Abstract
In the present study we show that luxS of Bifidobacterium breve UCC2003 is involved in the production of the interspecies signaling molecule autoinducer-2 (AI-2), and that this gene is essential for gastrointestinal colonization of a murine host, while it is also involved in providing protection against Salmonella infection in Caenorhabditis elegans. We demonstrate that a B. breve luxS-insertion mutant is significantly more susceptible to iron chelators than the WT strain and that this sensitivity can be partially reverted in the presence of the AI-2 precursor DPD. Furthermore, we show that several genes of an iron starvation-induced gene cluster, which are downregulated in the luxS-insertion mutant and which encodes a presumed iron-uptake system, are transcriptionally upregulated under in vivo conditions. Mutation of two genes of this cluster in B. breve UCC2003 renders the derived mutant strains sensitive to iron chelators while deficient in their ability to confer gut pathogen protection to Salmonella-infected nematodes. Since a functional luxS gene is present in all tested members of the genus Bifidobacterium, we conclude that bifidobacteria operate a LuxS-mediated system for gut colonization and pathogen protection that is correlated with iron acquisition.
Introduction
Various beneficial or probiotic effects have been attributed to strains belonging to the genera Bifidobacterium and Lactobacillus. Probiotic bacteria have been used to treat, among others, antibiotic-associated diarrhea, food allergies, atopic eczema, inflammatory bowel disease and arthritis [1]–[6]. In addition, several studies have inferred a role for probiotic bacteria as antagonists of pathogenic bacteria [7], [8]. Proposed mechanisms of action include competition for the same attachment sites as pathogenic bacteria, competition for nutrients, production of growth-inhibitory compounds and stimulation of the immune system [9]–[12]. Whether probiotics need to adhere to epithelial cells of the human gut in order to exert their beneficial effect is still a matter of debate, but close contact between the two is required at some stage [13]. Bacterial adhesion to the gut epithelium is a complex process in which host, bacterial and environmental factors interact, and it is reasoned that adhesion and associated probiotic activities are regulated by bacterial cell-to-cell communication systems.
Quorum sensing is a cell-to-cell communication system which allows (pathogenic) bacteria to coordinate gene expression and regulate virulence factor production in a cell density-dependent manner [14]–[17]. Many Gram-negative pathogens (e.g. Pseudomonas aeruginosa) use N-acylhomoserine lactones as signaling molecules [18]–[21], whereas some Gram-positive bacteria use species-specific oligopeptides [22], [23]. A third cell-to-cell signal molecule is autoinducer-2 (AI-2), produced by a variety of Gram-negative and Gram-positive bacteria. AI-2 is therefore often called an interspecies signaling molecule. A few well-known pathogens, including Vibrio spp. and Salmonella, use AI-2 as a cue to sense population density [24]–[27]. The key enzyme for AI-2 production is LuxS, which is an essential part of the activated methyl cycle, involved in recycling S-adenosylhomocysteine. More specifically, LuxS catalyzes the cleavage of S-ribosyl-homocysteine to homocysteine and 4,5-dihydroxy-2,3-pentanedione (DPD), which subsequently leads to the production of AI-2 [28]. A wide range of bacterial species produce AI-2, but evidence for the presence of signal reception and signal transduction pathways in organisms besides Escherichia coli, Vibrio spp. and Salmonella is lacking [29]. While this lack of evidence is sometimes used to question the role of AI-2 in interspecies signaling [30], an alternative explanation is that other types of receptors and signal transduction pathways are yet to be discovered [31].
Although the interspecies signaling molecule AI-2 is commonly linked to virulence and pathogenicity [24]–[27], it has recently been shown that the probiotic strain Lactobacillus acidophilus NCFM harbors a functional luxS gene and produces AI-2 [32]. Whether this signaling molecule plays a role in eliciting the beneficial traits of probiotic bacteria remains to be determined. Indeed, it was suggested that the ability to produce AI-2 affects attachment of L. acidophilus to intestinal epithelial cells, as a mutation in luxS was shown to result in decreased adherence to Caco-2 cells [32]. Additionally, luxS has been attributed a central metabolic role in Lactobacillus reuteri 100–23 and Lactobacillus rhamnosus GG, and has been shown to influence adherence, biofilm formation and exopolysaccharide production in the latter [33]–[35]. In a recent study [36] AI-2 production has been demonstrated for three strains of Bifidobacteria and overexpression of luxS enhanced biofilm formation by Bifidobacterium longum NCC2705.
In the present study we show that a functional luxS gene is widespread in the genus Bifidobacterium and that this gene in Bifidobacterium breve UCC2003 is involved in providing protection of Caenorhabditis elegans against Salmonella infection, a property which is linked to iron acquisition. Our data furthermore demonstrate that a functional luxS gene is required for murine gastrointestinal colonization by B. breve UCC2003.
Methods and Materials
Bacterial strains, plasmids and culture conditions
The Bifidobacterium strains used for the AI-2 biosensor assay are listed in Table 1. These were cultured anaerobically at 37°C in modified Columbia Broth (mCol). Galactose was used to replace glucose as a carbon source since the latter has been reported to possibly interfere with the AI-2 biosensor assay [37], [38]]. All strains were grown until they reached the stationary phase. All other bacterial strains, as well as the plasmids used in this study, are listed in Table 2. B. breve UCC2003 and derivative mutant strains were routinely cultured in Reinforced Clostridial Medium (RCM), supplemented with the appropriate antibiotics (10 µg ml−1 tetracycline or 3 µg ml−1 chloramphenicol). E. coli strains were cultured in LB broth at 37°C, while the V. harveyi BB170 biosensor strain was grown in Marine Broth at 25°C with agitation [38]. Where appropriate growth media contained tetracycline (Tet; 15 µg ml−1), chloramphenicol (Cm; 10 µg ml−1 for E. coli or 3 µg ml−1 for bifidobacteria), erythromycin (Em; 100 µg ml−1 for E. coli) or kanamycin (Km; 50 µg ml−1 for E. coli). Recombinant E. coli cells containing pORI19 were selected on LB agar containing Em, and supplemented with X-gal (5-bromo-4-chloro-3-indolyl-b-D-galactopyranoside) (40 µg ml−1) and 1 mM IPTG (isopropyl-b-D-galactopyranoside).
Table 1. List of Bifidobacterium strains, with additional information on the source of isolation.
| Species | Strain** | Source of isolation | AI-2 production | PCR* | |
| Mean (%) | SEM (%) | ||||
| B. adolescentis | LMG 10502T | Adult, intestine | 226 | 12 | + |
| LMG 10733 | Adult, intestine | 128 | 5 | + | |
| LMG 10734 | Adult, intestine | 155 | 9 | + | |
| LMG 11579 | Bovine, rumen | 138 | 7 | + | |
| LMG 18897 | Human, faeces | 126 | 5 | + | |
| LMG 18898 | Human, faeces | 135 | 16 | + | |
| B. angulatum | LMG 11039T | Human, faeces | 129 | 2 | - |
| LMG 11568 | Sewage | 126 | 12 | - | |
| B. animalis subsp. animalis | LMG 10508T | Rat, faeces | 237 | 4 | + |
| LMG 18900 | Rat, faeces | 250 | 11 | + | |
| B. animalis subsp. lactis | LMG 25734 | Yoghurt (Yogosan) | 242 | 7 | + |
| LMG 25755 | Yoghurt (Teddi) | 207 | 4 | + | |
| LMG 25756 | Food supplement (Hygiaflora) | 175 | 3 | + | |
| LMG 25757 | Food supplement (Friendly bifidus) | 320 | 16 | + | |
| LMG 11580 | Chicken, faeces | 124 | 6 | + | |
| LMG 18314T | Yoghurt | 165 | 3 | + | |
| LMG 18906 | Rabbit, faeces | 171 | 5 | + | |
| LMG 23512 | Human, faeces | 284 | 10 | + | |
| LMG 24384 | Milk | 272 | 23 | + | |
| B. bifidum | LMG 25758 | Pharmaceutical preparation (Infloran Berna) | 208 | 4 | + |
| LM 381 | Food supplement (Friendly bifidus) | 447 | 20 | + | |
| LM 588 | Food supplement (Biodophilus) | 216 | 4 | + | |
| LMG 11041T | Breast-fed infant, faeces | 149 | 3 | + | |
| LMG 11582 | Adult, intestine | 152 | 12 | + | |
| LMG 11583 | Adult, intestine | 210 | 7 | + | |
| LMG 13195 | Infant, intestine | 121 | 5 | + | |
| B. breve | LMG 25761 | Food supplement (Yakult bifiel) | 265 | 4 | + |
| UCC2003 (LMG 11040) | Nursing stool | 496 | 41 | + | |
| LMG 11084 | Blood | 218 | 4 | + | |
| LMG 11613 | Infant, intestine | 132 | 18 | + | |
| LMG 13194 | Infant, intestine | 447 | 37 | + | |
| LMG 13208T | Infant, intestine | 378 | 14 | + | |
| LMG 23729 | Infant, faeces | 224 | 4 | + | |
| B. catenulatum | LMG 11043T | Adult, intestine or faeces | 205 | 4 | - |
| LMG 18894 | Sewage | 173 | 20 | - | |
| B. dentium | LMG 10507 | Human, faeces | 126 | 7 | + |
| LMG 11045T | Dental caries | 140 | 3 | + | |
| LMG 11585 | Dental caries | 141 | 11 | + | |
| B. gallicum | LMG 11596T | Adult, intestine | 137 | 3 | + |
| B. longum subsp. infantis | LMG 25762 | Pharmaceutical preparation (Probiotical) | 209 | 4 | + |
| LMG 8811T | Infant, intestine | 248 | 7 | + | |
| LMG 11570 | Infant, intestine | 152 | 10 | + | |
| LMG 11588 | Infant, faeces | 180 | 5 | + | |
| LMG 13204 | Infant, intestine | 169 | 5 | + | |
| LMG 18901 | Infant, faeces | 119 | 5 | + | |
| LMG 23728 | Infant, faeces | 335 | 13 | + | |
| B. longum subsp. longum | LMG 25765 | Yoghurt (Lactoferrin) | 260 | 9 | + |
| LMG 25766 | Food supplement (Lola) | 269 | 9 | + | |
| LMG 11047 | Human | 121 | 4 | + | |
| LMG 11589 | Calf, faeces | 197 | 5 | + | |
| LMG 13196 | Infant, intestine | 120 | 7 | + | |
| LMG 13197T | Adult, intestine | 207 | 5 | + | |
| LMG 18899 | Adult, faeces | 189 | 19 | + | |
| B. pseudocatenulatum | LMG 10505T | Infant, faeces | 189 | 6 | + |
| LMG 11593 | Sewage | 169 | 17 | + | |
| LMG 18903 | Human, faeces | 134 | 4 | + | |
| LMG 18910 | Sewage | 161 | 7 | + | |
| B. scardovii | LMG 21589T | 50-year-old woman, blood | 257 | 5 | + |
| LMG 21590 | 44-year-old woman, hip | 260 | 6 | + | |
For each strain, the relative levels of AI-2 production (means +/- SEM) in the diluted supernatant are given compared to AI-2 levels produced by the biosensor itself ( = 100%). Data presented are mean +/- SEM from triplicate experiments. In addition, the results for the PCR assay with primers directed against luxS are shown. (* +, PCR positive result, -, PCR negative result; ** T, type strain of the species).
Table 2. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Relevant information | Reference or source |
| Strains | ||
| V. harveyi BB170 | AI-2 biosensor strain (luxN::Tn5) | [38] |
| E. coli | ||
| OP50 | C. elegans normal food source | Caenorhabditis Genetics Center, University of Minnesota, USA |
| EC101 | Cloning host | [44] |
| DH5α | AI-2 negative control strain | |
| B. breve | ||
| UCC2003 | Wild Type | |
| UCC2003PK1 | UCC2003 harbouring pPKCM | [43] |
| UCC2003-luxS | Insertion mutant in luxS (Bbr_0541) | This study |
| UCC2003-luxS [pBC1.2luxS] | Complemented strain | This study |
| UCC2003-bfeU | Insertion mutant in bfeU (Bbr_0221) | This study |
| UCC2003-bfeB | Insertion mutant in bfeB (Bbr_0223) | This study |
| S. enterica subsp. enterica serovar Typhimurium | ||
| NCTC 13348 | Infecting agent | Health Protecting Agency Culture Collections, UK |
| Plasmids | ||
| pPKCM | pCIBA089-ColE1-Cmr | [43] |
| pBC1.2 | pBC1-pSC101-Cmr | [45] |
| pORI19 | Emr, repA−, ori+, cloning vector | [44] |
| pAM5 | pBC1-puC19-Emr | [45] |
| pBC1.2luxS | pBC1.2 harboring luxS (for complementation) | This study |
| pORI19-luxS | pOR19 harboring internal fragment of luxS | This study |
| pORI19-luxS-tet | pORI19 harboring internal fragment of luxS + Tetr | This study |
| pORI19-bfeU | pOR19 harboring internal fragment of Bbr_0221 | This study |
| pORI19-bfeU-tet | pORI19 harboring internal fragment of Bbr_0221+ Tetr | This study |
| pORI19-bfeB | pOR19 harboring internal fragment of Bbr_0223 | This study |
| pORI19-bfeB-tet | pORI19 harboring internal fragment of Bbr_0223+ Tetr | This study |
Detection of AI-2 production by Bifidobacteria
A stationary phase culture of a given Bifidobacterium strain was centrifuged twice (5,000 g, 5 min, room temperature). Culture supernatant was neutralized (pH 7.0) with 5 M NaOH to exclude any possible pH effects, filter sterilized and subsequently diluted to final concentrations of 20% (v/v) with sterile deionised milliQ water. AI-2 levels were determined in a V. harveyi BB170 assay as described previously [38]. Briefly, an overnight culture of the reporter strain was diluted 1∶5000 into fresh sterile, double concentrated MB medium and 100 µl of this cell suspension was added to the wells of a black 96-well microtiter plate (Perkin Elmer). Subsequently, 100 µl of the appropriate sterile supernatant dilution was added to the wells, the microtiter plates were incubated at 30°C and bioluminescence was measured after 5 hours using the EnVision Multilabel Reader (Perkin Elmer). Bioluminescence was expressed as the fraction of bioluminescence measured in the positive control reaction.
Detection of luxS in bifidobacteria
An initial an extensive search of the NCBI Genome Project database (http://www.ncbi.nlm.nih.gov/sites/entrez?db=genome) provided the available sequences of luxS homologs in bifidobacteria. Subsequently, a nucleotide BLAST generated a series of additional sequences with high similarity to the sequences found. A set of degenerate primers (5′-CCC GGY TAC ACA TCG ACT GCT C-3′ and 5′-GTG GTC GCG RTA GTT GCC GC-3′) was then designed, using the ClustalX software package as an alignment tool. Extraction of total bifidobacterial bacterial DNA was performed as described previously [39] while PCRs were performed with the following conditions: initial denaturation at 94°C for 3 minutes was followed by 30 cycles of denaturation at 94°C for 30 s, primer annealing at 56°C for 30 s and elongation at 72°C for 30 s. The PCR reactions were terminated with a final elongation of 10 minutes at 72°C. The obtained products were separated by electrophoresis on 1.5% agarose gels and stained with GelRed (Biotium)
DNA manipulations
The general procedures used for DNA manipulation were essentially those described previously [40]. Restriction enzymes and T4 DNA ligase were obtained from Roche Diagnostics and used according to the manufacturer's instructions. PCRs were performed using Taq PCR master mix (Qiagen GmbH). Synthetic oligonucleotides were synthesized by MWG Biotech AG and are listed in Table S1. PCR products were purified by using a High-Pure PCR product purification kit (Roche). Plasmid DNA was introduced into E. coli and B. breve by electroporation and large-scale preparation of chromosomal DNA from Bifidobacterium spp. was performed as described previously [41]. Plasmid DNA was obtained from B. breve and E. coli using a QIAprep spin plasmid miniprep kit (Qiagen GmbH). An initial lysis step was performed using 30 mg/ml of lysozyme for 30 min at 37°C as part of the plasmid purification protocol for B. breve.
Construction of B. breve UCC2003 insertion mutants and complementation strains
Sequence data were obtained from the Artemis-mediated [42] annotations of the B. breve UCC2003 genome [43]. Internal fragments of luxS (Bbr_0540, 277-bp), bfeU (Bbr_0221, 440 bp) or bfeB (Bbr_0223, 457 bp) were amplified by PCR using B. breve UCC2003 chromosomal DNA as the template and the oligonucleotide primers luxS-277-f-hindIII and luxS-277-r-xbaI, 221IMhd3 and 221IMxba or 223IMhd3 and 223Imxba, respectively (Table S1). The generated PCR products were cloned into pORI19, an Ori+ RepA− integration plasmid [44] using the unique HindIII and XbaI restriction sites that were incorporated into the forward and reverse primer respectively. Ligations were introduced into E. coli EC101 by electroporation. The expected structure of the recombinant plasmids, designated pORI19-luxS, pORI19-bfeU and pORI19-bfeB, was confirmed by restriction mapping and sequencing. The tetW gene, amplified by PCR using pAM5 plasmid DNA as the template [45] and primers tetWf and tetWr (Table S1), thereby incorporating flanking SalI sites in the amplicon, was cloned into the SalI-cut pORI19-luxS, pORI19-bfeU and pORI19-bfeB plasmids to generate plasmid pORI19-tet-luxS, pORI19-tet-bfeU and pORI19-tet-bfeB. The latter plasmids were introduced into E. coli EC101 harboring pNZ-M.BbrII-M.BbrIII to facilitate methylation [41], and the resulting methylated pORI19-tet-luxS, pORI19-tet-bfeU and pORI19-tet-bfeB were then introduced into B. breve UCC2003 by electroporation and subsequent selection on RCA plates supplemented with the tetracycline. Site-specific recombination in potential tet-resistant mutant isolates was confirmed by colony PCR using primer combinations tetWFw and tetWRv to verify tetW gene integration, and primers luxS-Fw, Bbr_0221-Fw and Bbr_0223-Fw (upstream of the luxS, bfeU and bfeB gene fragments selected, respectively), each in combination with pORI19For to confirm integration at the expected chromosomal position. For the construction of the complementation construct pBC1.2luxS, a DNA fragment encompassing luxS, including its native promoter region was generated by PCR amplification from chromosomal DNA of B. breve UCC2003 using Pfu DNA polymerase (Agilent) and primer combination luxS-compl-f-xbaI and luxS-compl-r-xbaI (Table S1). The luxS-containing amplicon was digested with XbaI, and ligated to similarly digested pBC1.2. The ligation was introduced into E. coli EC101 by electroporation. For all cloning experiments, the plasmid content of a number of transformants was screened by restriction analysis and the integrity of positively identified clones was verified by sequencing.
Transcriptome analysis of B. breve UCC2003 and UCC2003-luxS during in vitro growth
In order to compare global transcription patterns of the B. breve UCC2003-luxS insertion mutant with the B. breve UCC2003 WT strain, cells grown to early exponential phase were collected and resuspended in DEPC-treated water. Cell disruption, RNA isolation, RNA quality control, cDNA synthesis and indirect labeling were performed as described previously [43]. DNA microarrays containing oligonucleotide primers representing each of the 1,864 annotated genes in the genome of B. breve UCC2003 were obtained from Agilent Technologies (Palo Alto). Labeled cDNA was hybridized using the Agilent gene expression hybridization kit (number 5188–5242) as described in the Agilent two-color microarray-based gene expression analysis v4.0 manual (publication number G4140-90050). Following hybridization, microarrays were washed as described in the manual and scanned using Agilent's DNA microarray scanner G2565A. The scans were converted to data files with Agilent's Feature Extraction software (version 9.5). DNA microarray data were processed as previously described [46], [47]. Differential expression tests were performed with the Cyber-T implementation of a variant of the t test [48]. A gene was considered differentially expressed between a mutant and the WT when a transcription ratio of 2 relative to the result for the WT was obtained, with a corresponding p-value equal to or less than 0.01. The transcriptional array data from a dye-swap biological replicate experiment has been deposited in the GEO database under accession number GSE49880.
Iron chelator assays
The MIC for the iron chelators 2,2-dipyridyl, ciclopirox olamine and phenanthroline was determined according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) standard broth microdilution protocol, with minor modifications [49]. Instead of Mueller-Hinton broth, RCM was used. A two-fold dilution series of 2,2-dipyridyl, ciclopirox olamine and phenanthroline, ranging from 1000 µM to 2 µM was tested. To assess whether DPD complementation could restore growth of the UCC2003-luxS insertion mutant in the presence of the iron chelators, MICs were also determined in the presence of 50 µM and 100 µM DPD.
Murine colonization experiments
Experiments with mice were approved by the University College Cork Animal Experimentation Ethics Committee and experimental procedures were conducted under license from the Irish Government (license number B100/3729). Seven-week-old female, BALB/c mice were housed in individually vented cages (Animal Care Systems) under a strict 12 h light cycle. Mice (n = 7 per group) were fed a standard polysaccharide-rich mouse chow diet and water ad libitum. Mice were inoculated by oral gavage (109 cfu of B. breve UCC2003PK1, B. breve UCC 2003-luxS or a mixture of B. breve UCC2003PK1 and B. breve UCC 2003-luxS in 100 µl of PBS). Fecal pellets were collected at intervals during 18 days to enumerate bacteria. Eighteen days after inoculation, mice were sacrificed and their intestinal tracts quickly dissected. The small intestine, cecum and large intestine were harvested for determination of colony forming units (cfu) (serial dilution plating on RCA agar plates with appropriate antibiotics).
C. elegans colonization experiments
C. elegans N2 (glp-4; sek-1) was propagated under standard conditions, synchronized by hypochlorite bleaching, and cultured on nematode growth medium using E. coli OP50 as food source [50], [51].To prepare conditioning plates, WT B. breve UCC2003, B. breve UCC2003-luxS, B. breve UCC2003-luxS [pBC1.2luxS], B. breve UCC2003-bfeU or B. breve UCC2003-bfeB were grown anaerobically RCM at 37°C until reaching stationary phase. 500 µl of the cell suspension was spread on a nematode growth medium (NGM) plate and dried for 3 h at 37°C [52]. Conditioning plates were used directly after preparation by transferring fresh hypochlorite-treated nematodes to these plates. After conditioning for 72 h, worms were washed three times with M9 buffer supplemented with 1 mM sodium azide to prevent expulsion of the intestinal load and to remove surface-attached bacteria [53]. The number of nematodes was then determined microscopically and nematodes were lysed in phosphate-buffered saline containing 400 mg 1.0 mm silicon carbide beads (BioSpec Products, Inc.) and mechanically disrupted using a pestle. Subsequently, the worm lysates were serially diluted, plated on RCA and incubated anaerobically at 37°C. After 48 h, CFU were determined and the number of bacteria per nematode was calculated [54].
In vivo gene expression analysis by qRT-PCR
A synchronized C. elegans nematode population was transferred to conditioning plates (as described above), and after 72 h incubation at 25°C, RNA isolation of in vivo grown WT B. breve UCC2003 was performed as described previously, with slight modifications [43]. After collection of the nematodes, total RNA was quickly isolated following the protocol of the TRIzol reagent (Invitrogen) and purified using the RNeasy minikit (Qiagen) including an on-column DNase digestion with RNase-free DNase (Qiagen). Next, qScript cDNA Supermix (Quanta Biosciences) was used to obtain cDNA. After development of forward and reverse primers for the reference genes and the genes of interest and after testing their specificity, real-time PCR (CFX96 Real Time 116 6 System, Bio-Rad) was performed using the iQ SYBR Green Supermix (Bio-Rad). The expression levels of the genes of interest were normalized using 5 reference genes, namely, atpD, rpoB, ldh, pdxS and gluC, by geometric averaging of multiple internal control genes with the GeNorm software package [55]. Primers were designed using Primer-Blast and are listed in Table S1. To ensure the specific amplification of Bifidobacterium RNA, primers were also BLASTed to the C. elegans and E. coli OP50 genome.
Infection assays in the C. elegans model
C. elegans survival experiments were performed as described earlier, with slight modifications [56]. Synchronized nematodes (L4 stage) were suspended in a medium containing 95% M9 buffer, 5% brain heart infusion broth, and 10 µg/ml cholesterol (Sigma-Aldrich). Then, 250 µl of this nematode suspension was transferred to the wells of a 24-well microtiter plate. Stationary phase cultures of E. coli OP50, B. breve UCC2003, B. breve UCC2003-luxS, B. breve UCC2003-luxS [pBC1.2luxS], B. breve UCC2003-bfeU or B. breve UCC2003-bfeB were centrifuged, resuspended in the assay medium, and standardized to 106 CFU/ml. Next, 250 µl aliquots of these standardized suspensions were added to each well. Subsequently, 500 µl of the assay medium was added to each well to obtain a final volume of 1 ml per well, and the microtiter plates were incubated at 25°C to allow colonization of the nematode gut. After 24 h incubation, an overnight culture of S. Typhimurium NCTC13348 was standardized as described above, and 250 µl of the suspension was added to the wells to establish gastro-intestinal infection. Nematodes not being administered any bifidobacteria were used as a control. Sterile assay medium was added to non-infected nematodes to correct for spontaneous mortality, not caused by S. Typhimurium infection. Finally, the plates were incubated at 25°C and the fraction of dead nematodes was determined after 24 h and 48 h by counting the number of dead worms and the total number of worms in each well, using a dissecting microscope.
Statistical data analysis
Statistical data analysis was carried out using the SPSS Statistics 17.0 software package. To assess if means were statistically significantly different from one another, a non-parametric Mann-Whitney U test was performed (significance level 0.05%).
Results
A functional luxS gene is widespread amongst bifidobacteria
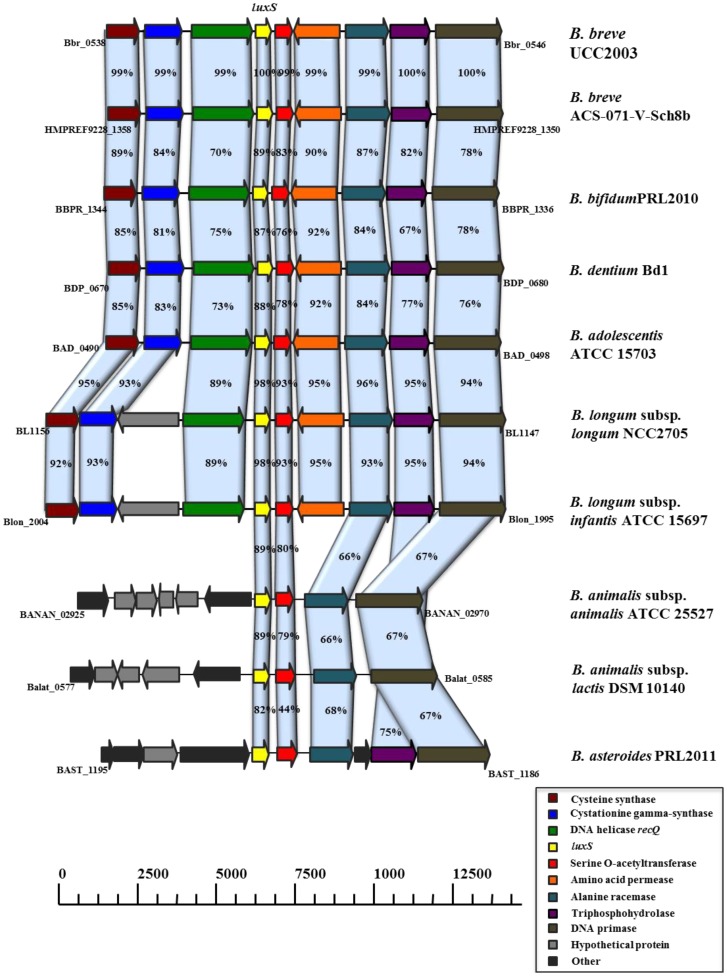
The availability of whole genome sequences of various Bifidobacterium spp. revealed the presence of the AI-2 synthase-encoding gene luxS in these strains. To verify if a functional luxS gene is widespread amongst bifidobacteria, phenotypic and genotypic experiments were performed. Using Vibrio harveyi BB170 as a biosensor, we demonstrated that all (n = 59) Bifidobacterium spp. strains tested produce AI-2 during stationary phase planktonic growth (Table 1). Using primers based on conserved regions of the luxS gene we showed that this gene is present in nearly all (n = 55) species investigated, except Bifidobacterium angulatum and Bifidobacterium catenulatum (Table 1). However, homologs of luxS have been found in all currently available genome sequences of Bifidobacterium spp. (img.jgi.doe.gov), including those of the two species mentioned above. Inspection of the luxS DNA sequence from the currently available genome sequences of Bifidobacterium angulatum DSM 20098 (JCM 7096) and Bifidobacterium catenulatum LMG 11043 suggests that negative PCR results were due to sequence differences at the PCR primer locations. A map comparing the genomic context of luxS in different Bifidobacterium genomes is provided in Figure 1 and shows that the organization of luxS and its neighboring genes is conserved in the genus Bifidobacterium. In particular, the presence and relative genomic position of four genes, encoding a serine O-acetyltransferase, alanine racemase, DNA primase and triphosphohydrolase, are highly conserved (Fig. 1).
Figure 1. Comparison of the luxS genetic loci of B. breve UCC2003 with corresponding luxS loci from other sequenced bifidobacteria.
Each solid arrow indicates an open reading frame. The lengths of the arrows are proportional to the length of the predicted open reading frame. The colour coding which is indicative of putative function, is indicated within the arrow. Orthologs are marked with the same colour while the amino acid identity of each predicted protein is indicated as a percentage relative to its equivalent protein encoded by B. breve UCC2003.
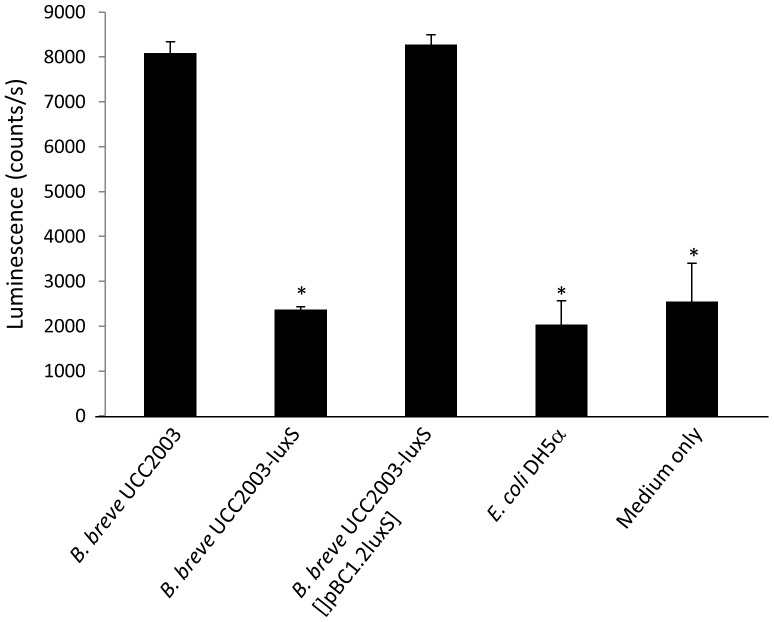
In B. breve UCC2003, inactivation of luxS by insertional mutagenesis (creating a strain designated as B. breve UCC2003-luxS) resulted in a drastic and significant decrease (p≤0.05) in AI-2 production (Fig. 2), thus providing further evidence that luxS is crucial for AI-2 production in wild type (WT) UCC2003. This was substantiated by providing a functional luxS gene on a plasmid in trans in the luxS-insertion mutant (strain B. breve UCC2003-luxS [pBC1.2luxS]), which restored AI-2 production to WT levels (Fig. 2).
Figure 2. Luminescence signal of the V. harveyi BB170 biosensor strain in the presence of sterile and neutralized supernatant of B. breve UCC2003, the insertion mutant B. breve UCC2003-luxS and the complemented strain B. breve UCC2003-luxS [pBC1.2luxS].
Data obtained with E. coli DH5α (a strain not producing AI-2) and a medium-only control are included as reference. Data shown are means ± SEM. (*, luminescence is significantly lower than than produced with supernatant of B. breve UCC2003, p<0.05, compared to WT; n = 3).
Impact of luxS inactivation on gene expression in B. breve UCC2003
To investigate the impact of luxS inactivation on gene expression in B. breve UCC2003, comparative transcriptome analysis between in vitro grown WT B. breve UCC2003 and B. breve UCC2003-luxS was carried out. The microarray analysis showed that 1.47% of the genes (27/1843) were significantly upregulated in B. breve UCC2003-luxS, while 5.70% (105/1843) were significantly downregulated (2-fold cut-off, p≤0.01) compared to the WT. Consistent with previous studies on the role of LuxS in lactobacilli [32]–[35] the microarray analysis suggest that the role of LuxS in B. breve is metabolic. Interestingly, the microarray analysis also showed that the transcription of a cluster of six genes, which encode a predicted iron-uptake system (see below) and which had previously been shown to be induced under iron-starvation conditions [57], is downregulated in B. breve UCC2003-luxS (Table 3), suggesting that the luxS mutation affects the mutant's ability to acquire iron.
Table 3. Relative normalized gene expression levels of a cluster of iron regulated genes as expressed in vitro in B. breve UCC2003-luxS compared to B. breve UCC2003.
| Locus tag and gene name | Annotation | Fold Downregulation |
| Bbr_0221 (bfeU) | Conserved hypothetical membrane spanning protein with iron permease FTR1 family domain | 3.321 |
| Bbr_0222 (bfeO) | Conserved hypothetical secreted protein | 5.384 |
| Bbr_0223 (bfeB) | Conserved hypothetical membrane spanning protein | 1.996 |
| Bbr_0224 | Permease protein of ABC transporter system | 5.389 |
| Bbr_0225 | Permease protein of ABC transporter system | 1.730 |
| Bbr_0226 | ATP-binding protein of ABC transporter system | 3.147 |
LuxS affects iron metabolism in B. breve UCC2003
To explore the possible link between luxS and iron metabolism, the tolerance of the B. breve strains towards iron chelators that specifically chelate ferrous iron (2,2-dipyridyl), ferric iron (ciclopirox olamine), or both (phenanthroline) was determined. Minimal inhibitory concentration (MIC) values were considerably higher for B. breve UCC2003 and B. breve UCC2003-luxS [pBC1.2luxS] than that obtained for B. breve UCC2003-luxS (Table 4), demonstrating that the insertion mutant is more susceptible to ferrous and ferric ion chelators than WT B. breve UCC2003. Addition of the AI-2 precursor DPD to the growth medium partially restored growth of the insertion mutant in the presence of the chelators, thereby supporting the notion that LuxS is directly or indirectly involved in iron acquisition (Table 4). In order to confirm the involvement of the predicted iron-uptake genes in iron acquisition two additional mutant strains, B. breve UCC2003-bfeU and B. breve UCC2003-bfeB, were constructed that harbor an insertion in the predicted iron-uptake genes, bfeU (Bbr_0221) and bfeB (Bbr_0223), respectively. As expected, both of these mutants were found to be more susceptible to the three iron chelators as compared to the parent strain B. breve UCC2003 (Table 4).
Table 4. MIC values of 2,2-dipyridyl, ciclopiroxolamine and phenanthroline for B. breve UCC2003 WT and various mutants, and for B. breve UCC2003-luxS supplemented with DPD (50 µM and 100 µM).
| Component (µM) | |||
| 2,2-dipyridyl | ciclopirox olamine | phenanthroline | |
| B. breve UCC2003 | 1000 | 125 | 250 |
| B. breve UCC2003-luxS | 250 | 31.25 | 62.5 |
| B. breve UCC2003-luxS [pBC1.2luxS] | 1000 | 125 | 250 |
| B. breve UCC2003-luxS + DPD (50 µM) | 500 | 62.5 | 125 |
| B. breve UCC2003-luxS + DPD (100 µM) | 500 | 62.5 | 125 |
| B. breve UCC2003-bfeU | 500 | 15.63 | 125 |
| B. breve UCC2003-bfeB | 500 | 31.25 | 125 |
The presence of luxS is required for murine gastrointestinal colonization
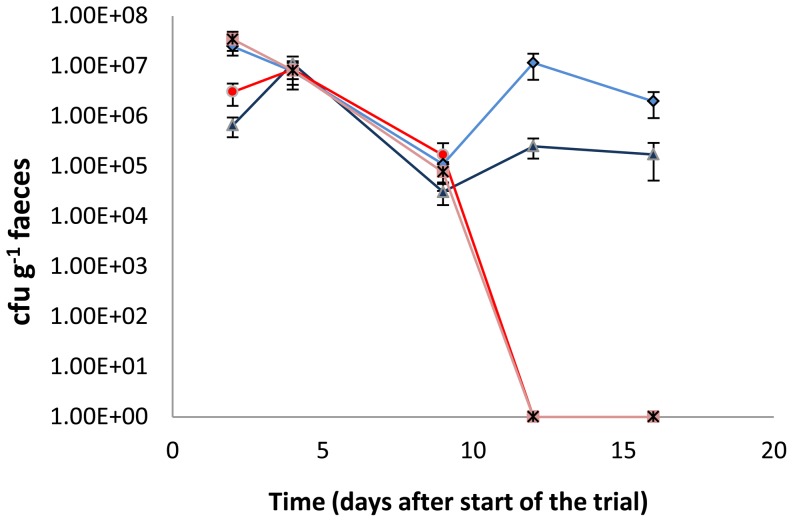
To verify whether a functional luxS gene is required for gut colonization in a competitive environment, the gut colonization capacity of B. breve UCC2003 and B. breve UCC2003-luxS was tested in BALB/c mice. In conventional BALB/c mice with a resident microbiota (i.e. in a competitive environment), WT B. breve UCC2003 was able to colonize the gastrointestinal tract, as was shown by plating of fecal samples (1×105 CFU/g feces retrieved 15 days after last administration; Fig. 3). Viable count determinations of the contents of the small intestine, large intestine and cecum of individual mice confirmed these findings (Table 5). In contrast, the luxS insertion mutant was unable to stably colonize the murine gastrointestinal tract. To examine whether the presence of the WT was able to rescue the luxS mutant's impaired ability to colonize, co-administration experiments were carried out. When equal numbers of B. breve UCC2003 and B. breve UCC2003-luxS were administered simultaneously, only the WT was able to colonize the gastrointestinal tract, once more confirming that a functional luxS gene is required for successful colonization of the gastrointestinal tract in a competitive environment (Fig 3).
Figure 3. Murine colonization trial.
CFU g−1 feces of B. breve UCC2003 (dark blue) and B. breve UCC2003-luxS (red) administered individually, or simultaneously {a mixture of equal numbers of B. breve UCC2003 (pale blue) and B. breve UCC2003-luxS (pink)}. Administration started at day 0 and was continued for 3 consecutive days. Data shown are mean ± SEM. (n = 7).
Table 5. Murine colonization experiments.
| LOG CFU retrieved after | ||||
| Single strain administration | Simultaneous administration | |||
| B. breve UCC2003 | B. breve UCC2003-luxS | B. breve UCC2003 | B. breve UCC2003-luxS | |
| Small intestine | 6.39±0.09 | 1.76±1.15 * | 5.94±0.43 | BDL |
| Cecum | 6.87±0.06 | 2.84±0.67 * | 6.96±0.43 | BDL |
| Large intestine | 6.51±1.34 | 0.85±1.15 * | 6.52±0.43 | BDL |
LOG CFU of B. breve UCC2003 or B. breve UCC2003-luxS, retrieved from the murine small intestine, cecum and large intestine (15 days after the last administration). Data are shown for single strain administration as well as for the simultaneous administration of equal numbers of both strains. Numbers shown are means ± SEM. BDL: below detection limit. (*, significantly lower than WT; p<0.05, n = 7).
B. breve UCC2003, B. breve UCC2003-luxS or B. breve UCC2003-luxS [pBC1.2luxS] colonize the C. elegans gut
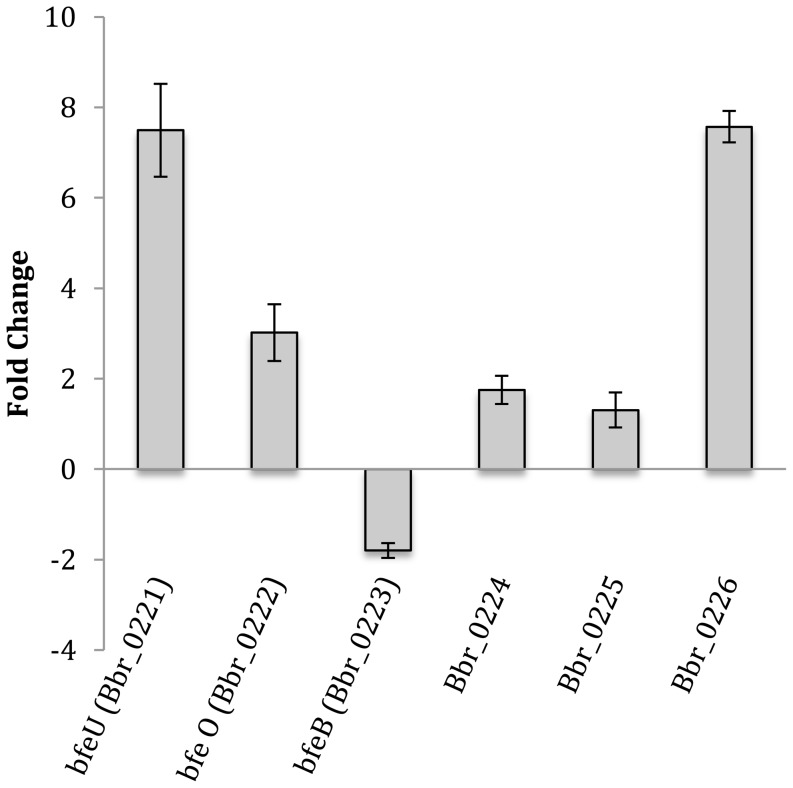
Having established that UCC2003-luxS could not colonize the murine gastrointestinal tract we sought another model to examine the potential role of luxS in providing a host-protecting effect against pathogen infection. We initially determined the colonization capacity of B. breve UCC2003, B. breve UCC2003-luxS or B. breve UCC2003-luxS [pBC1.2luxS] under monoxenic conditions in the gut of the nematode Caenorhabditis elegans [58]. When administered separately, the average number of WT B. breve UCC2003 able to colonize the C. elegans gut was about 1×103 CFU/nematode. The numbers for the insertion mutant B. breve UCC2003-luxS (1.20×103 CFU/nematode) and for the complemented strain B. breve UCC2003-luxS [pBC1.2luxS] (0.82×103 CFU/nematode) were not significantly different thereby identifying this model as appropriate for infection studies. However, simultaneous administration of a mixture of equal numbers of both WT and B. breve UCC2003-luxS revealed that the WT had a competitive advantage, as the average number of CFU recovered were approx. twice as high for the WT (0.81×103 CFU/nematode) as for B. breve UCC2003-luxS (0.43×103 CFU/nematode) (p<0.05, n = 3). In order to investigate the importance of the iron-regulated genes for B. breve UCC2003 when grown under gastrointestinal conditions in which iron is limited, qRT-PCR experiments were performed whereby the transcription levels of the iron regulated genes was compared between in vitro and in vivo (i.e. in the C. elegans gut) grown B. breve UCC2003. These experiments showed that bfeU (encoding a high affinity iron permease), bfeO (Bbr_0222, encoding a secreted protein with iron-binding domain) and Bbr_0226 (encoding an ABC-type transporter) exhibit a significantly higher level of transcription when B. breve UCC2003 was grown under in vivo conditions relative to in vitro conditions, indicating that in the C. elegans gut iron levels are limiting (Fig. 4). These findings are in full agreement with previously obtained in vivo transcriptome data (GEO database accession no. GSE27491), showing that six of the seven genes of this iron-regulated cluster also exhibit increased transcription in the murine gut relative to in vitro conditions [43].
Figure 4. Relative normalized expression levels (obtained with qRT-PCR) of the iron regulated genes in B. breve UCC2003 retrieved from C. elegans gut, compared to in vitro grown B. breve UCC2003.
Data shown are means ± SEM. (*, p<0.05, compared to in vitro expression levels; n = 3).
B. breve confers protection against Salmonella infection
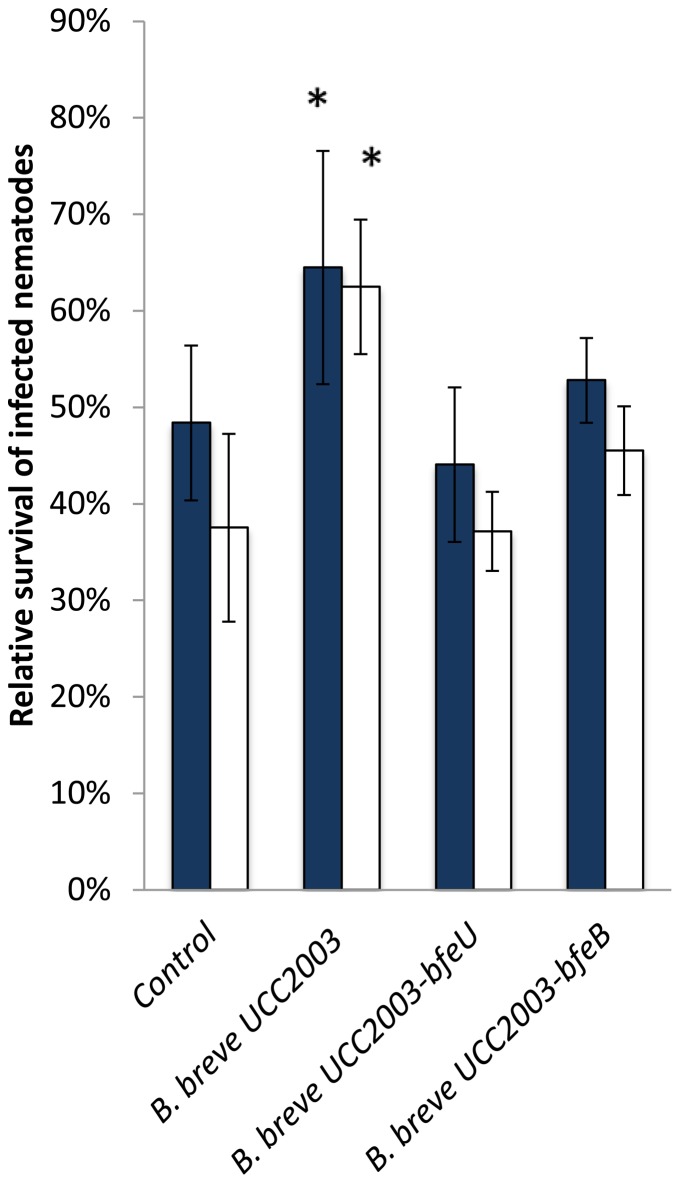
In order to examine the potential role of luxS in providing a host-protecting effect against pathogenic bacteria, an in vivo Salmonella infection experiment in the C. elegans model was adopted. A positive influence of bifidobacterial administration on the longevity of Salmonella-infected nematodes has previously been described [59]. The relative survival of Salmonella-infected C. elegans worms that were administered B. breve UCC2003, B. breve UCC2003-luxS or B. breve UCC2003-luxS [pBC1.2luxS] is shown in Figure 5. Relative survival of infected nematodes that were fed UCC2003 was significantly higher than survival of the infected nematodes that had not received B. breve UCC2003 (p≤0.05). Administration of the luxS insertion mutant resulted in a significantly decreased survival of infected nematodes compared to those that had received WT B. breve UCC2003, whereas administration of the complemented strain resulted in a significantly increased survival compared to those nematodes that had not received treatment and to those that had received the luxS mutant (p≤0.05). Similar to UCC2003-luxS, B. breve UCC2003-bfeU and B. breve UCC2003-bfeB each exhibit a significantly decreased ability to confer protection to Salmonella-infected nematodes as compared to the WT B. breve UCC2003, thereby confirming the importance of iron acquisition in gut pathogen protection (Fig. 6). Furthermore, as both B. breve UCC2003-bfeU and B. breve UCC2003-bfeB colonized C. elegans to a similar level as the WT, this decreased protective effect is not due to decreased ability to colonize the nematodes. Collectively this data indicates that luxS (and its gene product) is a prerequisite for B. breve UCC2003 to confer protection against Salmonella infection, and that this protection can be correlated with iron acquisition.
Figure 5. Relative survival of Salmonella-infected C. elegans nematodes to which B. breve UCC2003 WT and various mutants were administered (24 h{black bar} and 48 h{white bars} after Salmonella infection).
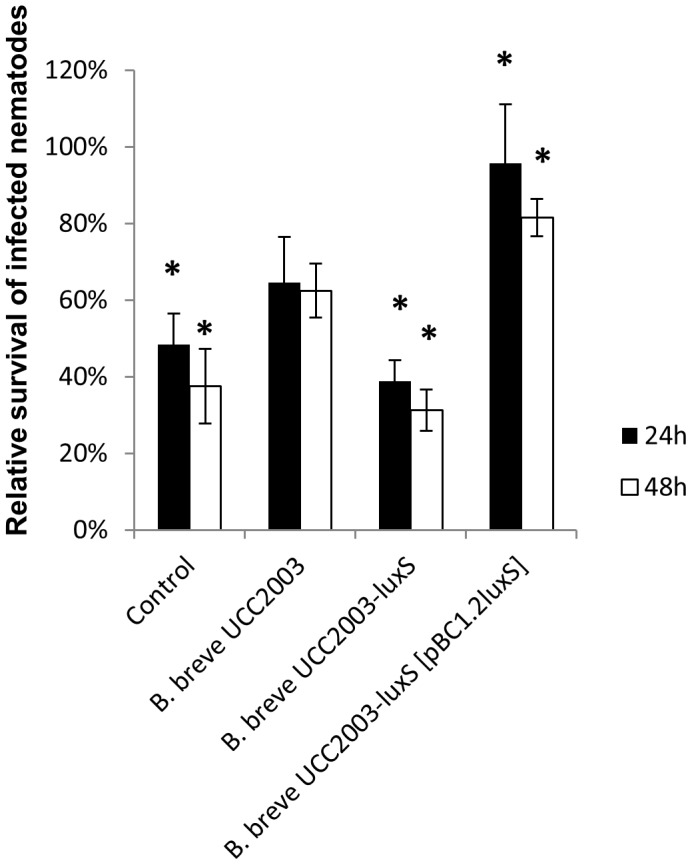
Data shown are means ± SEM. Control: infected nematodes that have not been administered any bifidobacteria. (* = significantly increased or decreased survival as compared to Salmonella-infected C. elegans nematodes to which B. breve UCC2003 WT was administered, p<0.05; n = 3).
Figure 6. Relative survival of Salmonella-infected C. elegans nematodes to which B. breve UCC2003 WT or mutants UCC2003-bfeU or UCC2003-bfeB were administered (24 h {blue bars} and 48 h {white bars} after Salmonella infection).
Data shown are means ± SEM. Control: infected nematodes that have not been administered any bifidobacteria. (* = significantly different as compared to the control, p<0.05; n = 3).
Discussion
Members of the genus Bifidobacterium are recognized as being numerically dominant representatives of the microbiota of healthy breast-fed infants [60], [61]. Colonization of the newborn infant gut commences during and after birth by microbes from the mother and the environment. A succession in the gut colonization of healthy vaginally-delivered infants has been reported, whereby initial colonization is by facultative anaerobes that include enterobacteria, staphylococci and streptococci. Once available oxygen in the gut is consumed the newly created anaerobic environment supports establishment of strict anaerobes of the genera Clostridium, Bacteroides and Bifidobacterium [62]. While this microbiota is recognized as optimal for healthy infants, an in depth understanding of molecular players involved in gut colonization and host protection by such bacteria, in particular Bifidobacterium sp., remains to be elucidated. The intricacies of host-microbe interactions in early life cannot be underestimated and the precise mechanisms by which elements of the infant gut microbiota contribute to health maintainence and promotion in early life are only beginning to emerge [63], [64].
The bacterial interspecies signaling molecule AI-2 is now well recognized for its role in the regulation of virulence factor production in pathogenic Gram-negative and Gram-positive bacteria [65]. These virulence-associated features include biofilm formation, toxin production, adherence to epithelial cells, motility as well as the metabolism of heavy metals and carbon [24]–[26]. In addition, the AI-2 synthase LuxS plays an important role in central metabolism, more specifically in the detoxification and recycling of S-adenosylhomocysteine [28]. Interestingly, all Bifidobacterium strains sequenced to date harbor a luxS gene and our investigations demonstrating that all tested bifidobacterial strains, representing 11 species of this genus, were capable of producing AI-2 is consistent with previous finding of [36] who detected AI-2 production for two Bifidobacterial species. In this respect it seemed intriguing that a gut commensal, dominant in the infant microbiota, would produce a molecule that potentially promotes the production of virulence factors in (opportunistic) gastrointestinal pathogens.
To further investigate this, B. breve UCC2003 was chosen as a representative of probiotic bifidobacteria. This strain, in addition to producing high levels of AI-2 in the biosensor assay, is a generally accepted model for the genus Bifidobacterium [8], [66]. Transcriptome analysis of B. breve UCC2003-luxS versus UCC2003, grown under in vitro conditions, revealed that the role of LuxS is primarily metabolic. These findings are supported by the fact that no AI-2 signal transduction pathways have previously been described in bifidobacteria and that a protein with high similarity to the known AI-2 receptor LuxP has not been identified from the genome of B. breve UCC2003. UCC2003-luxS was shown to be more sensitive to various iron chelators, and unable to colonize the murine gastrointestinal tract, while this mutant also conferred less protection against Salmonella infection in a C. elegans nematode model. These data demonstrate that LuxS plays a crucial role for bifidobacteria in their ability to establish themselves as gut commensals, which also includes their beneficial effect pertaining to pathogen protection/exclusion. Furthermore, our results show that LuxS activity is involved in iron acquisition, and we propose that this property gives B. breve UCC2003 a competitive advantage in iron-limited environments such as the gastrointestinal tract. The importance of iron acquisition mediated nutritional immunity in gut pathogen protection was further demonstrated by the fact that two additionally constructed mutants harboring insertions in either of two presumed iron-uptake genes proved to have a decreased ability to confer protection against Salmonella infection in the C. elegans model. In addition and as expected, these mutants were more susceptible to the iron chelators as compared to the parent strain B. breve UCC2003. It has previously been shown that LuxS affects genes involved in iron metabolism in Porphyromonas gingivalis [67], Vibrio vulnificus [68], Mannheimia haemolytica [69] and Actinobacillus pleuropneumoniae [70], while it was also demonstrated that iron availability increases the pathogenic potential of several gastrointestinal pathogens including S. Typhimurium, Citrobacter freundii, E. coli [71] and Listeria monocytogenes [72], [73]. Our results are consistent with the notion that bifidobacteria confer gut pathogen protection by nutritional immunity. This in turn suggests that LuxS/AI-2 can be versatile in various bacterial species and conditions. Since the colonization capacity of a (putative) probiotic bacterium is considered to be a prerequisite to exert its beneficial effects, the observation that AI-2-expressing B. breve UCC2003 outcompetes an isogenic derivative lacking this capacity contributes to the elucidation of molecular players and mechanisms of colonization requirements and probiotic effects [74].
Indeed, one application where administration of bifidobacterial strains may positively influence health is in the prevention of necrotizing enterocolitis (NEC) in premature infants. The precise causative agent of NEC is unknown; however, the preterm infant microbiota has been found to be dominated by pathogenic genera with Proteobacteria and Enterobacteriaceae dominating rather than characteristic species belonging to Bacteroidetes, Clostridium and Bifidobacterium [75]–[77]. The dominance of potentially pathogenic bacteria may increase the risk of infection in this vulnerable group. Neonatal nurseries in Finland, Italy and Japan have been routinely and successfully using probiotics as prophylaxis against NEC for over a decade, without ever reporting any adverse effects. Despite the safe use of practice and numerous randomized clinical trials that indicate that probiotics can reduce the incidence of NEC by at least 30% [78]–[80], clinical guidelines by the American Society for Parenteral and Enteral Nutrition (A.S.P.E.N) express the view that there is currently insufficient data to recommend the use of probiotics in infants at risk of NEC [81]. Integral to the resistance to adopt probiotics as a prophylaxis against NEC in premature infants is the lack of knowledge on the mechanism of action [82]. While the data presented here is merely one molecular mechanism and one probiotic attribute that is conserved among all bifidobacteria, this research provides a key insight into a mechanism of gut pathogen protection conferred by bifidobacteria that is of clinical relevance.
Supporting Information
Oligonucleotide primers used in this study
(DOCX)
Acknowledgments
The authors want to thank Nele Matthijs for excellent technical assistance.
Data Availability
The authors confirm that, for approved reasons, some access restrictions apply to the data underlying the findings. Identifying data could not be made freely available, but GenSalt may provide the association result for certain SNP and phenotypes. Requests for this data may be sent to Dr. Tanika N. Kelly.
Funding Statement
This work was financially supported by IWT Vlaanderen (Instituut voor Innovatie door Wetenschap en Technologie Vlaanderen), and the Interuniversity Attraction Poles Programme initiated by the Belgian Science Policy Office. This publication has emanated from research conducted with the financial support of a HRB postdoctoral fellowship (Grant no. PDTM/20011/9) awarded to MOCM and Science Foundation Ireland (SFI) awards under Grant Numbers 07/CE/B1368 and SFI/12/RC/2273; FB acknowledges financial assistance through an IRCSET postgraduate fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Isolauri E, Rautava S, Kalliomaki M, Kirjavainen P, Salminen S (2002) Role of probiotics in food hypersensitivity. Curr Opin Allergy Clin Immunol 2: 263–271. [DOI] [PubMed] [Google Scholar]
- 2. Butler CC, Duncan D, Hood K (2012) Does taking probiotics routinely with antibiotics prevent antibiotic associated diarrhoea? BMJ 344: e682. [DOI] [PubMed] [Google Scholar]
- 3. Gourbeyre P, Denery S, Bodinier M (2011) Probiotics, prebiotics, and synbiotics: impact on the gut immune system and allergic reactions. J Leukoc Biol 89: 685–695. [DOI] [PubMed] [Google Scholar]
- 4. Jonkers D, Penders J, Masclee A, Pierik M (2012) Probiotics in the management of inflammatory bowel disease: a systematic review of intervention studies in adult patients. Drugs 72: 803–823. [DOI] [PubMed] [Google Scholar]
- 5. Damman CJ, Miller SI, Surawicz CM, Zisman TL (2012) The microbiome and inflammatory bowel disease: is there a therapeutic role for fecal microbiota transplantation? Am J Gastroenterol 107: 1452–1459. [DOI] [PubMed] [Google Scholar]
- 6. de Vrese M, Schrezenmeir J (2008) Probiotics, prebiotics, and synbiotics. Adv Biochem Eng Biotechnol 111: 1–66. [DOI] [PubMed] [Google Scholar]
- 7. Corr SC, Gahan CG, Hill C (2007) Impact of selected Lactobacillus and Bifidobacterium species on Listeria monocytogenes infection and the mucosal immune response. FEMS Immunol Med Microbiol 50: 380–388. [DOI] [PubMed] [Google Scholar]
- 8. Fanning S, Hall LJ, Cronin M, Zomer A, MacSharry J, et al. (2012) Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc Natl Acad Sci U S A 109: 2108–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Siciliano RA, Mazzeo MF (2012) Molecular mechanisms of probiotic action: a proteomic perspective. Curr Opin Microbiol 15: 390–396. [DOI] [PubMed] [Google Scholar]
- 10. Reid G, Kim SO, Kohler GA (2006) Selecting, testing and understanding probiotic microorganisms. FEMS Immunol Med Microbiol 46: 149–157. [DOI] [PubMed] [Google Scholar]
- 11. Kim Y, Mylonakis E (2012) Caenorhabditis elegans immune conditioning with the probiotic bacterium Lactobacillus acidophilus strain NCFM enhances Gram-positive immune responses. Infect Immun 80: 2500–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Russell DA, Ross RP, Fitzgerald GF, Stanton C (2011) Metabolic activities and probiotic potential of bifidobacteria. Int J Food Microbiol 149: 88–105. [DOI] [PubMed] [Google Scholar]
- 13. Lebeer S, Vanderleyden J, De Keersmaecker SC (2010) Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat Rev Microbiol 8: 171–184. [DOI] [PubMed] [Google Scholar]
- 14. Bassler BL (1999) How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr Opin Microbiol 2: 582–587. [DOI] [PubMed] [Google Scholar]
- 15. Hodgkinson JT, Welch M, Spring DR (2007) Learning the language of bacteria. ACS Chem Biol 2: 715–717. [DOI] [PubMed] [Google Scholar]
- 16. Turovskiy Y, Kashtanov D, Paskhover B, Chikindas ML (2007) Quorum sensing: fact, fiction, and everything in between. Adv Appl Microbiol 62: 191–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Waters CM, Bassler BL (2005) Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21: 319–346. [DOI] [PubMed] [Google Scholar]
- 18. Pollumaa L, Alamae T, Mae A (2012) Quorum sensing and expression of virulence in pectobacteria. Sensors 12: 3327–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schuster M, Greenberg EP (2006) A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa . Int J Med Microbiol 296: 73–81. [DOI] [PubMed] [Google Scholar]
- 20. Sjoblom S, Brader G, Koch G, Palva ET (2006) Cooperation of two distinct ExpR regulators controls quorum sensing specificity and virulence in the plant pathogen Erwinia carotovora . Mol Microbiol 60: 1474–1489. [DOI] [PubMed] [Google Scholar]
- 21. Winson MK, Camara M, Latifi A, Foglino M, Chhabra SR, et al. (1995) Multiple N-acyl-L-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa . Proc Natl Acad Sci U S A 92: 9427–9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gordon CP, Williams P, Chan WC (2013) Attenuating Staphylococcus aureus virulence gene regulation: a medicinal chemistry perspective. J Med Chem 56: 1389–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Otto M (2012) Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu Rev Med 64: 175–188. [DOI] [PubMed] [Google Scholar]
- 24. Choi J, Shin D, Kim M, Park J, Lim S, et al. (2012) LsrR-mediated quorum sensing controls invasiveness of Salmonella typhimurium by regulating SPI-1 and flagella genes. PLoS One 7: e37059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Soni KA, Jesudhasan PR, Cepeda M, Williams B, Hume M, et al. (2008) Autoinducer AI-2 is involved in regulating a variety of cellular processes in Salmonella Typhimurium . Foodborne Pathog Dis 5: 147–153. [DOI] [PubMed] [Google Scholar]
- 26. Surette MG, Bassler BL (1999) Regulation of autoinducer production in Salmonella typhimurium . Mol Microbiol 31: 585–595. [DOI] [PubMed] [Google Scholar]
- 27. Kim SY, Lee SE, Kim YR, Kim CM, Ryu PY, et al. (2003) Regulation of Vibrio vulnificus virulence by the LuxS quorum-sensing system. Mol Microbiol 48: 1647–1664. [DOI] [PubMed] [Google Scholar]
- 28. Xavier KB, Bassler BL (2003) LuxS quorum sensing: more than just a numbers game. Curr Opin Microbiol 6: 191–197. [DOI] [PubMed] [Google Scholar]
- 29. Xavier KB, Bassler BL (2005) Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli . J Bacteriol 187: 238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rezzonico F, Duffy B (2008) Lack of genomic evidence of AI-2 receptors suggests a non-quorum sensing role for luxS in most bacteria. BMC Microbiol 8: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sun J, Daniel R, Wagner-Dobler I, Zeng AP (2004) Is autoinducer-2 a universal signal for interspecies communication: a comparative genomic and phylogenetic analysis of the synthesis and signal transduction pathways. BMC Evol Biol 4: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Buck BL, Azcarate-Peril MA, Klaenhammer TR (2009) Role of autoinducer-2 on the adhesion ability of Lactobacillus acidophilus . J Appl Microbiol 107: 269–279. [DOI] [PubMed] [Google Scholar]
- 33. Lebeer S, Claes IJ, Verhoeven TL, Shen C, Lambrichts I, et al. (2008) Impact of luxS and suppressor mutations on the gastrointestinal transit of Lactobacillus rhamnosus GG. Appl Environ Microbiol 74: 4711–4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lebeer S, De Keersmaecker SC, Verhoeven TL, Fadda AA, Marchal K, et al. (2007) Functional analysis of luxS in the probiotic strain Lactobacillus rhamnosus GG reveals a central metabolic role important for growth and biofilm formation. J Bacteriol 189: 860–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wilson CM, Aggio RB, O'Toole PW, Villas-Boas S, Tannock GW (2012) Transcriptional and metabolomic consequences of LuxS inactivation reveal a metabolic rather than quorum-sensing role for LuxS in Lactobacillus reuteri 100-23. J Bacteriol 194: 1743–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sun Z, He X, Brancaccio VF, Yuan J, Riedel CU (2014) Bifidobacteria exhibit LuxS-dependent autoinducer 2 activity and biofilm formation. PLoS One9: e88260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Janssens JC, Steenackers H, Metzger K, Daniels R, Ptacek D, et al. (2007) Interference with the quorum sensing systems of Salmonella enterica serovar typhimurium: possibilities and implications. Commun Agric Appl Biol Sci 72: 35–39. [PubMed] [Google Scholar]
- 38. Bassler BL, Wright M, Showalter RE, Silverman MR (1993) Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol 9: 773–786. [DOI] [PubMed] [Google Scholar]
- 39. Masco L, Huys G, Gevers D, Verbrugghen L, Swings J (2003) Identification of Bifidobacterium species using rep-PCR fingerprinting. Syst Appl Microbiol 26: 557–563. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J. 2001 Molecular Cloning: a laboratory manual (.Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY) 3rd Ed [Google Scholar]
- 41. O'Connell Motherway M, O'Driscoll J, Fitzgerald GF, van Sinderen D (2009) Overcoming the restriction barrier to plasmid transformation and targeted mutagenesis in Bifidobacterium breve UCC2003. Microb Biotechnol 2: 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, et al. (2000) Artemis: sequence visualization and annotation. Bioinformatics 16: 944–945. [DOI] [PubMed] [Google Scholar]
- 43. O'Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, et al. (2011) Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci U S A 108: 11217–11222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Law J, Buist G, Haandrikman A, Kok J, Venema G, Leenhouts K (1995) A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J Bacteriol 177: 7011–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Alvarez-Martin P, O'Connell-Motherway M, van Sinderen D, Mayo B (2007) Functional analysis of the pBC1 replicon from Bifidobacterium catenulatum L48. Appl Microbiol Biotechnol 76: 1395–1402. [DOI] [PubMed] [Google Scholar]
- 46. van Hijum SA, de Jong A, Baerends RJ, Karsens HA, Kramer NE, et al. (2005) A generally applicable validation scheme for the assessment of factors involved in reproducibility and quality of DNA-microarray data. BMC Genomics 6: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Long A D, Mangalam HJ, Chan BY, Tolleri L, Hatfield GW, Baldi P (2001) Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J Biol Chem. 276: 19937–19944. [DOI] [PubMed] [Google Scholar]
- 49. EUCAST (2003) Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin Microbiol Infect 9: 1–7. [DOI] [PubMed] [Google Scholar]
- 50. Cooper VS, Carlson WA, LiPuma JJ (2009) Susceptibility of Caenorhabditis elegans to Burkholderia infection depends on prior diet and secreted bacterial attractants. PLoS One 4: e7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stiernagle T (2006) Maintenance of C. elegans. WormBook: 1–11. [DOI] [PMC free article] [PubMed]
- 52. Garsin DA, Sifri CD, Mylonakis E, Qin X, Singh KV, et al. (2001) A simple model host for identifying Gram-positive virulence factors. Proc Natl Acad Sci U S A 98: 10892–10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Portal-Celhay C, Blaser MJ (2012) Competition and resilience between founder and introduced bacteria in the Caenorhabditis elegans gut. Infect Immun 80: 1288–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sivamaruthi BS, Ganguli A, Kumar M, Bhaviya S, Pandian SK, et al. (2011) Caenorhabditis elegans as a model for studying Cronobacter sakazakii ATCC BAA-894 pathogenesis. J Basic Microbiol 51: 540–549. [DOI] [PubMed] [Google Scholar]
- 55. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Brackman G, Cos P, Maes L, Nelis HJ, Coenye T (2011) Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo . Antimicrob Agents Chemother 55: 2655–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cronin M, Zomer A, Fitzgerald GF, van Sinderen D (2012) Identification of iron-regulated genes of Bifidobacterium breve UCC2003 as a basis for controlled gene expression. Bioeng Bugs 3: 157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Clark LC, Hodgkin J (2014) Commensals, probiotics and pathogens in the Caenorhabditis elegans model. Cell Microbiol 16: 27–38. [DOI] [PubMed] [Google Scholar]
- 59. Ikeda T, Yasui C, Hoshino K, Arikawa K, Nishikawa Y (2007) Influence of lactic acid bacteria on longevity of Caenorhabditis elegans and host defense against Salmonella enterica serovar enteritidis. Appl Environ Microbiol 73: 6404–6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, et al. (2012) Diversity of Bifidobacteria within the Infant Gut Microbiota. PLoS One 7: e36957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. O'Toole PW, Claesson MJ (2010) Gut microbiota: Changes throughout the lifespan from infancy to elderly. Int Dairy J 20: 281–291. [Google Scholar]
- 62. Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, et al. (2011) Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 108 Suppl 14578–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Turroni F, Ventura M, Buttó LF, Duranti S, O'Toole PW, et al. (2014) Molecular dialogue between the human gut microbiota and the host: a Lactobacillus and Bifidobacterium perspective. Cell Mol Life Sci 71: 183–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ventura M, Turroni F, O'Connell Motherway M, MacSharry J, van Sinderen D (2012) Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol 20: 467–476. [DOI] [PubMed] [Google Scholar]
- 65. Pereira CS, Thompson JA, Xavier KB (2013) AI-2-mediated signalling in bacteria. FEMS Microbiol Rev 37: 156–181. [DOI] [PubMed] [Google Scholar]
- 66. Fanning S, Hall LJ, van Sinderen D (2012) Bifidobacterium breve UCC2003 surface exopolysaccharide production is a beneficial trait mediating commensal-host interaction through immune modulation and pathogen protection. Gut Microbes 3: 420–425. [DOI] [PubMed] [Google Scholar]
- 67. James CE, Hasegawa Y, Park Y, Yeung V, Tribble GD, et al. (2006) LuxS involvement in the regulation of genes coding for hemin and iron acquisition systems in Porphyromonas gingivalis . Infect Immun 74: 3834–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kim SY, Lee SE, Kim YR, Kim CM, Ryu PY, et al. (2003) Regulation of Vibrio vulnificus virulence by the LuxS quorum-sensing system. Mol Microbiol 48: 1647–1664. [DOI] [PubMed] [Google Scholar]
- 69. van der Vinne AN, Lo RY, Shewen PE (2005) Construction and analysis of a Mannheimia haemolytica A1 luxS mutant. Vet Microbiol 110: 53–66. [DOI] [PubMed] [Google Scholar]
- 70. Li L, Xu Z, Zhou Y, Li T, Sun L, et al. (2011) Analysis on Actinobacillus pleuropneumoniae LuxS regulated genes reveals pleiotropic roles of LuxS/AI-2 on biofilm formation, adhesion ability and iron metabolism. Microb Pathog 50: 293–302. [DOI] [PubMed] [Google Scholar]
- 71. Kortman GA, Boleij A, Swinkels DW, Tjalsma H (2012) Iron availability increases the pathogenic potential of Salmonella typhimurium and other enteric pathogens at the intestinal epithelial interface. PLoS One 7: e29968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Brown JS, Holden DW (2002) Iron acquisition by Gram-positive bacterial pathogens. Microbes Infect. 4: 1149–1156. [DOI] [PubMed] [Google Scholar]
- 73. Gray MJ, Freitag NE, Boor KJ (2006) How the bacterial pathogen Listeria monocytogenes mediates the switch from environmental Dr. Jekyll to pathogenic Mr. Hyde. Infect Immun 74: 2505–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rautava S, Luoto R, Salminen S, Isolauri E (2012) Microbial contact during pregnancy, intestinal colonization and human disease. Nat Rev Gastroenterol Hepatol 9: 565–576. [DOI] [PubMed] [Google Scholar]
- 75. Mshvildadze M, Neu J (2010) The infant intestinal microbiome: Friend or foe? Early Hum Dev 86: S67–S71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Carlisle EM, Morowitz MJ (2013) The intestinal microbiome and necrotizing enterocolitis. Curr Opin Pediatr 25: 382–387. [DOI] [PubMed] [Google Scholar]
- 77. Barrett E, Kerr C, Murphy K, O'Sullivan O, Ryan CA, et al. (2013) The individual-specific and diverse nature of the preterm infant microbiota. Arch Dis Childhood Fet Neonatal Ed 98: F334–340. [DOI] [PubMed] [Google Scholar]
- 78. Li D, Rosito G, Slagle T (2013) Probiotics for the prevention of necrotizing enterocolitis in neonates: an 8-year retrospective cohort study. J Clin Pharm Ther 38: 445–449. [DOI] [PubMed] [Google Scholar]
- 79. Chen CC, Allan Walker W (2013) Probiotics and the mechanism of necrotizing enterocolitis.Semin Pediatr Surg. 22: 94–100. [DOI] [PubMed] [Google Scholar]
- 80. Ofek Shlomai N, Deshpande G, Rao S, Patole S (2014) Probiotics for preterm neonates: what will it take to change clinical practice? Neonatology 105: 64–70. [DOI] [PubMed] [Google Scholar]
- 81. Lin HY, Chang JH, Chung MY, Lin HC (2013) Prevention of necrotizing enterocolitis in preterm very low birth weight infants: Is it feasible? J Form Med Ass http://dx.doi.org/10.1016/j.jfma.2013.03.010 [DOI] [PubMed] [Google Scholar]
- 82. Cabana MD (2010) Evidence increasing that probiotics reduce incidence of necrotizing enterocolitis in very low birth weight infants. J Pediatr 157: 864–865. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligonucleotide primers used in this study
(DOCX)
Data Availability Statement
The authors confirm that, for approved reasons, some access restrictions apply to the data underlying the findings. Identifying data could not be made freely available, but GenSalt may provide the association result for certain SNP and phenotypes. Requests for this data may be sent to Dr. Tanika N. Kelly.