Abstract
Environment-wide association studies (EWAS) provide a way to uncover the environmental mechanisms involved in complex traits in a high-throughput manner. Genome-wide association studies have led to the discovery of genetic variants associated with many common diseases but do not take into account the environmental component of complex phenotypes. This EWAS assesses the comprehensive association between environmental variables and the outcome of type 2 diabetes (T2D) in the Marshfield Personalized Medicine Research Project Biobank (Marshfield PMRP). We sought replication in two National Health and Nutrition Examination Surveys (NHANES). The Marshfield PMRP currently uses four tools for measuring environmental exposures and outcome traits: 1) the PhenX Toolkit includes standardized exposure and phenotypic measures across several domains, 2) the Diet History Questionnaire (DHQ) is a food frequency questionnaire, 3) the Measurement of a Person's Habitual Physical Activity scores the level of an individual's physical activity, and 4) electronic health records (EHR) employs validated algorithms to establish T2D case-control status. Using PLATO software, 314 environmental variables were tested for association with T2D using logistic regression, adjusting for sex, age, and BMI in over 2,200 European Americans. When available, similar variables were tested with the same methods and adjustment in samples from NHANES III and NHANES 1999-2002. Twelve and 31 associations were identified in the Marshfield samples at p<0.01 and p<0.05, respectively. Seven and 13 measures replicated in at least one of the NHANES at p<0.01 and p<0.05, respectively, with the same direction of effect. The most significant environmental exposures associated with T2D status included decreased alcohol use as well as increased smoking exposure in childhood and adulthood. The results demonstrate the utility of the EWAS method and survey tools for identifying environmental components of complex diseases like type 2 diabetes. These high-throughput and comprehensive investigation methods can easily be applied to investigate the relation between environmental exposures and multiple phenotypes in future analyses.
1. Introduction
Computational methods to assess environmental exposures are essential to elucidate the complex nature of common human phenotypes. Genome-wide association studies (GWAS) have allowed for greater understanding of the genetic component of complex traits and identification of numerous loci associated with these traits [1]. They have provided a high-throughput approach for comprehensive testing of variants across the genome. However, this approach fails to consider the richly diverse and complex environment with which humans interact throughout the life course.
While GWAS have uncovered thousands of single nucleotide polymorphisms (SNPs) associated with disease, much remains unclear about the heritability and mechanisms that lead to common, complex human diseases [1,2]. It is likely that environmental exposure greatly impacts the genetic and cellular systems at play for many complex traits [2]. Environment-wide association studies (EWAS) [3] provide a method to test a variety of exposures across the human environment in a high-throughput, unbiased manner, much like GWAS tests for genetic effects. The utility of the EWAS approach was demonstrated for type 2 diabetes (T2D) using an array of laboratory measurements to identify a diverse number of exposures associated with T2D [3]. Such comprehensive laboratory measurements are rare and only assess exposures at a fixed time point without consideration of the various exposures throughout an individual's lifetime. Thus, there is a need to evaluate comprehensive and standardized survey tools that enable assessment of exposures and lifestyle choices over time and comparison of results across multiple studies.
The PhenX (consensus measures for Phenotypes and eXposures) toolkit (https://www.phenxtoolkit.org/) was developed as a resource for collecting standardized measures of phenotypes and environmental exposures [4]. Measures are available across 27 domains covering alcohol, tobacco, and other substance use; demographics; mental health; environmental exposures; diet; and disease, among others. In addition to providing information on traits, many of these measures can be used to ascertain information on environment, lifestyle, and environmental exposures. Other valuable resources for environmental measures include 1) the Measurement of a Person's Habitual Physical Activity, a questionnaire measuring a person's work, leisure, and sport activity level [5] (Baecke), and 2) the Dietary History Questionnaire (http://riskfactor.cancer.gov/DHQ/), a food frequency questionnaire [6,7] (DHQ).
Electronic health records (EHR) are a growing resource for measuring health outcomes in individuals, as they contain vast amounts of medical data including records of diagnoses, procedures, and clinical laboratory measurements [8]. These data can be used, with electronic algorithms, to systematically define cases and controls for numerous phenotypes of interest, such as type 2 diabetes. The Electronic Medical Records and Genomics (eMERGE) Network combines EHR data from sites across the United States and currently utilizes electronic phenotyping algorithms for over a dozen phenotypes [9]. The Marshfield Personalized Medicine Research Project Biobank (Marshfield PMRP) [10], part of the eMERGE Network, is one site currently employing EHR phenotyping as well as the PhenX Toolkit, the Measurement of a Person's Habitual Physical Activity (Beacke), and the Dietary History Questionnaire (DHQ). Taken together, the PMRP is a rich phenotypic resource for genomic and environmental association analyses to dissect the architecture of complex traits.
Here, we present the results of an EWAS for type 2 diabetes using survey questions from the PhenX Toolkit, DHQ, and Beacke surveys from the Marshfield PMRP. To seek replication of these results with similar survey questions when available, we used data from the National Health and Nutrition Examination Surveys (NHANES) [11]. To the authors’ knowledge, this is the first EWAS performed using EHR data. Environment-wide association studies provide a methodology to test environmental measures in a comprehensive, high-throughput manner. Integration of EWAS with phenome-wide association studies (PheWAS) [12-14] and genome-wide association studies (GWAS) [1] will further elucidate the complex interplay of gene and environment in common traits as well as the ways in which exposures modulate pleiotropy. Using multiple exposure and outcome variables to assess environment and lifestyle factors using EWAS will provide a richer understanding of the architecture of complex traits.
2. Methods
2.1. Marshfield PMRP and Type 2 Diabetes Case Identification
The Marshfield PMRP is a population based biobank with ~20,000 subjects, aged 18 years and older, enrolled in the Marshfield Clinic healthcare system in central Wisconsin [10]. DNA, plasma, and serum samples are collected at the time the enrollee completes a written informed consent document, with allowance for ongoing access to the linked electronic health records (EHR). PMRP participants also complete questionnaires, including responses regarding smoking history, occupation, physical activity, diet, and a variety of other PhenX measures. A subset of the Marshfield PMRP subjects completed the PhenX survey, the DHQ, and/or the Measurement of a Person's Habitual Physical Activity (Table 1).
Table 1.
Marshfield Type 2 Diabetes Case/Control Sample Size for Each Questionnaire
| Questionnaire | Total Sample Size | # Cases T2D | # Controls | |
|---|---|---|---|---|
| Total | PhenX | 2,243 | 433 | 1,810 |
| DHQ | 2,606 | 559 | 2,047 | |
| Activity | 2,571 | 552 | 2,018 | |
| Male | PhenX | 898 | 204 | 694 |
| DHQ | 1,051 | 260 | 791 | |
| Activity | 1,035 | 257 | 778 | |
| Female | PhenX | 1,345 | 229 | 1,116 |
| DHQ | 1,555 | 299 | 1,256 | |
| Activity | 1,535 | 295 | 1,240 | |
| Age | All | > 50 | ||
| Ancestry | All | European | ||
The NHGRI funded eMERGE network (Electronic Medical Records and Genomics) has implemented robust electronic phenotyping algorithms to select cases and controls for a number of different phenotypes/outcomes [9]. Using an algorithm developed by eMERGE [15], T2D patients were diagnosed by their records from the Marshfield EHR. The Marshfield samples were originally selected for eMERGE based on their cataract case-control status; however, this is an example of the reusability of biobank samples for additional traits. T2D cases were defined as having the following in their EMR: a T2D ICD-9 medical billing code, information about insulin medication, abnormal glucose or HbA1c levels, or more than two diagnoses of T2D by a clinician. All T2D cases with an ICD-9 code for T1D were removed from further analyses. All control subjects had to have at least 2 clinical visits, at least one blood glucose measurement, normal blood glucose or HbA1c levels, no ICD-9 codes for T2D or any related condition, no history of being on insulin or any diabetes related medication, and no family history of T1D or T2D.
2.2. Environmental Variable Measurements
2.2.1 Phenx Toolkit
The PhenX Toolkit (www.phenxtoolkit.org) was accessed to develop a self-administered questionnaire to assess environmental and lifestyle factors. Some of the PhenX measures were chosen because of the potential for gene/environment associations with age related cataract -which is a primary disease of interest for PMRP (smoking, alcohol, ultraviolet light exposure), some were chosen because of the potential for validation against prior PMRP questionnaire data and medical history information (demographics, physical activity, family history of heart attack, history of stroke) and the rest were chosen because of the potential for future research and cross-site collaborations (hypomania/mania symptoms, hand dominance) within the network funded through administrative supplements to collect PhenX measures. The time to complete the questionnaire ranged from 20 to 40 minutes in pre-testing, depending on how many questions were logical skips. The 32-page self-administered questionnaire was mailed to all eligible subjects with a cover letter and return address envelope. A second mailing was employed to increase the response rate. Subjects were offered $10 for their time to complete the questionnaire.
PhenX survey data were entered and merged with prior PMRP questionnaire information from the Marshfield Clinic electronic medical record. For validation purposes, the electronic medical record was considered to be the gold standard where possible. Two hundred fifty-five measures from the PhenX Toolkit were included for our analysis. Questions included a range of topics from the following classes: alcohol use, smoking, demographics, depression, mania, activity, residential environment, and UV exposure.
2.2.2. Diet History Questionnaire
Food frequency questionnaires (FFQs) are widely used to assess dietary intake in epidemiologic studies because they are more representative of usual intake and less expensive to implement than other methodologies including weighed food records and 24-hour dietary recalls because they are usually self-administered. Inclusion of aids to estimate portion sizes is essential to improve the accuracy and validity of FFQs [7]. Self-administered food frequency questionnaires (FFQ) are available on approximately 2/3 of the PMRP cohort to quantify usual dietary intake of all major nutrients. The selected FFQ, the Diet History Questionnaire (DHQ) (http://riskfactor.cancer.gov/DHQ/), was developed by researchers at the National Cancer Institute (NCI) and has been shown to be superior to the commonly used Willett FFQ and similar to the Block FFQ for estimating absolute nutrient intakes [7]. All three FFQs produce similar results after statistical adjustment for total energy intake. The list of foods and portion sizes on the DHQ was developed from nationally representative data, the USDA's 1994-1996 Continuing Survey of Food Intakes by Individuals, and is therefore most appropriate for use with this study population. The DHQ comprises 124 separate food items and asks about portion sizes for most foods. In addition, there are 10 questions about nutrient supplement intake. The DHQ was printed and scanned by National Computer Systems as has been done for all recent studies conducted at the NCI using the DHQ. The completed DHQ was mailed to National Computer Systems for scanning. After scanning, the data from the questionnaires are stored in ASCII format and then uploaded into the nutrient analysis software package. Diet*Calc software, available from the National Institutes of Health, is used for the nutrient analyses of the DHQ data (http://riskfactor.cancer.gov/DHQ/dietcalc/). This is the software package that was used for analysis of the DHQ for the Eating at America's Table Study. The DHQ is mailed to participants with their appointment reminders so that they can complete it prior to their appointment to save them time. The Research Project Assistants reviews all DHQs to ensure that they have been completed. Fifty-six measures of dietary intake were assessed for this EWAS that covered the following domains: vitamin, fat, protein, carbohydrate, fiber, cholesterol, caloric, grain, vegetable, caffeine, and alcohol intake.
2.2.3 Measurement of a Person's Habitual Physical Activity
As with measurement of dietary intake for epidemiologic studies, there are a number of different validated tools that have been used in the past. The agreement between physical activity questionnaire and gold standard tends to be somewhat lower than for dietary intake, but is reasonable for ranking relative activity levels in groups. The researchers preferred to use a previously developed physical activity assessment tool to allow comparison with results from other study populations. Requirements of the selected tool included: 1) self-administered, 2) previously validated, and 3) validated for use in a similar study population across a range of ages. The selected physical activity questionnaire, the ARIC/Baecke questionnaire, is self-administered, validated for use in both men and women, and currently being used in a large, prospective study in the US [16]. The questionnaire has been shown to have high reliability and accurate assessment of both high intensity activity and light intensity activity such as walking. It comprises 16 questions and generates three indices of activity: 1) a work index, 2) a sport index, and 3) a leisure-time index. This one-page self-administered physical activity questionnaire is mailed along with appointment reminders and the Diet History Questionnaire (DHQ). Information from the completed physical activity questionnaires are entered twice into a Microsoft Access database. The two entries are compared to ensure accuracy of the data entry. The three physical activity indices (work, sport, and leisure-time) are calculated and the data merged with anthropometric, dietary, and demographic data for subsequent analyses.
2.2.4. National Health and Nutrition Examination Surveys (NHANES)
NHANES III Phase 2, conducted between 1991-1994, and NHANES 1999-2002 measures the health and nutritional habits of participants by collecting medical, dietary, demographic, laboratory, lifestyle, and environmental exposure data using questionnaire and laboratory measures. The data of NHANES were collected by the National Center on Health Statistics (NCHS) at the Centers for Disease Control and Prevention (CDC). All participants were consented by the CDC at the time of the survey and sample collection.
To seek replication of the Marshfield results, we identified measures similar to the most significant Marshfield PMRP EWAS results in NHANES III and NHANES 1999-2002. Because different survey methods were utilized between Marshfield PMRP and the NHANES, measures were chosen when they matched a significant broad environmental “class”. For example, many smoking measures were included in the most significant EWAS results and any smoking measure found in either NHANES was included for replication. T2D case/control status was defined using an algorithm previously described [17].
2.3. Statistical Analysis
A total of 314 environmental variables were included in our analysis of the Marshfield data. Logistic regression was used, adjusting for age, sex, and body mass index (BMI), with PLATO [18]. Control was coded as 1 and case as 2. All significant results were investigated to ensure that all top ranking associations had greater than 10 responses for both cases and controls. Results in figures 1 and 2 were plotted using PheWAS View [19].
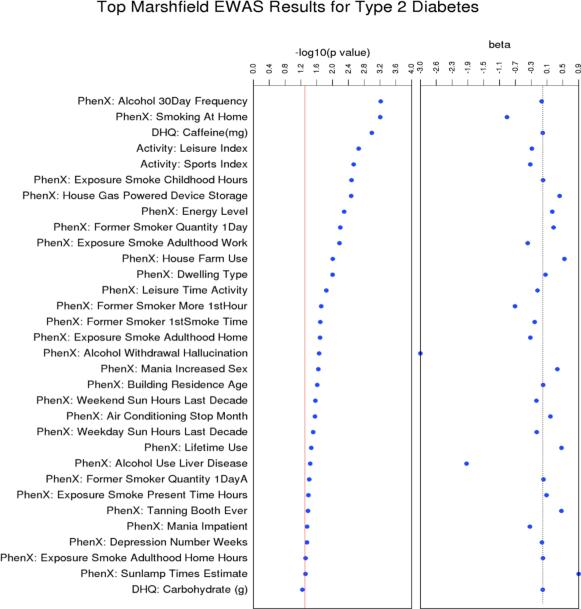
Figure 1. The most significant association results in the Marshfield sample using PhenX Toolkit, DHQ, and Measurement of a Person's Habitual Physical Activity surveys.
The PhenX variables are listed along the Y-Axis. The first track shows the results of our EWAS, with –log(10) of the p-value plotted from most significant result at the top and descending in order. The next track shows the magnitude and direction of the effect. Case/control status was coded as 1=Control, 2=Case.
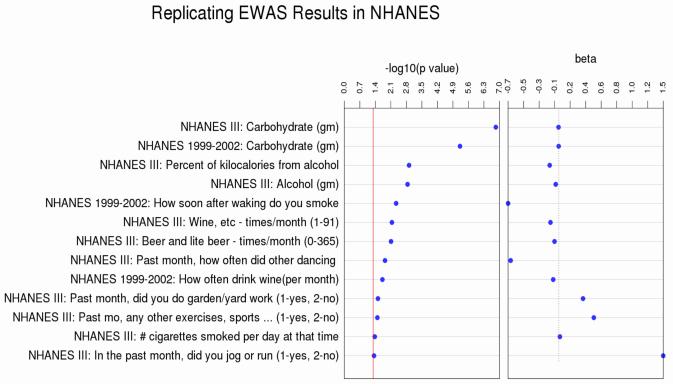
Figure 2. Replicating results of the most significant Marshfield EWAS associations from NHANES III and NHANES 1999-2002.
Results were considered a replication if the p-value was < 0.05 p-value and showed the same direction of effect as the Marshfield analyses. Controls were coded as 1 and Cases as 2. This figure is in the same format as Figure 1, with NHANES measurements on the y-axis ordered by descending association significance. The tracks show the p-value signficicance of the association in –log10(p-value) and the magnitude and direction of the effect.
For the NHANES data, logistic regression was used for all association testing, adjusting for age, sex, and BMI, in 46 to 3,964 samples (sample sizes varied for each measure) of European ancestry (self-identified non-Hispanic whites) for a total of 116 environmental variables from NHANES III (84) and NHANES 1999-2002 (32). All significant EWAS results were assessed to ensure sample size was greater than 10 for cases and controls for each variable.
3. Results
In this environment-wide association study of 314 variables for type 2 diabetes, we found 12 results with a p-value less than 0.01 in the Marshfield Clinic samples. Due to the exploratory and hypothesis generating nature of this method, we are presenting all the results with a p-value less than 0.05 (31 results). Figure 1 displays the most significant EWAS associations in the Marshfield sample.
All variables could be placed into seven broad environmental “classes”: smoking, alcohol use, mania, depression, activity, diet, UV exposure, and residence. Table 2 includes all results with a p-value less than 0.05 by environment class and displays the survey question for each measure from the PhenX Toolkit.
Table 2.
EWAS Variable Classes, Specific PhenX Toolkit Questions, and the EWAS Marshfield PMRP results
| Class | Survey: Variable | PhenX Toolkit Question | P- value | Beta |
|---|---|---|---|---|
| Alcohol | PhenX: Alcohol 30Day Frequency | Think specifically about the past 30 days, from [DATEFILL], up to and including today. During the past 30 days, on how many days did you drink one or more drinks of an alcoholic beverage? | 6E-04 | −0.03 |
| PhenX: Alcohol Withdrawal Hallucination | When you stopped, cut down or went without drinking, did you ever experience any of the following problems for most of the day for 2 days or longer? Did you see or hear things that weren't there? (Yes=1, No=2) | 0.022 | −3.041 | |
| PhenX: Lifetime Use | In your entire life, have you had at least 1 drink of any kind of alcohol, not counting small tastes or sips? (Yes=1, No=2) | 0.035 | 0.4655 | |
| PhenX: Alcohol Use Liver Disease | There are several health problems that can result from long stretches of drinking. Did drinking ever cause you to have liver disease or yellow jaundice? (Yes=1, No=2) | 0.037 | −1.894 | |
| Smoking Exposure | PhenX: Smoking At Home | Does anyone who lives here smoke cigarettes, cigars, or pipes anywhere inside this home? (Yes=1, No=2) | 6E-04 | −0.889 |
| PhenX: Exposure Smoke Childhood | How many hours were you exposed to smoke from other people's cigarettes or tobacco products during childhood per day? | 0.003 | 0.0064 | |
| PhenX: Former Smoker Quantity 1DayB | Former smokers who did not ever smoke every day for the at least 6 months: when you last smoked every day, on average how many cigarettes did you smoke each day? | 0.006 | 0.2683 | |
| PhenX: Exposure Smoke Work | Were you exposed to smoke from other people's cigarettes or tobacco products during adulthood at work? (Yes=1, No=2) | 0.007 | −0.375 | |
| PhenX: Former Smoker More 1stHour | Did you smoke more frequently during the first hours after waking than during the rest of the day? (Yes=1, No=2) | 0.019 | −0.689 | |
| PhenX: Former Smoker | 1stSmoke Time How soon after you wake up do/did you smoke your first cigarette? | 0.02 | −0.202 | |
| PhenX: Exposure Smoke Home | Were you exposed to smoke from other people's cigarettes or tobacco products during adulthood at home? (Yes=1, No=2) | 0.021 | −0.309 | |
| PhenX: Former Smoker Quantity 1DayA | Former smokers who smoked cigarettes every day for at least 6 months: when you last smoked every day, on average how many cigarettes did you smoke each day? | 0.039 | 0.0171 | |
| PhenX: Exposure Smoke Present Time Hours | At the present time, how many hours per day are you exposed to the smoke of others? | 0.041 | 0.0943 | |
| PhenX: Exposure Smoke Adulthood Home Hours | How many hours per day were you exposed to smoke from other people's cigarettes or tobacco products during adulthood at home? | 0.048 | 0.0046 | |
| Diet | DHQ: Caffeine(mg) | NA | 0.001 | −0.0005 |
| Activity | Activity: Leisure Index | NA | 0.002 | −0.27 |
| Activity: Sports Index | NA | 0.003 | −0.31 | |
| PhenX:Leisure Activity | Please check the box next to the one statement which best describes the way you spent your leisure-time during most of the last year. | 0.014 | −0.132 | |
| Residence | PhenX: House Gas Powered Device | Are any gas powered devices stored in any room, basement, or attached garage in this (house/apartment)? (Yes=1, No=2) | 0.003 | 0.4187 |
| PhenX: House Farm | Is this property actively used as a farm or ranch? (Yes=1, No=2) | 0.01 | 0.5382 | |
| PhenX: Dwelling Type | What is the type of dwelling? (1=Detached house, 2=Duplex/Triplex, 3=Row house, 4=Low rise apartment (1-3 floors), 5=High rise apartment (>3 floors), 6=Mobile home / Trailer7=Other) | 0.01 | 0.0684 | |
| PhenX: Building Residence Age | When did you start living there? | 0.024 | 0.0072 | |
| PhenX: Air Conditioning Stop Month | During which month (do you usually/would you) stop using air conditioning? | 0.028 | 0.1891 | |
| Depression | PhenX: Energy Level | Please indicate the one response that best describes your energy level for the past seven days. (0 = There is no change in my usual level of energy. 1 = I get tired more easily than usual. 2 = I have to make a big effort to start or finish my usual daily activities (for example,shopping, homework, cooking or going to work). 3 = I really cannot carry out most of my usual daily activities because I just don't have the energy.) | 0.005 | 0.2365 |
| PhenX: Depression Number Weeks | About how many weeks altogether did you feel this way? Count the weeks before, during and after the worst two weeks. The total period of depression/loss of interest was: | 0.044 | −0.022 | |
| Mania | PhenX: Mania Increased Sex | Please try to remember a period when you were in a “high” state. In such a state: I am more interested in sex, and/or have increased sexual desire (Yes=1, No=2) | 0.023 | 0.3615 |
| PhenX: Mania Impatient | Please try to remember a period when you were in a “high” state. In such a state: I am more impatient and/or get irritable more easily (Yes=1, No=2) | 0.044 | −0.321 | |
| UV Exposure | PhenX: Weekend Sun Hours Last Decade | On a typical weekend day in the summer, about how many hours did you generally spend in the mid-day sun in the past ten years? | 0.027 | −0.158 |
| PhenX: Weekday Sun Hours Last Decade | On a typical weekday in the summer, about how many hours did you generally spend in the mid-day sun in the past ten years? | 0.031 | −0.151 | |
| PhenX: Tanning Booth | Have you ever used a tanning booth? (Yes=1, No=2) | 0.042 | 0.4621 | |
| PhenX: Sunlamp Times | About how many times have you used a sunlamp in your life? | 0.048 | 0.8917 |
When available, similar questions from NHANES that fell into one of the above phenotype classes were included to seek replication. Measures were available in alcohol use, smoking exposure, diet, activity, depression, and mania but not in residence and UV exposure. Seven of the results were significant at p<0.01 and thirteen at p <0.05 with the same direction of effect as the related Marshfield associations (Figure 3).
The most significant survey questionnaire result in the Marshfield EWAS was alcohol frequency in the last 30 days, which was inversely associated with type 2 diabetes status. This relationship was also observed for two related measures in NHANES III: alcohol consumption questions beer and lite beer -times/month and wine, etc - times/month and one in NHANES 1999-2002: alcohol consumption question: How often drink wine (per month). Never having alcohol was associated with T2D status in Marshfield and did not replicate in either NHANES, though a similar, but not exact, measure was available and tested. Experiencing excessive alcohol use symptoms like hallucination due to alcohol withdrawal and liver disease from excess alcohol use was associated with having T2D in the Marshfield sample. Neither of these measures were available in either NHANES for comparison.
A number of significant results in Marshfield included measurements of first and second hand smoking exposure. Cigarette or other tobacco smoke exposure at home or at work, and for a greater number of hours during childhood, adulthood, and present time were all associated with T2D status. Additionally, for former smokers, greater number of cigarettes per day, smoking more frequently during the first hours of the day, and smoking earlier in the day were also associated with having T2D. Two of the smoking measures replicated in NHANES III: number of cigarettes smoked/day when smoked and NHANES 1999-2002: how soon after waking do you smoke? with the same direction of effect.
The two most significant results from the DHQ for the EWAS in Marshfield were a metric of caffeine consumption: caffeine (mg), which was inversely associated with T2D status and a metric of the consumption of carbohydrates (g). The caffeine measurement did not replicate in either NHANES, though increased coffee intake has been previously reported as having an association with lowered risk of T2D [20]. Carbohydrate intake did not meet the significance threshold of p-value less than 0.05 in Marshfield, but was included in the replication analysis because it was the second most significant DHQ result. When this association was investigated in NHANES III and NHANES 1999-2002 it was the most significant result for both studies.
4. Discussion
Using a systematic, high-throughput EWAS method, we identified and replicated novel as well as established associations between environmental exposures and T2D. The replicating results of the association between less alcohol use per month and T2D status is consistent with prior research that demonstrates that moderate alcohol use is associated with decreased risk of T2D [21,22]. The association between T2D status and the specific symptoms of hallucination and liver disease has not been observed in the literature, to the best of the authors’ knowledge. However, prior research has indicated that binge drinking and high levels of alcohol use are associated with increased risk of T2D [21,22]. It is possible that these results are spurious, or that there may be some mechanism at play by which these extreme alcohol-related measures are related to T2D. Comparison with other studies for this measure is necessary before conclusions can be drawn.
The relationship between increased smoking exposure and having T2D is also well established [23-25]. Activity level also has a well-documented link with T2D [26-28]. Here we observed a number of results from both Marshfield and NHANES III that demonstrate this association. Work activity was not significantly associated with T2D in the PhenX or Baecke measures. However, lower amounts of leisure and sports activity was associated with T2D status in Marshfield. This relationship was validated with similar measures in NHANES III: dancing, gardening/yard work, sports, and running or jogging in the past month.
A number of associations from the residence, depression, mania, and UV exposure classes in Marshfield did not replicate in either NHANES. This could indicate that these were false positive findings, or it could also be due to differences in measures that were used, deviation in survey question wording, or low sample sizes for a given question. Additionally, many of these results could not be evaluated for replication in either NHANES because they were not available. This demonstrates the need for standardized measures of environmental exposures, as the utilization of these measures will allow the validation of significant results across multiple studies.
Another limitation to this EWAS design is the difficulty in determining whether associations occurred simply due to T2D diagnosis. For instance, the activity questions measured activity for the past month and did not include information on activity level during childhood or if activity level changed when T2D symptoms were experienced. It is possible that the individuals with T2D participated in less leisure and sport activity due to symptoms but had greater activity levels earlier in life. Similarly, the inverse association observed between T2D and carbohydrate intake may be reflective of individuals who are restricting carbohydrate intake due to T2D diagnosis, a common dietary treatment for the disease [29]. This issue indicates the importance of gathering environmental variables that measure multiple points of an individual's lifetime. Additionally, this approach does not currently consider the full spectrum of environmental exposures. Limitations in the types of exposures assessed, and when they are collected, restricts thorough understanding of all the environmental components involved in the development of complex diseases such as T2D. Future incorporation of biological exposure data such as toxins [30] and nutrients [31] will provide additional data on the exposures associated with complex traits.
Environment-wide association studies allow the testing of multiple environmental exposures for association with common disease. Here, we demonstrate the utility of this approach for research using health record data, a novel use for this type of resource. Using this systematic EWAS approach, exposures will be identified as potential causative agents for complex traits. Significant associations can be investigated for gene-environment interactions [32,33]. Incorporating genetic data will lead to a more complete understanding of the mechanisms that lead to complex phenotypes, such as T2D Similar to the PheWAS [12-14] method, the EWAS approach can be used to test for association between a diverse array of exposures and numerous phenotypes to discover the types of exposure that are associated with multiple traits. The search for interactions between environmental variables and genetic loci, as well as the independent exposures involved in multiple traits, will further elucidate the genetic and environmental architecture of complex human phenotypes.
Acknowledgements
This work was supported in part by NIH U01 HG004798 and its ARRA supplements and by NIH grants HG006389 and HG006385 in addition to an administrative supplement from PhenX RISING (NOT-HG-11-009). We would like to thank Dr. Geraldine McQuillan and Jody McLean for their help in accessing the Genetic NHANES data. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Center for Disease Control and Prevention.
Contributor Information
MOLLY A. HALL, Center for Systems Genomics, Department of Biochemistry and Molecular Biology, The Pennsylvania State University, 512 Wartik Lab, University Park, PA 16802, USA mah546@psu.edu
SCOTT M. DUDEK, Center for Systems Genomics, Department of Biochemistry and Molecular Biology, The Pennsylvania State University, 512 Wartik Lab, University Park, PA 16802, USA sud23@psu.edu
ROBERT GOODLOE, Center for Human Genetics Research, Vanderbilt University, Nashville TN, 37232, USA robert.goodloe@chgr.mc.vanderbilt.edu.
DANA C. CRAWFORD, Center for Human Genetics Research, Vanderbilt University, Nashville TN, 37232, USA crawford@chgr.mc.vanderbilt.edu
SARAH A. PENDERGRASS, Center for Systems Genomics, Department of Biochemistry and Molecular Biology, The Pennsylvania State University, 503 Wartik Lab, University Park, PA 16802, USA sap29@psu.edu
PEGGY PEISSIG, The Marshfield Clinic Marshfield, WI, USA Peissig.Peggy@securityhealth.org.
MURRAY BRILLIANT, The Marshfield Clinic Marshfield, WI, USA BRILLIANT.MURRAY@mcrf.mfldclin.edu.
CATHERINE A. MCCARTY, Essentia Institute of Rural Health, Duluth, MN, USA CMcCarty@eirh.org
MARYLYN D. RITCHIE, Center for Systems Genomics, Department of Biochemistry and Molecular Biology, The Pennsylvania State University, 512 Wartik Lab, University Park, PA 16802, USA Marylyn.ritchie@psu.edu
References
- 1.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maher B. Personal genomes: The case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 3.Patel CJ, Bhattacharya J, Butte AJ. An Environment-Wide Association Study (EWAS) on type 2 diabetes mellitus. PLoS One. 2010;5:e10746. doi: 10.1371/journal.pone.0010746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamilton CM SL, Pratt JG, Maiese D, Hendershot T, et al. The PhenX Toolkit: get the most from your measures. Am J Epidemiol. 2011:253–260. doi: 10.1093/aje/kwr193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baecke JABJ, Frijters JE. A short questionnaire for the measurement of habitual physical activity in epidemiological studies. Am J Clin Nutr. 1982;36:936–942. doi: 10.1093/ajcn/36.5.936. [DOI] [PubMed] [Google Scholar]
- 6.Thompson FE, Subar AF, Brown CC, Smith AF, Sharbaugh CO, et al. Cognitive research enhances accuracy of food frequency questionnaire reports: results of an experimental validation study. J Am Diet Assoc. 2002;102:212–225. doi: 10.1016/s0002-8223(02)90050-7. [DOI] [PubMed] [Google Scholar]
- 7.Subar AF, Thompson FE, Kipnis V, Midthune D, Hurwitz P, et al. Comparative validation of the Block, Willett, and National Cancer Institute food frequency questionnaires : the Eating at America's Table Study. Am J Epidemiol. 2001;154:1089–1099. doi: 10.1093/aje/154.12.1089. [DOI] [PubMed] [Google Scholar]
- 8.Kohane IS. Using electronic health records to drive discovery in disease genomics. Nat Rev Genet. 2011;12:417–428. doi: 10.1038/nrg2999. [DOI] [PubMed] [Google Scholar]
- 9.McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCarty CAWRGP, Wesbrook SD, Caldwell MD. Marshfield Clinic Personalized Medicine Research Project (PMRP): design, methods and recruitment for a large population-based biobank. Personalized Medicine. 2005:49–79. doi: 10.1517/17410541.2.1.49. [DOI] [PubMed] [Google Scholar]
- 11.(CDC) CfDCaP . National Center for Health Statistics (NCHS). National Health and Nutrition Examination Survey Questionnaire (or Examination Protocol, or Laboratory Protocol) Department of Health and Human Services, Centers for Disease Control and Prevention; Hyattsville, MD: U.S: 2013. [Google Scholar]
- 12.Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pendergrass SA, Brown-Gentry K, Dudek SM, Torstenson ES, Ambite JL, et al. The use of phenome-wide association studies (PheWAS) for exploration of novel genotype-phenotype relationships and pleiotropy discovery. Genet Epidemiol. 2011;35:410–422. doi: 10.1002/gepi.20589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pendergrass SA, Brown-Gentry K, Dudek S, Frase A, Torstenson ES, et al. Phenome-wide association study (PheWAS) for detection of pleiotropy within the Population Architecture using Genomics and Epidemiology (PAGE) Network. PLoS Genet. 2013;9:e1003087. doi: 10.1371/journal.pgen.1003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kho AN, Hayes MG, Rasmussen-Torvik L, Pacheco JA, Thompson WK, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J Am Med Inform Assoc. 2012;19:212–218. doi: 10.1136/amiajnl-2011-000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baecke JA, Burema J, Frijters JE. A short questionnaire for the measurement of habitual physical activity in epidemiological studies. Am J Clin Nutr. 1982;36:936–942. doi: 10.1093/ajcn/36.5.936. [DOI] [PubMed] [Google Scholar]
- 17.Haiman CA, Fesinmeyer MD, Spencer KL, Buzkova P, Voruganti VS, et al. Consistent directions of effect for established type 2 diabetes risk variants across populations: the population architecture using Genomics and Epidemiology (PAGE) Consortium. Diabetes. 2012;61:1642–1647. doi: 10.2337/db11-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grady BJ, Torstenson E, Dudek SM, Giles J, Sexton D, et al. Finding unique filter sets in PLATO: a precursor to efficient interaction analysis in GWAS data. Pac Symp Biocomput. 2010:315–326. [PMC free article] [PubMed] [Google Scholar]
- 19.Pendergrass SA, Dudek SM, Crawford DC, Ritchie MD. Visually integrating and exploring high throughput Phenome-Wide Association Study (PheWAS) results using PheWAS-View. BioData Min. 2012;5:5. doi: 10.1186/1756-0381-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Dam RM, Willett WC, Manson JE, Hu FB. Coffee, caffeine, and risk of type 2 diabetes: a prospective cohort study in younger and middle-aged U.S. women. Diabetes Care. 2006;29:398–403. doi: 10.2337/diacare.29.02.06.dc05-1512. [DOI] [PubMed] [Google Scholar]
- 21.Carlsson S, Hammar N, Grill V, Kaprio J. Alcohol consumption and the incidence of type 2 diabetes: a 20-year follow-up of the Finnish twin cohort study. Diabetes Care. 2003;26:2785–2790. doi: 10.2337/diacare.26.10.2785. [DOI] [PubMed] [Google Scholar]
- 22.Pietraszek A, Gregersen S, Hermansen K. Alcohol and type 2 diabetes. A review. Nutr Metab Cardiovasc Dis. 2010 doi: 10.1016/j.numecd.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Willi C, Bodenmann P, Ghali WA, Faris PD, Cornuz J. Active smoking and the risk of type 2 diabetes: a systematic review and meta-analysis. JAMA. 2007;298:2654–2664. doi: 10.1001/jama.298.22.2654. [DOI] [PubMed] [Google Scholar]
- 24.Yeh HC, Duncan BB, Schmidt MI, Wang NY, Brancati FL. Smoking, smoking cessation, and risk for type 2 diabetes mellitus: a cohort study. Ann Intern Med. 2010;152:10–17. doi: 10.7326/0003-4819-152-1-201001050-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie XT, Liu Q, Wu J, Wakui M. Impact of cigarette smoking in type 2 diabetes development. Acta Pharmacol Sin. 2009;30:784–787. doi: 10.1038/aps.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu G, Qiao Q, Silventoinen K, Eriksson JG, Jousilahti P, et al. Occupational, commuting, and leisure-time physical activity in relation to risk for Type 2 diabetes in middle-aged Finnish men and women. Diabetologia. 2003;46:322–329. doi: 10.1007/s00125-003-1031-x. [DOI] [PubMed] [Google Scholar]
- 27.Helmrich SP, Ragland DR, Leung RW, Paffenbarger RS., Jr. Physical activity and reduced occurrence of non-insulin-dependent diabetes mellitus. N Engl J Med. 1991;325:147–152. doi: 10.1056/NEJM199107183250302. [DOI] [PubMed] [Google Scholar]
- 28.Laaksonen DE, Lindstrom J, Lakka TA, Eriksson JG, Niskanen L, et al. Physical activity in the prevention of type 2 diabetes: the Finnish diabetes prevention study. Diabetes. 2005;54:158–165. doi: 10.2337/diabetes.54.1.158. [DOI] [PubMed] [Google Scholar]
- 29.Nielsen JV, Joensson EA. Low-carbohydrate diet in type 2 diabetes: stable improvement of bodyweight and glycemic control during 44 months follow-up. Nutr Metab (Lond) 2008;5:14. doi: 10.1186/1743-7075-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rappaport SM, Smith MT. Epidemiology. Environment and disease risks. Science. 2010;330:460–461. doi: 10.1126/science.1192603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tzoulaki I, Patel CJ, Okamura T, Chan Q, Brown IJ, et al. A nutrient-wide association study on blood pressure. Circulation. 2012;126:2456–2464. doi: 10.1161/CIRCULATIONAHA.112.114058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel CJ, Chen R, Butte AJ. Data-driven integration of epidemiological and toxicological data to select candidate interacting genes and environmental factors in association with disease. Bioinformatics. 2012;28:i121–126. doi: 10.1093/bioinformatics/bts229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel CJ, Chen R, Kodama K, Ioannidis JP, Butte AJ. Systematic identification of interaction effects between genome- and environment-wide associations in type 2 diabetes mellitus. Hum Genet. 2013;132:495–508. doi: 10.1007/s00439-012-1258-z. [DOI] [PMC free article] [PubMed] [Google Scholar]