Abstract
RNA interference (RNAi) mediated by small interfering RNAs (siRNAs) is a powerful new tool for analyzing gene knockdown phenotypes in living mammalian cells. To facilitate large-scale, high-throughput functional genomics studies using RNAi, we have developed a microarray-based technology for highly parallel analysis. Specifically, siRNAs in a transfection matrix were first arrayed on glass slides, overlaid with a monolayer of adherent cells, incubated to allow reverse transfection, and assessed for the effects of gene silencing by digital image analysis at a single cell level. Validation experiments with HeLa cells stably expressing GFP showed spatially confined, sequence-specific, time- and dose-dependent inhibition of green fluorescence for those cells growing directly on microspots containing siRNA targeting the GFP sequence. Microarray-based siRNA transfections analyzed with a custom-made quantitative image analysis system produced results that were identical to those from traditional well-based transfection, quantified by flow cytometry. Finally, to integrate experimental details, image analysis, data display, and data archiving, we developed a prototype information management system for high-throughput cell-based analyses. In summary, this RNAi microarray platform, together with ongoing efforts to develop large-scale human siRNA libraries, should facilitate genomic-scale cell-based analyses of gene function.
Genomic sequencing and gene expression profiling have generated and continue to generate extensive information on the structure, sequence variation, and expression levels of human genes. However, high-throughput methods for investigating the significance of human genes in specific cell functions are not available at present (Mousses et al. 2002). RNA interference (RNAi) is a natural mechanism for sequence-specific, posttranscriptional gene silencing triggered by double-stranded RNA (dsRNA; Fire et al. 1998; Kennerdell and Carthew 1998; Misquitta and Paterson 1999). In Caenorhabditis elegans, RNAi triggered by long dsRNAs (over ∼80 bp) has been applied in large-scale studies to generate parallel “somatic knockdowns” for the functional analysis of thousands of genes (Fraser et al. 2000; Gönczy et al. 2000; Piano et al. 2000, 2002; Ashrafi et al. 2003; Kamath et al. 2003; Lee et al. 2003; Pothof et al. 2003). For example, in initial screens of >4000 transcripts, a phenotypic endpoint, including effects on embryonic viability, developmental delay, and sterility, was assigned to ∼500 genes with no previous known function (Fraser et al. 2000; Gönczy et al. 2000). A subsequent study of >16,000 transcripts (nearly 90% of the predicted genes of C. elegans) assigned 10% of these transcripts, the majority of which had not been previously identified using traditional approaches, to a definable phenotype (Kamath et al. 2003). Additional, more specific studies have investigated sets of 750 to 5000 genes associated with embryogenesis, mitochondrial function, fat metabolism, and suppression of mutagenesis (Piano et al. 2000, 2002; Ashrafi et al. 2003; Lee et al. 2003; Pothof et al. 2003). A genome-wide screen using RNAi is also presently underway in Drosophila (http://genetics.med.harvard.edu/~perrimon/RNAiGenomeProject.html; RNAi Genome Project).
In mammalian cells, shorter synthetic 20-23-nt dsRNA molecules or small interfering RNAs (siRNAs; Tuschl et al. 1999) can induce RNAi, without activation of the nonspecific dsRNA-dependent pathways triggered by longer dsRNAs in these cells (Caplen et al. 2001; Elbashir et al. 2001). Chemically or enzymatically synthesized siRNAs, as well as plasmids expressing short hairpin RNAs (shRNAs), have been extensively applied to the study of function of individual mammalian genes (for review, see McManus and Sharp 2002). Systematic, large-scale analyses of gene function using RNAi in mammalian cells is also likely to be highly informative. Efforts are now under way to create large siRNA-based libraries targeting human genes on a genomic scale using RNAi (Hannon 2002; Paddison and Hannon 2002; Frankish 2003). However, technologies, strategies, and information management systems compatible with high-throughput cell-based analyses will be required to make full use of these libraries. Here, we have developed and validated a system for efficient, uniform, and miniaturized analysis of multiple RNAi events in cultured human cells in a microarray format. siRNAs were first arrayed on glass slides, overlaid with a monolayer of cells, and incubated to allow transfection. The phenotypic effects of gene silencing were then quantified by digital image analysis at a single-cell level. A prototype Web-based information management system for cell-based microarray analysis was also developed. The RNAi microarray format provides potent, sequence-specific, and spatially confined gene silencing, with the potential to enable efficient cell-based analyses of gene function on a genome scale.
RESULTS
To test if it is possible to achieve measurable RNAi-induced gene knockdowns and phenotypes in mammalian cells on a microarray platform, we arrayed siRNAs corresponding to the enhanced Green Fluorescent Protein (GFP) gene (egfp) on microscope slides, and plated HeLa cells permanently expressing a destabilized version of eGFP (HeLa/d2eGFP cells) on top of the slides. Figure 1 describes the RNAi microarray process (Fig. 1A), and shows the cellular uptake, “reverse transfection” (Ziauddin and Sabatini 2001), of rhodamine-tagged siRNA (rh-egfp), and silencing of GFP expression in HeLa/d2eGFP cells (Fig. 1B,C). The uptake of rh-egfp siRNA led to a spatially confined silencing of the GFP expression, as visualized by “black holes”, where green fluorescence is completely missing in the monolayer of the green HeLa/d2eGFP cells (Fig. 1B). The affected cells within the rh-egfp siRNA microarray spot were localized by DAPI (4′6-diamidine-2-phenyl indole) staining of their nuclei and showed no indication of cytotoxicity. The siRNAs were printed in a cationic lipid/Matrigel mixture, which helped to confine the spread of the siRNA, leading to a clearly defined edge to the spot (Fig. 1D), as indicated by potent inhibition of green fluorescence within the spot (Fig. 1E) and little or no inhibition surrounding the spot (Fig. 1F). Together, these observations show that arrayed siRNAs on the surface of a glass slide can enter cells via transfection, and mediate potent inhibition of the target protein expression in a spatially confined manner.
Figure 1.
The RNAi microarray platform. (A) HeLa cells expressing a destabilized version of eGFP (HeLa/d2eGFP) cells were plated on slides arrayed with an egfp siRNA (2.5 ng) tagged with fluorescent rhodamine (rh-egfp). HeLa/d2eGFP cells grown on an rh-egfp siRNA microarray slide for 72 h were fixed with 2% paraformaldehyde and counterstained with DAPI. (B) An rh-egfp spot (2.5 ng siRNA) showing the localized inhibition of green fluorescence (25×, green channel only). (C) The uptake of rh-egfp siRNA-lipoplexes showing cellular accumulation of the siRNA in silenced cells (1000×). (D) A clear edge to the arrayed rh-egfp siRNA spot was present only in the green channel (200×, overlaid green and blue images), with (E) inhibition of eGFP fluorescence occurring within the spot (400×, overlaid green and blue images). (F) No inhibition of eGFP fluorescence in cells was observed around the rh-egfp siRNA spot (400×, overlaid green and blue images).
The assessment of the functional effects of RNAi in a microarray format requires quantitative image analysis with single-cell resolution for thousands of array elements. Conventional microarray laser scanners have insufficient resolution (maximum resolution of 5 μm) for the visualization of cellular and subcellular details. We therefore developed an automated, high-resolution, microscope-based digital image acquisition system for RNAi microarrays as well as custom algorithms for quantitative image analysis. Three images for three different fluorescent channels were acquired from one area within each microarray element/spot. The blue channel was used for DAPI counterstaining (cell enumeration and defining the nuclear compartment), the green for a phenotypic marker (such as for eGFP expression in this study), and the red channel for a second marker (such as another phenotypic marker or uptake of rhodamine-tagged siRNAs in this study). The population of cells in the fluorescent image was segmented to identify all the individual cell areas, and the images were analyzed to extract multiple parameters from each individual cell area (including the minimum, maximum, mean, median, and total fluorescence intensity; see Methods and Supplemental Material for additional details). Separate extraction and quantification of nuclear and cytoplasmic staining was also developed based on segmentation of the nuclear area.
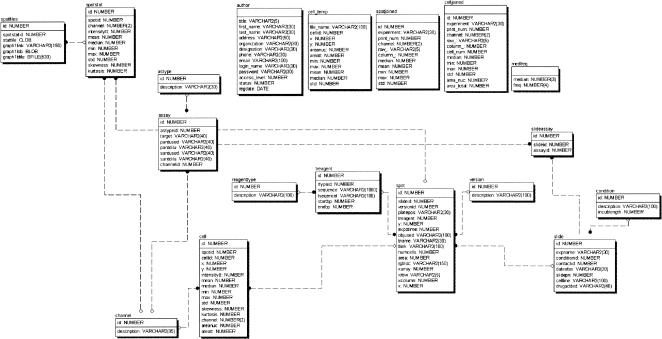
Summary statistics for these parameters were calculated for all the cells in each image from an array element. A pilot version of an information management system was created to integrate a wide range of information and data (Fig. 2). This Web-based database warehoused and helped to manage images from each spot and various levels of data including the experimental conditions and assays in each fluorescent channel, siRNA sequence contained in each microarray element, image archiving with file annotation (indicating, e.g., resolution, exposure time, and filter), image processing, segmentation and analysis, statistical analysis, and visualization of quantitative data (Fig. 2). Representative images, image processing, results from the statistical analysis of the data, and comparison from parallel experiments using flow cytometry data are shown in Figure 3 for two different siRNA microarray spots arrayed on the same microscope slide. The percentage of GFP-positive HeLa/d2eGFP cells declined from 79% (in the negative control spot) to 19% (egfp siRNA spot).
Figure 2.
The entity relationship diagram shows the relational model applied to tracking data on individual experiments. The fields and their relationships for the various levels of information are shown by connecting lines. System users are able to load, retrieve, and update experiment information in the database via a Web interface.
Figure 3.
Single-cell level quantification of RNAi on a microarray platform. The imaging and quantification of fluorescence in HeLa/d2eGFP cells plated on two representative siRNA spots (a negative control cat siRNA spot [A-F] and an experimental rh-egfp siRNA spot [G-L], 2.5 ng of siRNA per spot) on a microarray slide. To quantify the effect of the siRNAs on GFP silencing, green fluorescence DAPI-stained nuclei (A,G) and “watershed” boundaries (defined using the green fluorescence images, B and H) were used to locate and define the area for the cells (white) in each of the two images (C and I; see Supplemental Fig. A). These segmented images were used to measure single-cell parameters used in subsequent data analysis (Supplemental Material, data sets 1 and 2). (D,J) For each spot, the distribution of the median green fluorescence (after local background fluorescence subtraction) in every segmented cell was plotted. A cell was considered to be positive for eGFP green fluorescence when its median fluorescence intensity was >4 units in the green channel. The distributions of the median green fluorescence intensities obtained by microarray analysis (D,J) were compared with traditional flow cytometric measurements of Hela/d2eGFP cells (10,000 events) transfected with the same siRNAs (E,K). The vertical line indicates the estimated level of background fluorescence. The degree of gene silencing detected by flow cytometry was consistent with that detected by the RNAi microarray imaging system. Statistical analysis (F,L) indicated a decrease in the green fluorescence intensities when the data fromthe cat siRNA and an rh-egfp siRNA spot were compared, whereas other parameters, such as the total cell area and nuclear area were similar in the two spots.
To demonstrate the sensitivity and specificity of RNAi microarray platform and quantitative image analysis, we conducted the following validation experiments. First, a significant time-dependent reduction in eGFP fluorescence was seen (p < 0.001) in spots containing the rh-egfp siRNA (Fig. 4A). Second, a modest effect of dose, consistent with previous observations by other methods (Caplen et al. 2001), was observed at both time points within rh-egfp siRNA spots, but not within the negative control siRNA spots (Fig. 4B,C). Third, other parameters, such as mean cell areas (Fig. 4D), were not affected by either of the two siRNAs. Fourth, three siRNAs corresponding to different sequences within the egfp gene showed differential effectiveness in reducing eGFP expression (Fig. 4E). Finally, the RNAi microarray results for these three egfp siRNAs produced similar results to those obtained using parallel flow cytometry measurements (Fig. 4F). Given this variability in efficacy of different siRNAs targeting the same gene, the data demonstrate that RNAi microarrays can also be used to quantitatively assess and optimize siRNA design.
Figure 4.
Assessment of the sensitivity and specificity of the RNAi microarray platform. (A) An analysis of eGFP expression (median green fluorescence intensity) in HeLa/d2eGFP cells over time following exposure to either arrayed egfp (black bars) or negative control (cat) siRNAs (gray bars; 2.5 ng siRNA per spot). The inhibition of eGFP expression by arrayed egfp siRNAs (black bars) is dose-dependent (0.25 ng, 1 ng, or 2.5 ng of siRNAs), and this effect can be observed over time. (B) 24 h; (C) 72 h. No change was seen in the fluorescence intensity of Hela/d2eGFP cells exposed to the negative control cat siRNAs (gray bars). (D) Cells exposed to arrayed siRNAs (negative control, gray bars; experimental siRNAs, black bars) were of the same size and were otherwise comparable; see Supplemental Material for additional statistics. Three different siRNAs against egfp were compared for their ability to inhibit eGFP expression using RNAi microarray (2.5 ng siRNA) (E) and flow cytometry (2 μg siRNA was transfected using Lipofectin into ∼2 × 105 HeLa/d2eGFP cells) (F). (A-E) The average median green fluorescence of triplicate siRNA spots is plotted with the error bar representing 1 SD; (F) the average geometric mean green fluorescence of three independent transfections is plotted with the error bar representing 1 SD.
DISCUSSION
The experiments described here have shown sequence-, time-, and dose-dependent inhibition of gene expression using a reverse transfection RNAi microarray. The RNAi microarray is flexible, robust, and has a higher capacity than presently available well-based platforms. Coupled with the cellular-level quantitative digital image analysis and the information management system developed here, this RNAi microarray technology has the potential to facilitate and accelerate the screening of the emerging genome-scale siRNA libraries for phenotypic effects of gene silencing in cultured mammalian cells.
One of the key advantages of the RNAi microarray for genome-scale screening is that it permits miniaturization far beyond what is physically possible in present well-based systems. At present, most cell-culture-based assays of gene function make use of 6-, 12-, 24-, or 96-well plate formats. Although RNAi experiments can be conducted in a 384-well plate format, a “wellless” microarray platform will be significantly more effective. Even though the size and density of the spots are limited by the need to have at least several dozens if not hundreds of cells on each spot, it should be feasible to array and analyze as many as 5000 to 10,000 individual siRNAs spots per slide. In these pilot experiments, we used spots that were 100-700 μm in diameter. Another advantage of the array-based miniaturization is the reduction in reagent costs. In particular, chemically synthesized siRNAs are expensive and are produced in relatively small quantities. We estimate that at least 50 times less siRNA can be used in a microspot on an RNAi microarray compared with experiments in a typical 96-well plate. The RNAi microarray format will also reduce the cost of the phenotypic assays because only a single microscope slide needs to be subjected to analysis. The well-less nature of the platform also allows for improving the level of uniformity in the experimental culturing conditions and assay conditions, because all processes take place on the same surface. In this study, we used a fluorescent endpoint, but any assay that can be conducted on cells growing on a glass slide can be applied to the RNAi microarray. Additional means of assaying mRNA, protein, and other biomolecules include chemiluminescent, colorimetric, or radioactive detection. This should enable analysis of RNAi knockdowns using a wide range of molecular and phenotypic endpoints such as toxicity, differentiation, proliferation, and cell cycle effects in fixed or live cells. These types of assays would be used to directly evaluate gene function. For example, gene silencing resulting in cytotoxicity, as assayed by detecting and quantifying changes in cell number, cell morphology, as well as more specific markers of cell death, could lead to the identification of genes with a functional role in cell survival.
To achieve measurable and quantitative phenotypes in a microarray format, it was necessary to overcome a number of technical problems. These included optimizing the conditions required for efficient transfection, while retaining cell attachment and restricting the lateral spread of the siRNA-lipid complex. A limitation intrinsic to the RNAi microarray platform is that only adherent cells can be used. As with any other platform using cationic lipid agents for transfection of siRNAs, this RNAi microarray system has the disadvantage that the specific agents and conditions for transfection may be very cell-type-specific, and thus optimization of the formulation may be required for each cell type. The lipid choice in this study was based on optimization experiments with the same cell line using traditional plate-based transfection. For optimization on glass slides, multiple parameters including dose of lipid and ratio of lipid to siRNA were also evaluated for their effect on transfection from printed spots. The protocol presented was the most consistent for maximal transfection efficiency off the glass slide in the HeLa cell line used in this study. Further development of more efficient transfection systems and polymers with suitable diffusion characteristics could potentially improve this technology. In addition, procedures such as freeze drying or desiccation could be used to increase the potential for long-term storage of RNAi microarray slides.
To enable the analysis of the effects of gene silencing, we developed image analysis tools that resolved phenotypic changes at a single cell (and even subcellular) level. Furthermore, a prototype information management system was developed that enabled storage and analysis of potentially hundreds of gigabytes of images for each experiment. The system was designed with a Web-based interface, with capabilities for tracking experimental data, image uploading, algorithms for off-line image analysis, histogram data display, and statistical analyses, as well as tracking of data on a cell-by-cell, image-by-image, and microarray spot-by-spot basis. This kind of sophisticated infrastructure will be needed for future large-scale experimentation, whereas most laboratories can begin analyses with regular quantitative image analysis systems. Our information management prototype illustrates the challenges involved in the development of capabilities for processing, archiving, analyzing, and integrating image-based data of cell biological experiments in a genomic scale. Such online, Web-based database capabilities will be required for integrating molecular databases with phenotypic and morphological information of cell functions.
In conclusion, we have developed and validated RNAi microarray technology for high-throughput cellular analysis of gene function and demonstrated its capabilities in measuring the effects of transient gene silencing mediated by siRNAs. The RNAi microarray platform is flexible and expandable for parallel RNAi-mediated functional characterization of genes on a genome scale when combined with automated digital image analysis systems and an integrated information management system as described here. We expect that large human siRNA libraries akin to those developed for C. elegans and Drosophila will soon be widely available, making genomic-scale analyses of gene functions possible in living mammalian cells using the RNAi microarray platform.
METHODS
Nucleic Acids
Single-stranded RNA (ssRNA) oligonucleotides were chemically synthesized and HPLC-purified by Xeragon Inc. Duplex RNA molecules were generated as previously described (Caplen et al. 2001). The integrity of the siRNAs was confirmed by gel electrophoresis. To assess cellular localization of siRNAs, an egfp siRNA was synthesized with a rhodamine (TAMRA)(rh-egfp siRNA) added to the 3′ end of the sense strand of the RNA duplex; this addition has no effect on the efficacy with which the dsRNA mediates gene silencing (N.J. Caplen, unpubl.). The sequences of the RNA oligonucleotides used to form the siRNAs are: cat (CAT, chloramphenicol acetyl transferase) sense, 5′-PO4r(gagugaauaccacgacgauuuc)-3′; cat antisense, 5′-PO4r(aaucgucgugguauucacucca)-3′; egfp sense, 5′-PO4r(gcaagcugacccugaaguucau)-3′; egfp antisense, 5′-PO4r(gaacuucagggucagcuugccg)-3′; egfpB sense, 5′-PO4r(cccugaaguucatcugcaccac)-3′; egfpB antisense, 5′-PO4r(ggugcagaugaacuucagggtc)-3′; egfpC sense, 5′-PO4r(caucugcaccaccggcaagcug)-3′; egfpC antisense, 5′-PO4r(gcuugccgguggugcagaugaa)-3′. Plasmid pEGFPd2-N3 (Clontech) expresses a mammalian enhanced version of green fluorescent protein with a defined protein half-life of 2 h.
Cell Culture
HeLa cells, a human epithelial cell line derived from a cervical adenocarcinoma (American tissue culture collection, ATTC, Manassas, VA), were grown in Dulbecco's Modified Eagle's Medium (Life Technologies) supplemented with 10% fetal bovine serum (FBS; Gemini BioProducts Inc.)(D10) at 37°C/5% CO2. HeLa/d2eGFP cells stably expressing the destabilized version of EGFP were generated by cationic lipid transfection of the pd2EGFP-N3 (Clontech) plasmid and selection with 1 mg/mL G418. To obtain a population of GFP-positive cells with a narrow range of fluorescence intensity, cells were subjected to flow cytometric sorting.
RNAi Microarray
For transfection on a microarray platform, a lipoplex of siRNA (0.25 μg to 2.5 μg) and lipid (Lipofectin, Life Technologies; 1:2 wt:wt RNA:lipid) was formed, at room temperature, over a period of 15 min in OptiMEM (Life Technologies) supplemented with 0.2 M sucrose in a final volume of 100 μL. After formation of the lipoplex, 0.5-1 μL of the basement membrane matrix Matrigel (BD Biosciences Discovery Labware) was added, and between 0.1 and 1 μL (0.25 ng-2.5 ng siRNA) of the mixture was arrayed on to poly-lysine coated plates (Mousses et al. 2000). We have successfully arrayed and analyzed siRNA spots between 100 and 500 μM in diameter and with a center-center distance of 300-1000 μM apart. Slides were pretreated in OptiMEM at 37°C/5% CO2 for 1 h to remove surface siRNAs. Slides were transferred to fresh sterile dishes (10 cm2) and cells in OptiMEM medium (10 mL) were plated onto the slides at a concentration of ∼5 × 105 cells/mL. Approximately 18 h after cell plating, an equal volume of growth medium supplemented with 20% FBS was added. Cells were assayed 24 to 72 h after the plating of cells.
Imaging, Quantification, Database, and Statistical Analysis
An AxioPlan2 microscope fitted with a Marchauser 8 position stage and an Axiocam CCD digital camera (Carl Zeiss GmbH) at 25- to 1000-fold magnification was applied for automated image acquisition (100-200-fold magnification) using a custom macro written in the scripting language of the KS400 (Zeiss) software package. A multistep process was used to generate binary images for measuring fluorescence intensities in each channel for segmented cells. The steps in the image analysis for the present application were: (1) the DAPI (blue channel) images and the green image were regionally thresholded to generate binary images. (2) These two binary images were combined to produce a merged image of the union of the two thresholds. (3) The green image was analyzed to determine the graduations in the green signal intensity gradients, and “watershed” lines were placed at the inflection points in the valleys of this green signal using the DAPI-derived binary image as a marker. (4) The merged binary image in step 2 was now combined with the green image subjected to watershedding to locate and define the precise area of every cell in the image. (5) Partial cells and noise were subtracted to produce a fully segmented image suitable for extraction of multiparametric data (see Supplemental Material for images reflecting each step in this process). Statistical analysis of the multiparametric data including generation of global statistics of cell populations in each image was performed automatically with the help of MATLAB 6 (The MathWorks, Inc.). Local background was calculated and subtracted from the global statistics. All statistical tests in this study were performed using a two-tailed Student's t-test. A live-cell RNAi microarray Web-based database schema was developed using a J2EE architecture and Oracle 8i database.
Acknowledgments
We thank Stacie Anderson and Martha Kirby for assistance with the sorting of the HeLa/d2EGFP cells, Ken Pratt (Zeiss) for assistance in coding the image acquisition and autofocusing routines, Darryl Leja for assistance with graphics, and Max Muenke for useful discussion and encouragement.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
[Supplemental Material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1478703.
References
- Ashrafi, K., Chang, F.Y., Watts, J.L., Fraser, A.G., Kamath, R.S., Ahringer, J., and Ruvkun, G. 2003. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421: 268-272. [DOI] [PubMed] [Google Scholar]
- Caplen, N.J., Parrish, S., Imani, F., Fire, A., and Morgan, R.A. 2001. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. 98: 9742-9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494-498. [DOI] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806-811. [DOI] [PubMed] [Google Scholar]
- Frankish, H. 2003. Consortium uses RNAi to uncover genes' function. Lancet 361: 584. [DOI] [PubMed] [Google Scholar]
- Fraser, A.G., Kamath, R.S., Zipperlen, P., Martinez-Campos, M., Sohrmann, M., and Ahringer, J. 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408: 325-330. [DOI] [PubMed] [Google Scholar]
- Gönczy, P., Echeverri, G., Oegema, K., Coulson, A., Jones, S.J., Copley, R.R., Duperon, J., Oegema, J., Brehm, M., Cassin, E., et al. 2000. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408: 331-336. [DOI] [PubMed] [Google Scholar]
- Hannon, G.J. 2002. RNA interference. Nature 418: 244-251. [DOI] [PubMed] [Google Scholar]
- Kamath, R.S., Fraser, A.G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231-237. [DOI] [PubMed] [Google Scholar]
- Kennerdell, J.R. and Carthew, R.W. 1998. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95: 1017-1026. [DOI] [PubMed] [Google Scholar]
- Lee, S.S., Lee, R.Y., Fraser, A.G., Kamath, R.S., Ahringer, J., and Ruvkun, G. 2003. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat. Genet. 33: 40-48. [DOI] [PubMed] [Google Scholar]
- McManus, M.T. and Sharp, P.A. 2002. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 3: 737-747. [DOI] [PubMed] [Google Scholar]
- Misquitta, L. and Paterson, B.M. 1999. Targeted disruption of gene function in Drosophila by RNA interference (RNA-i): A role for nautilus in embryonic somatic muscle formation. Proc. Natl. Acad. Sci. 96: 1451-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousses, S., Bittner, M.L., Chen, Y., Dougherty, E.R., Baxevanis, A., Meltzer, P.S., and Trent, J.M. 2000. Gene expression analysis by cDNA microarrays. In Functional genomics (ed. F.J.H. Livesey), pp. 113-137. Oxford University Press, Oxford.
- Mousses, S., Kallioniemi, A., Kauraniemi, P., Elkahloun, A., and Kallioniemi, O.P. 2002. Clinical and functional target validation using tissue and cell microarrays. Curr. Opin. Chem. Biol. 6: 97-101. [DOI] [PubMed] [Google Scholar]
- Paddison, P.J. and Hannon, G.J. 2002. RNA interference: The new somatic cell genetics? Cancer Cell 2: 17-23. [DOI] [PubMed] [Google Scholar]
- Piano, F., Schetterdagger, A.J., Mangone, M., Stein, L., and Kemphues, K.J. 2000. RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr. Biol. 10: 1619-1622. [DOI] [PubMed] [Google Scholar]
- Piano, F., Schetter, A.J., Morton, D.G., Gunsalus, K.C., Reinke, V., Kim, S.K., and Kemphues, K.J. 2002. Gene clustering based on RNAi phenotypes of ovary-enriched genes in C. elegans. Curr. Biol. 12: 1959-1964. [DOI] [PubMed] [Google Scholar]
- Pothof, J., Van Haaften, G., Thijssen, K., Kamath, R.S., Fraser, A.G., Ahringer, J., Plasterk, R.H., and Tijsterman, M. 2003. Identification of genes that protect the C. elegans genome against mutations by genome-wide RNAi. Genes & Dev. 17: 443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuschl, T., Zamore, P.D., Lehmann, R., Bartel, D.P., and Sharp, P.A. 1999. Targeted mRNA degradation by double-stranded RNA in vitro. Genes & Dev. 13: 3191-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziauddin, J. and Sabatini, D.M. 2001. Microarrays of cells expressing defined cDNAs. Nature 411: 107-110. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://genetics.med.harvard.edu/~perrimon/RNAiGenomeProject.html; RNAi Genome Project.