuts and 68 missing fragments (out of 565 fragments in total). Normally, optical maps do not report restriction fragments <500 bp and attenuated recording of fragments <1 kb (Meng et al. 1995). Accordingly, further analysis showed that four out of five missing fragments for the HindIII optical map, and 42 out of 68 missing fragments for EcoRI optical map of chromosome I were <1 kb.
The alignment of the HindIII optical map with the corresponding chromosome I in silico maps (Fig. 3A) revealed several differences—two false cuts (present in the optical map but not in the in silico map), four missing cuts (present in the in silico map but not in the optical map), and five missing fragments (out of a total of 186 fragments).
A similar analysis was then performed between the optical maps and chromosome II sequence data. Here, the alignment of the EcoRI optical map (Fig. 3B) showed three false cuts, nine missing cuts, and 19 missing fragments (out of a total of 167 fragments), while the HindIII optical map (Fig. 3B) showed no false cuts, no missing cuts, and one missing fragment (out of a total of 40 fragments) in the optical map. The only fragment missing in the HindIII optical map of chromosome II was 0.49 kb, and 10 out of 19 of the missing fragments in the EcoRI optical map were <1 kb.
The Resolution of Locus 4
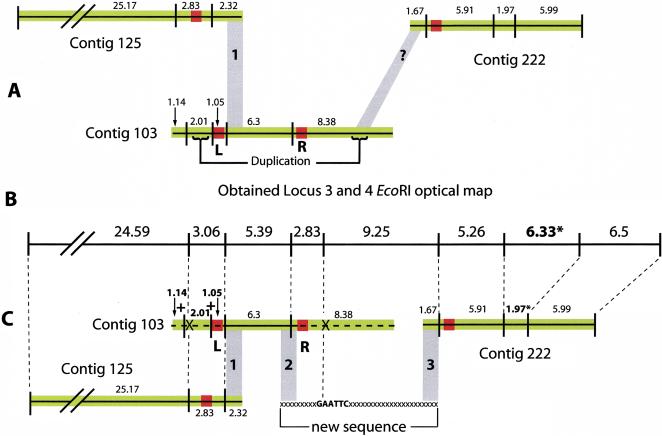
Closure of CI required the joining of three contigs that genome scaffold data suggested were in the order: 125–103–222. The gaps between the contigs were thought to be <3 kb; however, PCR across these gaps had been a consistent failure. The assembler had been unable to put these three contigs together but had suggested that a region at the 3′ end of contig 125 and the 5′ end of contig 222 showed strong, high-quality sequence alignment with contig 103 (Fig. 4A). Although contigs 125 and 222 had regions of poor sequence at their 3′ and 5′ ends respectively, their in silico maps were overall in agreement (Table 2 and Fig. 4B,C) with their optical maps. This suggested that the PCR failure did not reside with the primers designed to the ends of these contigs. Why then had PCR been a failure? Contig 103 had two regions of poor quality sequence flanked by regions encoding in silico EcoRI sites that were present/absent from the optical map, suggesting misassembly of contig 103. After paring back the sequences at the 3′ and 5′ ends of contig 103 to EcoRI sites that existed in both the in silico and the optical map, it assembled cleanly with contig 125. The subsequent acquisition of new sequences between contigs 103 and 222 permitted CI closure. The homology between contig 222 and contig 103 represented by the angled grey area in Figure 4A was a “red herring” that fulfilled our preconceptions of what this region “should look like”. The high resolution of the EcoRI optical map suggested that our assumptions were wrong and indicated that a large gap (∼10 kb) must be reexamined before closure could be achieved. This proved to be the case.
Figure 4.
The melding of contigs 125, 103 and 222 after the resolution of contig 103 misassembly by optical mapping, see also Locus 3 and 4 in Table 2. (A) The three contigs 125, 103 and 222 are shown in the order suggested by sequence scaffold data. Contigs are represented by black horizontal lines. In silico EcoRI sites are represented by vertical lines and between them the predicted in silico EcoRI fragment sizes in kb. Green areas indicate regions of high sequence quality (10–22 -fold coverage), red areas of poor sequence coverage (2–4 fold) and/or poor quality sequence data. Grey block 1 indicates a region of high sequence identity between the 3′ end of contig 125 and a region lying 4.5 kb from the 5′ end of contig 103. The angled grey block indicated by a question mark (?) shows a region of high sequence identity between the 5′ end of contig 222 and a region towards the 3′ end of contig 103. This match later proved to be spurious. The 3′ region of contig 103 is duplicated internally towards its 5′ end. (B) The EcoRI optical map covering part of the region for loci 3 and 4. The sizes in this panel are optical map fragment sizes in kb. (C) The solution to the resolution of this region. EcoRI sites considered equivalent on the optical and in silico maps are joined by fine dotted lines. Optical mapping indicated that poor quality region L had resulted in a misassembly at the 5′ end of contig 103. This was concluded from: 1) the presence in a region of high quality sequence of two additional EcoRI sites (indicated by + signs); and 2) the absence of a restriction site (indicated by X). These were in conflict with the optical map data. Removal of the sequences 5′ of and inclusive of, region L (indicated by a dotted horizontal line) permitted the melding of contig 125 and 103 at grey region 1. The absence of an EcoRI site 3′ of poor quality region R (represented by X) suggested that region R may have given rise to a misassembly of the 3′ end of contig 103. Removal of this region (also indicated by a dotted line) followed by resequencing in this area resulted in the acquisition of a missing EcoRI site and permitted the assembly of the new sequence and joining of contigs 103 and 222 at grey regions 2 and 3. Note that contig 222 has a 1.97 kb EcoRI fragment; however, optical mapping suggests this fragment should be 6.33 kb (these fragments are marked *). To resolve this size discrepancy additional sequencing of this region is currently under way.
DISCUSSION
Two whole-genome shotgun optical maps (EcoRI and HindIII) were constructed for the phototrophic bacterium, R. sphaeroides 2.4.1 for the purposes of validating sequence assemblies and simplification of the gap closure procedures, which were ongoing activities within the parallel genome sequencing project. Both maps were used to check nascent sequence contigs by the alignment of the in silico maps with the optical data, to assess sequence assembly errors, to validate, and to orient sequence contigs on the optical map scaffold. This process also readily characterized gaps between the sequence contigs. Given that our map and sequence comparisons were performed using unfinished sequence, a remarkable degree of correspondence was noted. The availability of two independent optical restriction maps enabled us to confidently identify several discrepancies at the distal ends of several chromosome I sequence contigs, and such aberrancies may often be attributed to sequence assembly errors. As such, assembly efforts involving the garnering of additional reads, PCR analysis or new assembly schemes can be critically focused on these sequence loci.
The estimated genome size of R. sphaeroides 2.4.1 strain is 4.13 Mb (3.19 Mb for chromosome I, and 938 kb for chromosome II) based on the HindIII optical maps, which is very close to the sum of all sequence contigs (4.14 Mb), meaning a mere 0.24% difference or map error. Compared to the genome size estimate of R. sphaeroides 2.4.1 determined by pulsed field gel electrophoresis (PFGE; 3.05 Mb +/–95 kb for chromosome I and 914 kb +/–17 kb for chromosome II; Suwanto and Kaplan 1989b), the genome sizing error was smaller for optical mapping than for PFGE (1.5%–4.25% error). The EcoRI optical maps of chromosome I and II of R. sphaeroides 2.4.1 were also highly congruent with the in silico EcoRI maps constructed from the sequence data. The genome size estimated from the EcoRI maps was 3.99 Mb (3.1 Mb for chromosome I and 890 kb for chromosome II), which is still closer (3.62% error) to the size of genome sequence data than the genome size estimated by PFGE. The differences between the two optical maps stemmed mainly from the absence of small restriction fragments, which are <2 kb in the EcoRI optical map.
The final maps showed some variation in contig depth (the number of molecules aligned to a given genome locus); both the HindIII and EcoRI optical maps have less coverage for chromosome II compared to chromosome I. A possible explanation is that because chromosome I is a much larger circular DNA molecule than chromosome II, it was more frequently sheared to produce linear molecules, which were mapped; circular molecules may have existed in a supercoiled state, which were imaged as bright “balls,”—unsuitable for mapping.
Improvements in DNA mounting, image collection, processing, and map assembly software have made it possible to generate a high resolution map (the average fragment size <15 kb) using our new optical mapping system as shown before (Zhou et al. 2002). However, when the map resolution approaches 5–7 kb, as evidenced by the EcoRI maps presented here, a significant proportion of restriction fragments in the map will be <1 kb, and thus poorly represented in the final consensus map using our current DNA mounting approaches. Although high resolution maps have proven utility for validating nascent sequence contigs and guiding whole-genome shotgun sequence assemblies, further uses for such high-resolution maps are in the realm of comprehensive genotyping and comparative genomics, as genes are highly conserved amongst different strains of the same bacterial species or even closely related species (Moore and Kaplan 1992; Perna et al. 2000). To meet this challenge, our laboratory is developing newer approaches to address the need for ultra-high resolution maps.
In summary, we have presented here two whole-genome shotgun optical maps of R. sphaeroides using the optical mapping system. These maps were used by the concurrent genome sequencing effort for the validation of sequence contigs by the detection of misassemblies. Such common errors, inherent to modern whole-genome sequencing approaches are rapidly and confidently discerned when shotgun sequencing is combined with optical mapping.
Acknowledgments
This work was supported by a grant from the DOE DE-FC02–01ER63175 to D.C.S. and NIH 15590 and DOE DE-FG02–01ER63232 grants to S.K. Special thanks to Thomas Anantharaman for algorithmic efforts for map assembly.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1128803.
References
- Anantharaman, T.S., Mishra, B., and Schwartz, D.C. 1997. Genomics via optical mapping 2. Ordered restriction maps. J. Comput. Biol. 4: 91–118. [DOI] [PubMed] [Google Scholar]
- ____. 1998. Genomics via optical mapping III: Contiging genomic DNA and variations. In Courant technical report, p. 760. Courant Institute, New York University, NY.
- ____. 1999. Genomics via optical mapping III: Contiging genomic DNA and variations. The Seventh International Conference on Intelligent Systems for Molecular Biology 7: 18–27. [PubMed] [Google Scholar]
- Aston, C., Hiort, C., and Schwartz, D.C. 1999a. Optical mapping: An approach for fine mapping. Meth. Enzymol. 303: 55–73. [DOI] [PubMed] [Google Scholar]
- Aston, C., Mishra, B., and Schwartz, D.C. 1999b. Optical mapping and its potential for large-scale sequencing projects. Trends Biotech. 17: 297–302. [DOI] [PubMed] [Google Scholar]
- Barber, R.D. and Donohue, T.J. 1998. Function of a glutothione-dependent formaldehyde dehydrogenase in Rhodobacter sphaeroides, formaldehyde oxidation and assimilation. Biochemistry 37: 530–537. [DOI] [PubMed] [Google Scholar]
- Cai, W., Jing, J., Irvin, B., Ohler, L., Rose, E., Shizuya, H., Kim, U., Simon, M., Anantharaman, T., Mishra, B., et al. 1998. High-resolution restriction maps of bacterial artificial chromosomes constructed by optical mapping. Proc. Natl. Acad. Sci. 95: 3390–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary, M., Mackenzie, C., Nereng, K.S., Sodergren, E., Weinstock, G.M., and Kaplan, S. 1994. Multiple chromosomes in bacteria: Structure and function of chromosome II of Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 176: 7694–7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary, M., Mackenzie, C., Nereng, K.S., Sodergren, E., Weinstock, G.M., and Kaplan, S. 1997. Low-resolution sequencing of Rhodobacter sphaeroides 2.4.1T: Chromosome II is a true chromosome. Microbiol. 143: 3085–3099. [DOI] [PubMed] [Google Scholar]
- Clayton, R.K. and Sistrom, W.R. 1978. The photosynthetic bacteria. Plenum Press, New York.
- Fleischmann, R.D., Adams, M.D., White, O., Clayton, R.A., Kirkness, E.F., Kerlavage, A.R., Bult, C.J., Tomb, J.F., Dougherty, B.A., Merrick, J.M., et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269: 496–512. [DOI] [PubMed] [Google Scholar]
- Gordon, D., Albanian, C., and Green, P. 1998. Consed: a graphical tool for sequence finishing. Genome Research. 8: 195–202. [DOI] [PubMed] [Google Scholar]
- Jing, J., Lai, Z., Aston, C., Lin, J., Carucci, D.J., Gardner, M.J., Mishra, B., Anantharaman, T., Tettelin, H., Cummings, L.M., et al. 1999. Optical mapping of Plasmodium falciparum chromosome 2. Genome Res. 9: 175–181. [PMC free article] [PubMed] [Google Scholar]
- Lai, Z., Jing, J., Aston, C., Clarke, V., Apodaca, J., Dimalanta, E.T., Carucci, D.J., Gardner, M.J., Mishra, B., Anatharaman, T.S., et al. 1999. A shotgun optical map of the entire Plasmodium falciparum genome. Nat. Genet. 23: 309–313. [DOI] [PubMed] [Google Scholar]
- Lim, A., Dimalanta, E.T., Potamousis, K.D., Yen, G., Apodoca, J., Tao, C., Lin, J., Qi, R., Skiadas, J., Ramanathan, A., et al. 2001. Shotgun optical maps of the whole Escherichia coli O157:H7 Genome. Genome Res. 11: 1584–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J., Qi, R., Aston, C., Jing, J., Anatharaman, T.S., Mishra, B., White, O., Daly, M.J., Minton, K.W., Venter, J.C., et al. 1999. Whole-genome shotgun optical mapping of Deinococcus radiodurans. Science 285: 1558–1562. [DOI] [PubMed] [Google Scholar]
- Lueking, D.R., Fraley, R.T., and Kaplan, S. 1978. Intracytoplasmic membrane synthesis in synchronous cell populations of Rhodopseudomonas sphaeroides. J. Biol. Chem. 253: 451–457. [PubMed] [Google Scholar]
- Mackenzie, C., Choudhary, M., Larimer, F.W., Predki, P.F., Stilwagen, S., Armitage, J.P., Barber, R.D., Donohue, T.J., Hosler, J.P., Newman, J.E., et al. 2001. The home stretch, a first analysis of the nearly completed genome of Rhodobacter sphaeroides 2.4.1. Photosynthesis Research 70: 19–41. [DOI] [PubMed] [Google Scholar]
- Marra, M.A., Kucaba, T.A., Dietrich, N.L., Green, E.D., Brownstein, B., Wilson, R.K., McDonald, K.M., Hillier, L.W., McPherson, J.D., and Waterston, R.H. 1997. High throughput fingerprint analysis of large-insert clones. Genome Res. 7: 1072–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, X., Benson, K., Chada, K., Huff, J.E., and Schwartz, D.C. 1995. Optical mapping of λ bacteriophage clones using restriction endonucleases. Nat. Genet. 9: 432–438. [DOI] [PubMed] [Google Scholar]
- Moore, M.D. and Kaplan, S. 1992. Identification of intrinsic high-level resistance to rare-earth oxides and oxyanions in members of the class Proteobacteria: Characterization of tellurite, selenite and rhodium sesquioxide reduction in Rhodobacter sphaeroides. J. Bacteriol. 174: 1505–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouncey, N.J., Choudhary, M., and Kaplan, S. 1997. Characterization of genes encoding dimethylsulfoxide reductase of Rhodobacter sphaeroides 2.4.1T: An essential metabolic gene function encoded on chromosome II. J. Bacteriol. 179: 7617–7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidle, E. and Kaplan, S. 1993. Expression of the Rhodobacter sphaeroides 2.4.1 hemA and hemT gebes encoding two aminolevulinic acid synthase isozymes. J. Bacteriol. 175: 2292–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna, N.T., Plunkett III, G., Burland, V., Mau, B., Glasner, J.D., Rose, D.J., Mayhew, G.F., Evans, P.S., Gregor, J., Kirpatrick, H.A., et al. 2000. Genome sequence of entrohemorrhagic Escherichia coli O157: H7. Nature 409: 529–533. [DOI] [PubMed] [Google Scholar]
- Schwartz, D.C. and Cantor, C.R. 1984. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell 37: 67–75. [DOI] [PubMed] [Google Scholar]
- Sensen, C.W. 1999. Sequencing microbial genomes. In Organization of the prokaryotic genome (ed. R.L. Charlebois), pp. 1–9. ASM Press, Washington, D.C.
- Soderlund, C., Longden, I., and Mott, R. 1997. FPC: A system for building contigs from restriction fingerprinted clones. Comput. Appl. Biosci. 13: 523–535. [DOI] [PubMed] [Google Scholar]
- Suwanto, A. and Kaplan, S. 1989a. Physical and genetic mapping of the Rhodobacter sphaeroides 2.4.1 genome: Presence of two unique circular chromosomes. J. Bacteriol. 171: 5850–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1989b. Physical and genetic mapping of the Rhodobacter sphaeroides 2.4.1 genome: Genome size, fragment identification, and gene localization. J. Bacteriol. 171: 5840–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1992. Chromosome transfer in Rhodobacter sphaeroides: Hfr formation and genetic evidence for two unique circular chromosomes. J. Bacteriol. 174: 1135–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, D., Oyaizu, Y., Oyaizu, H., Olsen, G.J., and Woese, C.R. 1985. Mitochondrial origins. Proc. Nat. Acad. Sci. 82: 4443–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeliseev, A.A. and Kaplan, S. 1995. A sensory transducer homologous to the mammalian peripheral-type benzodiazepine receptor regulates photosynthetic membrane complex formation in Rhodobacter sphaeroides 2.4.1. J. Biol. Chem. 270: 21167–21175. [DOI] [PubMed] [Google Scholar]
- Zhou, S., Deng, W., Anatharaman, T.S., Lim, A., Dimalanta, E.T., Wang, J., Wu, T., Tao, C., Creighton, R., Kile, A., et al. 2002. A whole-genome shotgun optical map of Yersinia pestis strain KIM. Appl. Environ. Microbiol. 68: 6321–6331. [DOI] [PMC free article] [PubMed] [Google Scholar]