Abstract
RNA interference is a powerful tool for studying gene function and for drug target discovery in diverse organisms and cell types. In mammalian systems, small interfering RNAs (siRNAs), or DNA plasmids expressing these siRNAs, have been used to down-modulate gene expression. However, inefficient transfection protocols, in particular, for primary cell types, have hampered the use of these tools in disease-relevant cellular assays. To be able to use this technology for genome-wide function screening, a more robust transduction protocol, resulting in a longer duration of the knock-down effect, is required. Here, we describe the validation of adenoviral vectors that express hairpin RNAs that are further processed to siRNAs. Infection of cell lines, or primary human cells, with these viruses leads to an efficient, sequence-specific, and prolonged reduction of the corresponding target mRNA, resulting in a reduction of the encoded protein level in the cell. For knock-down of one of the targets, GαS, we have measured inhibition of ligand-dependant, G-protein-coupled signaling. It is expected that this technology will prove to be of great value in target validation and target discovery efforts.
RNA interference (RNAi) is a sequence-specific posttranscriptional gene-silencing process initiated by double-stranded RNA (dsRNA) in animals and plants (for reviews, see Sharp 1999; Tuschl 2001; Hannon 2002; McManus and Sharp 2002). The dsRNA is rapidly processed by the RNase III-type enzyme Dicer (Bernstein et al. 2001; Ketting et al. 2001) into shorter (21-23 nt) RNA duplexes with distinct 2-nt overhangs at their 3′ termini called small interfering RNAs (siRNAs; Elbashir et al. 2001a). These siRNAs assemble into a partially defined complex, called the RISC (RNA-induced silencing complex). The RISC is responsible for the sequence-specific degradation of target RNAs that contain homologous sequences. The resulting down-modulation of the encoded proteins can subsequently lead to induction of a specific phenotype. Since its discovery in Caenorhabditis elegans (Fire et al. 1998), RNAi has been used as a genetic tool to study gene function in various systems, initially using longer dsRNA fragments, particularly in C. elegans and Drosophila. In C. elegans, RNAi has been used to identify gene functions in screens that cover the entire genome (Ashrafi et al. 2003; Kamath et al. 2003). The discovery of siRNAs allowed the use of RNAi in mammalian cells (Elbashir et al. 2001b).
Recently, siRNA expression systems were described based on the intracellular production of siRNA molecules using various promoter systems such as those derived from U6 snRNA, H1 RNA, and CMV (Brummelkamp et al. 2002a; Lee et al. 2002; McManus et al. 2002; Miyagishi and Taira 2002; Paddison et al. 2002; Paul et al. 2002; Sui et al. 2002; Yu et al. 2002; Zeng et al. 2002). These systems are based on the expression of siRNAs either as two separate strands or as a single short hairpin RNA. Plasmid transfection strategies as well as permanent expression by the generation of stable cell lines were used in these studies. However, the application of siRNA expression systems in primary cell types, which are the cell types of choice for disease-related functional studies, is difficult because of inefficient gene transfer methods. In addition, functional cellular effects of mRNA knockdown often require a sufficiently long period of target mRNA reduction to repress protein expression, which cannot be achieved by plasmid transfections. Adenoviral vectors offer the advantage of a broad tropism and high gene transfer efficiency, including various primary cell types. Furthermore, they allow long-term gene expression (Michiels et al. 2002) and therefore are favored over DNA or RNA transfections.
Here, we show that adenoviral expression of small hairpin RNAs mediated by the U6 promoter results in reduction of target mRNA levels in a sequence-specific manner in primary human cells. In combination with capsid-modified vectors, gene expression can be specifically suppressed in a wide array of disease-relevant human cells, for target discovery using arrayed libraries, as well as target validation.
RESULTS
Our aim was to develop a robust mammalian knock-down system in an adenoviral vector that would allow the study of gene function in disease-relevant cell types on a genome-wide scale. Such a loss-of-function gene collection would provide an ideal complement to our arrayed adenoviral gain-of-function libraries (Michiels et al. 2002). We started with plasmid-based expression systems targeting a luciferase reporter to measure gene suppression. Different cassettes for expression of small hairpin RNAs were analyzed. One was based on a genomic fragment (338 bp) of the human Let-7 microRNA region of Chromosome 22. This expression cassette showed sequence-specific repression of the luciferase reporter containing the target sequence and could be reprogrammed to knock down a reporter with a different target sequence (data not shown). A systematic analysis to obtain shorter functional loops was based on the Let-7 RNA loop sequence as well as loops derived from other microRNAs (Lagos-Quintana et al. 2001). This resulted in a functional loop of 12 nt derived from the Let-7 microRNA (see Fig. 1B) that was used henceforward.
Figure 1.
Adenoviruses efficiently deliver and express siRNAs with multiple target sequences. (A) Schematic representation of the U6-promoter-based siRNA expression cassette integrated in the adenoviral vector. The starting G and the stretch of Ts present for efficient transcription purposes are indicated. The replication-competent adenoviral molecule based on serotype 5 (Ad5), lacking the E1 and the E2A regions, is generated in Per.C6/E2A packaging cells by homologous recombination. Not drawn to scale. (B) Predicted secondary structures of hairpin RNAs derived from the adenoviral U6-driven expression constructs. The structure with GNAS (guanine nucleotide-binding protein α stimulating activity polypeptide 1) target sequence CGATGTGACTGCCATCATC (corresponding to the lower strand in the hairpin structure) is given as example, as well as that of the mutant version GNASm with a point mutation at the central position (lower case) of the target sequence (CGATGTGACaGCCATCATC). The variable sequence in the hairpin RNA that is dependent on the target sequence is underlined. (C) Expression analysis of small RNAs by Northern blotting using denaturing 15% polyacrylamide gels. Samples were isolated 3 d postinfection from U2OS cells that were infected at m.o.i. 3000 with the various adenoviral constructs as indicated. The blots were hybridized with labeled oligonucleotides recognizing either the antisense strand of the target sequence (upper panel, sense probe) or sense strand of the target sequence (lower panel, antisense probe). (D) Real-time reverse transcriptase (RT)-PCR expression analyses measuring the endogenous mRNA levels for GNAS, IKBKB (IKKβ, inhibitor of κ light polypeptide gene enhancer in B-cells, kinase β), or M6PR (mannose-6-phosphate receptor) of samples derived from U2OS cells 3 d after infection at m.o.i. 3000 with the indicated siRNA expression viruses. Values are plotted relative to samples derived from cells infected with control virus Ad-siRNA-GL2 and are normalized to GAPD (GAPDH, glyceraldehyde-3-phosphate dehydrogenase) mRNA as an internal reference. Values are obtained from three independent infection experiments; error bars show standard deviation. (E) Comparison of Ad-siRNA-GNAS versus Ad-siRNA-GNASm vectors by real-time RT-PCR analyses on samples derived from U2OS cells infected at m.o.i. 3000 6 d postinfection. See Figure 1B for sequences. Shown are the representative results from two independent experiments. Error bars indicate high-low values.
We designed an siRNA expression cassette based on the human U6 promoter, which is a strong, ubiquitously active promoter that lacks essential promoter elements within the transcribed region (Medina and Joshi 1999). The U6 promoter-based expression cassette was cloned in an adenoviral adapter plasmid at the position of the E1 region. The adenoviral vectors were based on adenovirus serotype 5 with deleted E1 and E2A regions (Fig. 1A; Michiels et al. 2002). Predicted secondary structures of expressed hairpin RNAs are exemplified in Figure 1B.
Adenoviruses were generated containing U6-promoter-based siRNA expression cassettes with sequences directed against the cellular target mRNAs GNAS (GαS, guanine nucleotide-binding protein α-stimulating activity polypeptide 1), IKBKB (IKKβ, inhibitor of κ light polypeptide gene enhancer in B-cells, kinase β), and M6PR (mannose-6-phosphate receptor). The expression levels of the siRNAs of samples derived from U2OS cells infected with the appropriate adenoviruses at a multiplicity of infection (m.o.i.) of 3000 as based on viral particles (Ma et al. 2001) were determined by polyacrylamide gel Northern blot analysis 3 d postinfection. The blots were probed with labeled oligonucleotides recognizing the sense or antisense sequences as shown in Figure 1C. The transcripts were processed into smaller RNA species of ∼21 nt with either the sense or antisense sequence. Longer precursor transcripts could be detected, albeit very weakly. Higher expression levels of these RNA species were seen when increased m.o.i.s were used (data not shown). Although some variations were observed, siRNA expression levels and hairpin RNA processing efficiency into mature siRNA strands were not dramatically affected by the selected target sequences.
Next, we determined whether infection with the adenoviral knock-down constructs resulted in reduction of the target mRNA levels. A quantitative mRNA expression analysis using real-time PCR was performed on samples infected with the adenoviruses expressing the siRNAs against GNAS, IKBKB, and M6PR. The data were normalized for the mRNA expression levels of GAPD (GAPDH, glyceraldehyde-3-phosphate dehydrogenase). Knockdown of the mRNA was reproducibly obtained for all constructs tested (Fig. 1D). The Ad-siRNA-GNAS virus showed the strongest effect. The reduction of the GNAS mRNA was sequence-specific because introduction of a single point mutation at the central position of the target sequence (in Ad-siRNA-GNASm) abolished the knock-down activity (Fig. 1E). The data were plotted relative to values obtained from cells infected with a control adenoviral siRNA expression construct targeting an unrelated sequence (here, Ad-siRNA-GL2, targeting luciferase GL2). Moreover, in a luciferase reporter assay, Ad-siRNA-GL2 showed efficient and sequence-specific repression of the luciferase GL2 reporter activity, but not that of the GL3 reporter, which differs only 3 nt from GL2 in the target sequence (data not shown).
To obtain optimal knock-down activity, different sequences derived from the same target mRNA were tested, in this example for IKBKB (IKKβ) mRNA. The selection of sequences in the target mRNA was basically done using criteria described by the Tuschl lab (Elbashir et al. 2002), such as a preferred GC content ∼50%, >50-100 nt downstream of the start codon, and unique for target as verified by alignment to the expressed sequence databases. siRNA expression of all three IKBKB adenoviral constructs (cf. also Fig. 1C) resulted in reduction of the IKBKB mRNA to comparable levels (Fig. 1D). More extensive analyses with various target mRNAs led us to conclude that different target sequences in the same mRNA can, indeed, result in different degrees of knock-down activity (see also below). Therefore, the use of several independent constructs for a single target mRNA will increase the probability of obtaining a construct that results in an efficient knock-down. However, when different adenoviral constructs for the same target mRNA, IKBKB, were mixed, we did not obtain a higher knock-down efficiency (data not shown).
To study how the knock-down activity could be further improved, we started to monitor the kinetics of the knock-down effect of the adenoviral vector. The expressed siRNAs accumulate during the first days after infection of U2OS cells, as exemplified by adenoviruses expressing GNAS siRNAs (Ad-siRNA-GNAS) in Figure 2A. Their levels remained constant, at least up to 10 ds postinfection even under conditions in which the virus was removed from the cells after 3 d. Therefore, we conclude that the adenoviral vectors are able to express processed hairpin RNAs for at least 10 d. The same samples were analyzed for GNAS mRNA expression levels by quantitative real-time PCR analysis (Fig. 2B). Infection with Ad-siRNA-GNAS, but not with control virus, resulted in efficient reduction of GNAS mRNA that started already at day 1 and reached knock-down levels of >90% from day 3 on, up to day 10. The levels of GNAS mRNA knock-down can also be influenced by using different m.o.i.s for infection of U2OS cells as shown in Figure 2C for m.o.i.s 300, 1000, and 3000. Other adenoviral constructs targeting different mRNAs have been analyzed in a similar manner, and, as expected, in general the knockdown effect is enhanced by raising the m.o.i. because of an increase in the copy number per cell (data not shown). In summary, these data show that knock-down levels can be manipulated by varying the m.o.i. applied, the expression time of the adenoviral siRNA constructs, and the sequence in the target mRNA that was chosen.
Figure 2.
Adenoviral siRNA expression reduces target mRNA levels in a time- and m.o.i.-dependent manner. (A) Northern blot analysis of adenoviral expression of siRNAs against GNAS (Ad-siRNA-GNAS) in U2OS cells (m.o.i. 3000) in a time course from day 1 to 10 postinfection (p.i.). The viruses were removed from the cells at day 3, and medium was refreshed at days 3, 6, and 8. Cells reached confluency at day 4. From day 8, cultures still appeared viable, although some cells detached. In spite of these observations, the RNA yield as well as the absolute Ct values of the GAPD mRNA did not show large variations between these samples, indicating no significant adverse effects during this long experiment. As control for equal loading of the gel, the results of rehybridization with a probe against endogenous U6 snRNA is given at the bottom. (B) Real-time RT-PCR expression analyses for GNAS mRNA of the same samples as used in panel A. Data were plotted relative to samples derived from uninfected cells. The results of the infections of Ad-siRNA-GNAS (solid bars) as well as for control virus (open bars) are given. As control virus, Ad-siRNA-GL2 is used for the data point of day 3, and Ad-siRNA-empty (a construct lacking a hairpin sequence downstream of the U6 promoter) for the other data points. The GNAS mRNA expression of each individual sample was normalized for GAPD. (C) Real-time PCR expression analysis for GNAS mRNA of samples derived from U2OS cells infected with Ad-siRNA-GNAS at m.o.i. 300, 1000, or 3000 harvested at 1, 2, 3, or 6 d postinfection. The values are plotted relative to samples derived from cells infected with control virus Ad-siRNAGL2 harvested at the same time points. Shown are representative results from independent experiments.
The use of siRNA tools in primary cells has been hampered by inefficient transfection protocols. The adenoviral vectors we use have a broad tropism and very efficiently infect many different primary cell types. To confirm that our adenoviral siRNA expression system is, indeed, of use in primary cell types, mRNA knock-down efficiency was determined in primary keratinocytes and primary synoviocytes, two cell types that are notoriously difficult to transfect. An efficient knock-down was obtained in both keratinocytes and synoviocytes, as shown in Figure 3, A and B, respectively. Efficient transduction in keratinocytes was obtained using an adenoviral vector with a modified capsid.
Figure 3.
Adenoviral mediated knock-down in primary cell types. (A) Knock-down activity was measured for CHUK (IKKα, conserved helix-loop-helix ubiquitous kinase), IKBKB and GNAS at 3 and 6 d postinfection of primary keratinocytes (NHEK) at m.o.i. 500 using adenoviral siRNA expression constructs with a modified capsid. Analyses were performed by real-time RT-PCR as described. (B) Adenoviral knock-down activity was measured 6 d after infecting primary synoviocytes at m.o.i. 7500 for IKBKB and MMP2 (matrix metalloproteinase) using two constructs for each target. Analyses were performed as described. (C) Knock-down activity of siRNA-expressing adenoviruses (m.o.i. 1000) containing 13 different target sequences was analyzed in primary HUVEC cells (human umbilical vein endothelial cells). Relative mRNA expression levels were determined by real-time PCR analyses, normalized to internal GAPD values, and plotted relative to samples infected with Ad-siRNA-GL2 control viruses. Samples were obtained from HUVEC cells at two time points postinfection (3 or 6 d). For all eight mRNA targets, gene-specific real-time analysis was performed: PSEN1 (presenilin 1); CTSK (cathepsin K); ADAM10 (a disintegrin and metalloproteinase domain 10); CHUK; M6PR; BACE (β-site APP-cleaving enzyme); IKBKB; and GNAS.
Subsequently, to test the feasibility to do a whole-genome approach in primary cells, the analysis was extended to include more target mRNAs. Sequences were selected against various target mRNAs, and adenoviral constructs were generated. For 8 different target mRNAs (13 constructs in total), the knock-down efficiency was determined at the mRNA level by real-time PCR. For this, RNA was isolated from primary HUVEC cells (human umbilical vein endothelial cells) 3 and 6 d postinfection with adenoviral knock-down constructs (Fig. 3C). These data show that >50% knock-down efficiency can be obtained for all genes tested. Some constructs like GNAS-60 were very potent, whereas others, like PSEN1-5 that targets presenilin 1, did not reach such high knock-down efficiencies. Analysis of this set of target sequences by ranking them based on the efficiency of observed knock-down did not result in obvious rules to predict the efficacy of knock-down based on the characteristics of the target sequences. In this set, 9 out of 13 constructs yielded >75% reduction in the target mRNA levels. Thus, the level of mRNA knockdown varies depending on the target sequence of constructs as well as the target mRNA itself. Again, these data clearly demonstrate that adenoviral vectors are capable of inducing a specific knock-down effect for various targets, allowing their use in larger screens based on disease-relevant assays in primary cells.
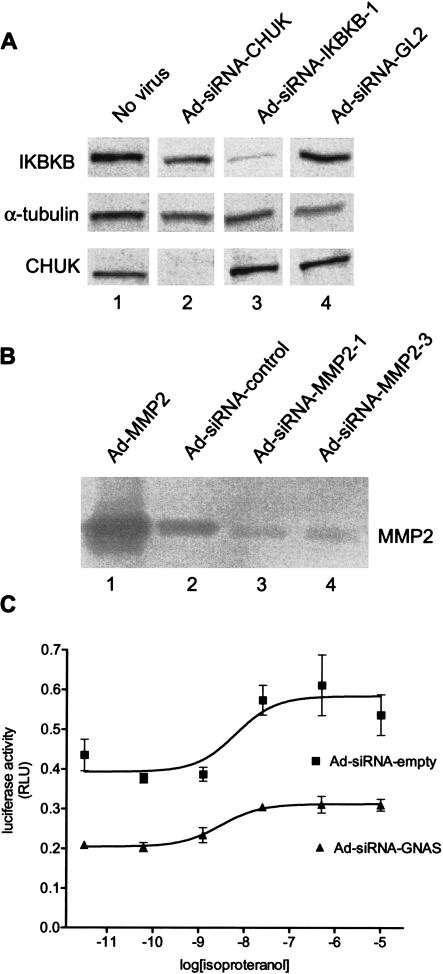
Besides the analysis at the RNA level, we studied how mRNA knock-down affects protein expression and protein activity. U2OS cells were infected with adenoviruses expressing siRNA to knock down CHUK (IKKα, conserved helix-loop-helix ubiquitous kinase) or IKBKB (IKKβ; Fig. 4A). As controls, we used uninfected cells (no virus), or Ad-siRNA-GL2. Western blot analysis was performed to determine protein levels of CHUK and IKBKB 6 d postinfection. Specific reduction of CHUK and IKBKB was obtained at the protein level with Ad-siRNA-CHUK and Ad-siRNAIKBKB-1, respectively, whereas the control virus Ad-siRNA-GL2 did not influence the expression levels of these genes. In another experiment, the constitutively expressed matrix metalloproteinase MMP2 (gelatinase A) was targeted for knock-down with two different adenoviral constructs (Ad-siRNA-MMP2-1 or Ad-siRNAMMP2-3) in primary human synoviocytes. Both constructs resulted in very efficient reduction (>95% knock-down) of MMP2 mRNA determined by real-time PCR analysis (Fig. 3B), as well as the MMP2 protein activity as determined by its gelatin-degrading activity using zymogram gels (Fig. 4B). Adenoviral overexpression of the MMP2 protein (Ad-MMP2) served to demonstrate the identity of the band visualized in this assay.
Figure 4.
Adenoviral siRNA expression leads to reduction of the target protein. (A) Protein expression levels for CHUK (IKKα), IKBKB (IKKβ), and α-tubulin were detected by Western analysis of samples derived from U2OS cells 6 d after infection with Ad-siRNA-CHUK, Ad-siRNA-IKBKB-1, control virus Ad-siRNA-GL2, or from uninfected cells (no virus). The protein levels of CHUK and IKBKB were specifically reduced by the corresponding adenoviral knock-down construct. (B) The effect of adenoviral knock-down was determined at the level of protein activity. Gelatin-degrading activity of MMP2 was measured using 10% gelatin-zymogram gels. Supernatants of primary synoviocytes were analyzed 6 d after infection at m.o.i. 7500 with Ad-siRNA-MMP2-1 and Ad-siRNA-MMP2-3, and compared with Ad-siRNA-control, a virus containing a target sequence against M6PR. For reference, an adenoviral cDNA expression construct encoding the MMP2 protein (Ad-MMP2) was used and demonstrated the identity of the MMP2 band. The inverted image of the zymogram is presented. (C) Knock-down of GαS (encoded by GNAS) by Ad-siRNAGNAS results in reduction of β2-adrenergic receptor signaling. HEK293 cells were infected (m.o.i. 150) with Ad-siRNA-GNAS or control virus Ad-siRNA-empty for 3 d. Subsequently, the medium was refreshed and the cells were reinfected (m.o.i. 300) with a reporter adenovirus encoding a luciferase gene under control of the cAMP-responsive element (CRE)-dependent promoter and incubated for 2 extra days. The endogenously expressed β2-adrenergic receptor is stimulated with increasing concentrations of the agonist isoproteranol. Signaling via the active GαS subunit results in increased cAMP levels and subsequently activates the CRE-dependent promoter. The luciferase activity (indicated on the Y-axis as relative light units; RLU) was measured 6 h after stimulation with the agonist. The isoproteranol concentration (in moles per liter) is plotted as logarithmic values on the X-axis. pEC50 values remain largely unaffected (values between 8.2 and 8.5). Error bars indicate the standard deviation values based on 3 data points.
Furthermore, the central function of GαS protein encoded by GNAS in the cAMP-dependent signal transduction pathway of the β2-adrenergic receptor was challenged. A reporter virus with the luciferase gene under control of a cAMP-dependent promoter was used and stimulated by increasing concentrations of isoproteranol, an agonist for the β2-adrenergic G-protein-coupled receptor expressed in HEK293. The results show a significant reduction of the promoter activity by the Ad-siRNA-GNAS virus compared with the control Ad-siRNA-empty virus (Fig. 4C). Reduction was also observed when different controls were used; for example, cells infected with only the luciferase reporter virus without an siRNA-expressing virus or with a virus expressing an siRNA with unrelated target sequence (data not shown). The remaining activity of the luciferase signal in the samples derived from Ad-siRNA-GNAS-infected cells are consistent with the incomplete knock-down that was seen at the mRNA level in this experiment (∼50%; data not shown). The reduction in luciferase activity follows closely the reduction in mRNA levels. pEC50 values were unaffected by the knockdown of GNAS. In this example, the knockdown of GNAS mRNA results in a significant reduction of the activity of a cellular pathway in which the GNAS-encoded protein GαS plays a crucial role.
Altogether, the data presented in this report clearly show that adenoviral vectors can efficiently introduce and express siRNAs to levels that result in a specific knock-down of the targeted transcript and the encoded protein in human cells, including primary human cells. In addition, we have shown that adenoviral delivery of a specific siRNA can be used to modulate a signal transduction pathway.
DISCUSSION
RNA interference has proven to be of great value as a genetic tool to study gene functions. In C. elegans, the entire genome has been screened, and biological functions have been assigned to genes that were only predicted with bioinformatics tools (Ashrafi et al. 2003; Kamath et al. 2003). In mammalian cells, the use of RNAi to study gene function was made available to scientists after the discovery that short siRNAs could introduce specific gene silencing (Elbashir et al. 2001b) and is widely used since (for review, see McManus and Sharp 2002). Several groups have reported the use of plasmids that drive expression of siRNAs by different promoter systems (Brummelkamp et al. 2002a; Lee et al. 2002; McManus et al. 2002; Miyagishi and Taira 2002; Paddison et al. 2002; Paul et al. 2002; Sui et al. 2002; Yu et al. 2002; Zeng et al. 2002). These could be used in drug target validation or discovery screens, but their application in primary cells, which are most relevant for disease-related assays, is very limited because of their poor transfection efficiencies. Virus-based knock-down systems have also been described, but they have only been used to target single genes (Brummelkamp et al. 2002b; Devroe and Silver 2002) or merely reporter genes (Xia et al. 2002). Therefore, their use for genome-wide discovery screens has not been established yet.
The U6-promoter-based expression cassettes in our adenoviral knock-down system give robust knock-down in cell lines as well as in primary cell types. The expressed hairpin RNA is efficiently processed into smaller RNA species of ∼21 nt containing either the sense or antisense sequence. In our hands, knock-down can be obtained using constructs with the antisense sequence positioned either upstream or downstream of the loop sequence (data not shown). In other expression systems, the orientation with the antisense sequence downstream of the loop was frequently used and in some cases even preferred (Paul et al. 2002). These different observations might be influenced by the sequence of the loop as observed in eight different microRNA-based loop sequences we have tested. That not all loop sequences are compatible with proper processing of the hairpin is in line with reports from others (Brummelkamp et al. 2002a).
The short, transient effects of siRNA- or plasmid-transfection approaches make their use in cellular assays that require longer incubation periods difficult. Moreover, transfection efficiencies in primary cell cultures are mostly poor and in general not robust enough for large target discovery programs. The adenoviral siRNA expression system described here overcomes all these limitations. It allows the use of cell lines and primary cell types, and gives knockdown effects over longer periods of up to 14 d postinfection (data not shown). Prolonged siRNA expression and activity can be obligatory for knock-down of stable protein targets or to study phenotypes that take significant time to develop. Examples of such disease-relevant assays are in vitro capillary formation, and osteoblast and adipocyte differentiation. We have shown that the adenoviral knock-down system works in the three primary cell types HUVEC, keratinocytes, and synoviocytes. In addition, we have successfully used PPARγ knock-down to modulate the differentiation to mature adipocytes (data not shown). We are presently expanding the number of primary cell cultures to cells relevant to other disease areas including mast cells, Th1/Th2 cells, and mesenchymal precursor cells. An actual siRNA-mediated phenotypic change is determined by factors other than only the efficiency of the mRNA knock-down, for example, the stability of the encoded protein, functional redundancy of proteins in the cell, or the regulatory role the protein plays in a pathway. The results of ongoing target discovery screens with a larger number of viruses as well as the use in target validation will give us more insight into the applicability of this technology.
Design of target sequences is done with the aid of generally accepted criteria (Elbashir et al. 2002). Additionally, we try to avoid positions where single nucleotide polymorphisms (SNPs) are known. Two different strategies can be used for alternatively spliced transcripts: Target sequences can be designed that are directed against a region shared by several transcript variants, or transcript variant-specific target sequences can be selected directed at the differences between the variants. We prefer the first strategy to limit the amount of target sequences in our knock-down library, but in some cases the selection of transcript-specific sequences is done to cover all the variants. We decided to design multiple constructs per target mRNA to ensure having at least one construct that results in a >75% knock-down efficiency. We are presently using the adenoviral vector for the construction of a library that covers all genes that belong to druggable gene classes (Hopkins and Groom 2002). This collection will allow screening of a nearly complete druggable genome in disease-relevant assays based on cell lines or primary human cell cultures, and, because of the knock-down format, mimics the use of antagonistic compounds. Therefore, a complete disease area can be screened for only those genes that are of interest to the pharmaceutical industry. Because the collection is arrayed, a wide range of assay formats can be used including molecular, biochemical, morphological, and cell proliferation assays such as those described (Michiels et al. 2002).
Besides a whole druggable genome approach, we are also exploring the technology for in vivo target validation as well as for the construction of libraries from transcripts that are up- or down-regulated in a particular disease, or that are derived from candidate disease genes as determined by genetic mapping. These collections of genes, albeit potentially biased from their mere selection, allow small disease-relevant knock-down collections that can be built quickly.
METHODS
Cloning
The genomic human U6 gene (M14486), spanning the region from nucleotide -265 upstream of the transcription start site until nucleotide +198 downstream of the transcription start site, was obtained by a PCR-based cloning strategy. The primers used were: hU6 (-265)-F1-XbaI, 5′-GcacgTTCTAGAAGGTCGGGCAG GAAGAGGGCCT-3′; hU6 (+198)-R1-HindIII, 5′-ccgtgcAAGCT TTGGTAAACCGTGCACCGGCGTA-3′.
The PCR fragments were digested with the enzymes XbaI and HindIII and cloned into AvrII-HindIII-digested pIPspAdapt6-deltaPolyA, thereby replacing the CMV promoter sequence. pIPspAdapt6-deltaPolyA was constructed from pIPspAdapt6 (Michiels et al. 2002) by digestion with XbaI, and excision of the 142-bp fragment containing the poly(A) signal. The U6 snRNA sequence from nucleotides +2 to +102 were subsequently replaced using an assembly PCR strategy by either a hairpin sequence for siRNA expression or by a linker sequence containing suitable cloning sites. This latter vector enabled us to directly clone synthetic oligonucleotides (56 nt) yielding U6-promoter-based knock-down constructs. In the final constructs, the starting 5′ G-nucleotide for efficient transcription initiation and the 3′ T-stretch as transcription termination signal were present.
For the endogenous target mRNAs, target sequences were selected by searching stretches of 19nt, ending with a C. The complementary sequence then starts with a G at position +1. For example, target sequences are: GNAS (NM_000516), CGATGTGA CTGCCATCATC; IKBKB (IKKβ; AF080158) IKBKB-1, CTTAAAGC TGGTTCATATC; IKBKB-2, AGAATCATCCATCGGGATC; IKBKB-3, CGTCGACTACTGGAGCTTC; and M6PR (NM_002355) GGAAGTAATTGGATCATGC.
Adenovirus Production
For the generation of the adenoviruses, the pIPspAdapt-based constructs were transiently transfected in a 96-well format together with adenoviral helper DNA into the producer cells Per.C6/E2A (Michiels et al. 2002; Schouten et al. 2002) The produced adenoviruses were propagated by reinfecting Per.C6/E2A. Titers of the crude lysates were determined by real-time PCR analysis as described (Ma et al. 2001). Typically, the ratio of virus particles over infectious units varied between 30 and 100.
Real-Time RT-PCR Analysis
Endogenous mRNA levels were measured by two-step real-time PCR analysis based on SYBR Green detection with the ABI Prism 7700 Sequence Detection system, according to the manufacturer's instructions. Briefly, RNA was isolated either using TRIzol Reagent (Invitrogen) or using the SV-96 Total RNA Isolation kit (Promega). cDNA synthesis was performed in larger volumes such that each sample could be tested in different subsequent PCR reactions with 5-100 ng of total RNA equivalent in each reaction; a typical reaction performed in 25 μL total volume consisted of 1× TaqMan buffer A (Applied Biosystems), 5 mM MgCl2, 500 μM total dNTPs, 2.5 μM random hexamers, 0.4 U/μL RNase Inhibitor, and 1.25 U/μL MultiScribe Reverse Transcriptase. The mixture was incubated at 25°C for 10 min, at 48°C for 30 min, and at 95°C for 5 min.
The subsequent real-time PCR reaction contained 5 μL of the RT reaction product in a total volume of 25 μL, consisting of 1× SYBR Green mix (Applied Biosystems), 300 nM forward primer, and 300 nM reverse primer. Each sample was analyzed in duplo. The PCR reaction was performed using the following program: 95°C for 10 min followed by 40 cycles of (95°C for 15 sec, 60°C for 1 min). After each PCR reaction, the products were analyzed by measuring the dissociation curve by incubating at 95°C for 15 sec, and at 60°C for 15 sec, followed by increasing the temperature to 95°C over a 20-min time period, ending with 95°C for 15 sec. The used primer sets are given in Supplemental Table 1.
Northern Blotting
For Northern blotting, RNA was isolated using TRIzol (Invitrogen) according to the manufacturer's instructions. The concentration of the RNA samples was determined by UV absorption (A260) or by RiboGreen (Molecular Probes) measurement. Then 0.5-15 μg of RNA was prepared for gel analysis. The RNAs were mixed with denaturing loading dye (TBE Urea sample buffer; Biorad) and denatured by heating at 90°C for 3 min just before loading on a 15% polyacrylamide Ready Gel (Biorad). The gel was run for 50 min at 200 V. The gel was blotted onto Hybond N+ filter (Amersham) using a Transblot semidry Electroblotter (Biorad) for 30 min at 0.2 A/blot with a maximum of 25 V. The blotted filter was hybridized with a 5′-end-labeled oligonucleotide probe using Rapid Hyb solution (Amersham) according to the manufacturer's instructions, and exposed to Hyperfilm MP (Amersham) for 1-3 d. The sequences of the 19-nt probes were identical (sense) or complementary (antisense) to the target sequences.
Cell Culture
Primary HUVEC cells and primary keratinocytes (NHEK-adults) were purchased from Cambrex, synoviocytes (RASF: human rheumatoid arthritis synovial fibroblast) from Cell Applications, Inc. All cells were cultivated according to the manufacturer's recommendations (HUVEC, in EBM-2 bullet kit medium; NHEK in KGM-2 medium; synoviocytes of passage number 8-10 in DMEM + 10% FBS). During infections, the synoviocytes were grown in synoviocyte growth medium (Cell Applications, Inc.), and 3 d after infection, the medium was changed to low-serum-containing medium (M199 + 0.5% FBS). Two to three days after medium switch, the medium was collected and subjected to zymography. U2OS and HEK293 cells were cultured in DMEM containing 10% FBS.
Protein Detection
Western analysis was done by standard procedures using antibodies recognizing IKBKB (Transduction Laboratories), CHUK (IKKα; Cell Signaling Technology), or α-tubulin (Abcam). The detection of the MMP2 activity in the supernatants was performed using 10% gelatin zymogram gels (Biorad). Renaturing and development buffers were from Biorad and were used according to the manufacturer's instructions. The incubation time for the development step was 16 h. Luciferase activity was determined using the luclite kit (PerkinElmer) used according to the manufacturer's instructions.
Acknowledgments
We thank Theo Verwoerd for some of the constructs, Mike Briggs (Pharmacia), Dirk Pollet and Robin Brown for discussions, as well as other Galapagos colleagues for interest in the project.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1332603. Article published online before print in September 2003.
[Supplemental Material is available online at www.genome.org.]
References
- Ashrafi, K., Chang, F.Y., Watts, J.L., Fraser, A.G., Kamath, R.S., Ahringer, J., and Ruvkun, G. 2003. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421: 268-272. [DOI] [PubMed] [Google Scholar]
- Bernstein, E., Denli, A.M., and Hannon, G.J. 2001. The rest is silence. RNA 7: 1509-1521. [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp, T.R., Bernards, R., and Agami, R. 2002a. A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550-553. [DOI] [PubMed] [Google Scholar]
- ____. 2002b. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2: 243-247. [DOI] [PubMed] [Google Scholar]
- Devroe, E. and Silver, P.A. 2002. Retrovirus-delivered siRNA. BMC Biotechnol. 2: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Lendeckel, W., and Tuschl, T. 2001a. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 15: 188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. 2001b. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494-498. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Weber, K., and Tuschl, T. 2002. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26: 199-213. [DOI] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806-811. [DOI] [PubMed] [Google Scholar]
- Hannon, G.J. 2002. RNA interference. Nature 418: 244-251. [DOI] [PubMed] [Google Scholar]
- Hopkins, A.L. and Groom, C.R. 2002. The druggable genome. Nat. Rev. Drug Discov. 1: 727-730. [DOI] [PubMed] [Google Scholar]
- Kamath, R.S., Fraser, A.G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231-237. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., and Plasterk, R.H. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 15: 2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853-858. [DOI] [PubMed] [Google Scholar]
- Lee, N.S., Dohjima, T., Bauer, G., Li, H., Li, M. J., Ehsani, A., Salvaterra, P., and Rossi, J. 2002. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 20: 500-505. [DOI] [PubMed] [Google Scholar]
- Ma, L., Bluyssen, H.A., De Raeymaeker, M., Laurysens, V., van der Beek, N., Pavliska, H., van Zonneveld, A.J., Tomme, P., and van Es, H.H. 2001. Rapid determination of adenoviral vector titers by quantitative real-time PCR. J. Virol. Methods 93: 181-188. [DOI] [PubMed] [Google Scholar]
- McManus, M.T. and Sharp, P.A. 2002. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 3: 737-747. [DOI] [PubMed] [Google Scholar]
- McManus, M.T., Petersen, C.P., Haines, B.B., Chen, J., and Sharp, P.A. 2002. Gene silencing using micro-RNA designed hairpins. RNA 8: 842-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina, M.F. and Joshi, S. 1999. RNA-polymerase III-driven expression cassettes in human gene therapy. Curr. Opin. Mol. Ther. 1: 580-594. [PubMed] [Google Scholar]
- Michiels, F., Van Es, H., Van Rompaey, L., Merchiers, P., Francken, B., Pittois, K., Van Der Schueren, J., Brys, R., Vandersmissen, J., et al. 2002. Arrayed adenoviral expression libraries for functional screening. Nat. Biotechnol. 20: 1154-1157. [DOI] [PubMed] [Google Scholar]
- Miyagishi, M. and Taira, K. 2002. U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol. 20: 497-500. [DOI] [PubMed] [Google Scholar]
- Paddison, P.J., Caudy, A.A., Bernstein, E., Hannon, G.J., and Conklin, D.S. 2002. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes & Dev. 16: 948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, C.P., Good, P.D., Winer, I., and Engelke, D.R. 2002. Effective expression of small interfering RNA in human cells. Nat. Biotechnol. 20: 505-508. [DOI] [PubMed] [Google Scholar]
- Schouten, G., Vogels, R., Bout, A., and Van Es, H. 2002. High-throughput screening of gene function using libraries for functional genomics applications. US 6,430,595.
- Sharp, P.A. 1999. RNAi and double-strand RNA. Genes & Dev. 13: 139-141. [PubMed] [Google Scholar]
- Sui, G., Soohoo, C., Affar el, B., Gay, F., Shi, Y., and Forrester, W.C. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. 99: 5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuschl, T. 2001. RNA interference and small interfering RNAs. Chembiochem. 2: 239-245. [DOI] [PubMed] [Google Scholar]
- Xia, H., Mao, Q., Paulson, H.L., and Davidson, B.L. 2002. siRNA-mediated gene silencing in vitro and in vivo. Nat. Biotechnol. 20: 1006-1010. [DOI] [PubMed] [Google Scholar]
- Yu, J.Y., DeRuiter, S.L., and Turner, D.L. 2002. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl. Acad. Sci. 99: 6047-6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Y., Wagner, E.J., and Cullen, B.R. 2002. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell 9: 1327-1333. [DOI] [PubMed] [Google Scholar]