Abstract
We used photolithographic microfabrication techniques to create very small stainless steel fountain pens that were installed in place of conventional pens on a microarray spotter. Because of the small feature size produced by the microfabricated pens, we were able to print arrays with up to 25,000 spots/cm2, significantly higher than can be achieved by other deposition methods. This feature density is sufficiently large that a standard microscope slide can contain multiple replicates of every gene in a complex organism such as a mouse or human. We tested carryover during array printing with dye solution, labeled DNA, and hybridized DNA, and we found it to be indistinguishable from background. Hybridization also showed good sequence specificity to printed oligonucleotides. In addition to improved slide capacity, the microfabrication process offers the possibility of low-cost mass-produced pens and the flexibility to include novel pen features that cannot be machined with conventional techniques.
DNA microarrays have proven to be powerful tools for the analysis of many biological and medical problems, from tumor typing (Golub et al. 1999; Alizadeh et al. 2000) to reverse engineering biological circuits and pathways (DeRisi et al. 1997; Chu et al. 1998; Lockhart and Winzeler 2000). Extremely high density microarrays that may fit an entire genome on a single substrate are desirable for a number of reasons, including sensitivity, cost, convenience, and controlling experimental error due to variation between slides. Arrays that could accommodate multiple replicates of each gene are also desirable to increase data quality—especially for genes expressed at very low levels (Jin et al. 2001)—but require densities beyond the reach of available printing methods.
High-density DNA microarrays are currently produced via one of three technologies: photolithographic DNA synthesis, modified ink-jet systems, or precisely controlled robotic pens. Although the photolithographic technique (Lipshutz et al. 1999) can produce feature sizes as small as 18 microns, it has drawbacks that include high cost and limited oligonucleotide length. Furthermore, because ∼10 to 20 different probes are needed for reliable detection of each gene, commercial arrays have thus far been limited to ∼19,500 transcripts on a 1.28 × 1.28-cm square chip (12,000 genes/cm2; http://www.affymetrix.com/support/technical/datasheets/hgu133_datasheet.pdf). It is anticipated that feature sizes can ultimately reach ∼10 microns with this technology, corresponding to a density of ∼39,000 transcripts/cm2. A recent variation of light-directed DNA synthesis uses a digital micromirror array in place of photomasks (Nuwaysir et al. 2002). The highest density commercial product based on this technology is a 17.4 × 13-mm microarray containing 39,000 transcripts (feature size of 16 μm, five probe pairs per gene), corresponding to a density of ∼17,000 genes/cm2 (http://www.nimblegen.com/products/human.html). More efficient chemistry is used in the ink-jet method for in situ synthesis, permitting longer oligos that have sufficient specificity to detect genes using fewer probes. Although feature sizes are larger, arrays of 25,000 spots on 25 × 75-mm glass slides have been reported (Shoemaker et al. 2001).
These synthetic methods require prior knowledge of the genome sequence, and when this is not available, one must resort to methods of arraying isolated genetic material, such as cDNA. Techniques for the deposition of cDNA (or synthetic oligonucleotides), including bubble jet printers and robotically controlled pens, are capable of producing features as small as 70-75 microns (Okamoto et al. 2000; http://www.majerprecision.com/pins.htm; http://arrayit.com/Products/Printing/Stealth/stealth.html). To our knowledge, the largest reported deposition array contains 82,944 spots in an 18 × 72-mm area, corresponding to a density of 6400 genes/cm2 (http://arrayit.com/Products/Printing/Stealth/stealth.html). Ceramic capillary tips from the microelectronics industry have also been used to print microarrays (George et al. 2001). Using the smallest available tips (50 μm), the highest demonstrated density is 10,000 spots/cm2. These pens are cheaper and more durable than their metal counterparts, but are not in widespread use for printing microarrays. Deposition methods are preferable to synthetic methods when printing DNA molecules with certain modifications and when printing molecules other than DNA.
Traditionally, the individual fountain pens for cDNA arrayers have been machined by hand: Stainless steel or titanium rods are first ground to a sharp tip, and then a slot is cut in the tip. Miniature grinding wheels and saws were used to cut early slots, but most commercial pen manufacturers now use wire electrical discharge machining (EDM) or laser cutting methods to achieve slots as small as 10-40 microns in width (http://www.majerprecision.com/pins.htm; http://arrayit.com/Products/Printing/Stealth/stealth.html; http://www.biorobotics.com/micspot.html). Because of the precision grinding and machining each pen requires, they are expensive. Cost is an important consideration in microarray systems because the pens are often used in multiplexed print heads of 16, 32, or even 48 pens.
The dominant factor in spot size tends not to be the slot width but rather the much larger contact area of the pen with the substrate (Reese 2001). However, as pens shrink, practical problems arise. Sharper tips are less durable and become dulled by repeated tapping, and narrower slots suffer from clogging and rapid sample evaporation. To address these issues, we used stainless steel microfabrication techniques (Matson et al. 1999; Dziurdzia et al. 2000) to make fountain pens with controlled features and geometry. The high precision and resolution of microfabrication allows one to design pens with small slot widths and contact areas, yet with large reservoirs to prevent evaporation. These pens can be manufactured cheaply in high volumes, and their resolution surpasses that of the best hand-machined pens, allowing a considerable increase in array density. We used our pens in a robotic arrayer system to deposit spots that are 10-30 microns wide and 20-140 microns long, a significant improvement over the current state of the art for deposition techniques. Arrays were created with densities as high as 25,000 spots/cm2. Carryover during array printing was tested with dye, labeled DNA, and hybridized DNA and was found to be indistinguishable from background. Successful hybridizations to printed oligonucleotides demonstrate that hybridization experiments are indeed possible on the droplets deposited, with negligible carryover and good sequence specificity.
RESULTS AND DISCUSSION
Pen Design
High-resolution photolithography is a popular method to create micromechanical devices. In the past, this has been done by using standard silicon fabrication technologies and resulted in the very powerful techniques known as microelectromechanical device (MEMS) technologies (Petersen 1982). Microplating through lithographic masks, commonly referred to as the LIGA process (Becker et al. 1986), has also been widely used to define metal microstructures. In this procedure, metals are electrodeposited through lithographically defined photoresist or x-ray resist masks, and very high aspect ratio features can be achieved. However, microelectroplating of alloys is often difficult to control, and heat treatment of the resulting metal structures can be impossible. Thus, alloys formed by typical LIGA processes suffer significant limitations in their mechanical properties, in particular resilience and tensile strength. In the present study, we decided to use a different approach, which rests on the use of chemical or electrochemical etching of metals in a subtractive procedure, in which the photolithographic resist on the surface of the metal also serves as a chemically resistant etch mask. This provides us with a very inexpensive and versatile technique to define arbitrary geometries into most metal alloys.
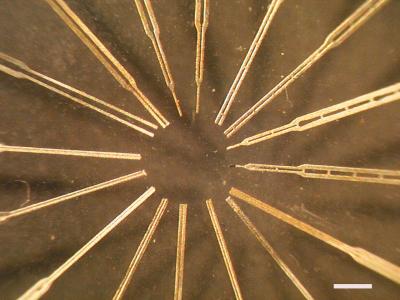
We fabricated the pens from stainless steel foil using optical lithography. Photomasks were made with a 3386-dpi laser printer on standard overhead transparencies. The minimum line spacing on these masks is roughly 25 microns, but the foil was only 12.7 microns thick, allowing us to produce pens with a rectangular cross-section in which one dimension is extremely small. Figure 1 shows a collection of pens of varying designs created using this technique, including features such as reservoirs and mechanical support struts. Conventional pens work by capillary action, which requires that the length of the slot be greater than the width (Dreyer 1994). Because this was impossible to achieve with the printer resolution, we designed a different geometry in which the two-walled slot was replaced with a three-walled trench. When the pen is coated with hydrophilic polyurethane, the trench provides enough capillary action to trap the liquid. The unique design of this pen creates a surprising result: The total tip size is no longer the dominant property in determining the droplet size. Instead, the trench size determines the droplet width. The length of each rectangular droplet is controlled by pen flexure.
Figure 1.
A collection of microfabricated stainless steel pens. Various features have been incorporated in different pens, including reservoirs, support struts, and trenches to increase capillation forces. The ability to lithographically create features of arbitrary shape and small size gives significant design flexibility. Although the resolution of our lithography was limited to 30 microns, more sophisticated equipment should easily approach 1 micron. Bar, 1 mm.
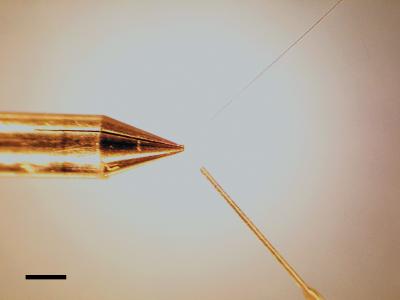
The trench was etched to a depth of 6 microns in the 12.7-micron-thick stainless steel. At the tip, the side walls of the trench are 30 microns wide, and the trench itself has a width of 30 microns. Away from the tip, the trench width and the width of the side walls increase to 90 microns and 120 microns, respectively, to increase the sturdiness of the pen. Figure 2 shows a comparison of the microfabricated trench pen with a conventionally machined slot pen. Higher-resolution photomasks will allow further reduction of pen features. Indeed, smaller channel widths will increase capillation, thus making the pens even more effective.
Figure 2.
A comparison of our trench pen with a conventionally machined slot pen. Although the tips are comparable in size in the lateral dimension, the trench pen (shown head on and in profile) is significantly thinner. The slot pen is a Majer Precision MicroQuill pen. Bar, 1 mm.
Although delicate, there is no reason to expect mechanical failure of the pens during normal spotting or cleaning. Stainless steel is an excellent mechanical material, and we have observed no plastic deformation from the slight deflections the pens undergo during printing and sonication. When printing, the pens contact the printing surface at an angle of 20-30 degrees from perpendicular. Using a nonperpendicular angle serves two purposes. First, this allows greater predictability of pen tip positioning due to flexion of the tip. Second, it serves as a way of crudely managing height variations in the slide, because the pen itself bends as a cantilever beam. Although previous systems used springs as shock absorbers to manage height control, in this case the pen itself acts as a shock absorber. One benefit of this approach is that the pen does not dull; it bends but does not “break.” In the experiments described here, the pen deflects <5% of its length. A second result from using a flexible pen is the characteristic rectangular shape of the footprints of the pen, which can be lengthened or shortened based on the amount of deflection.
Pen Testing
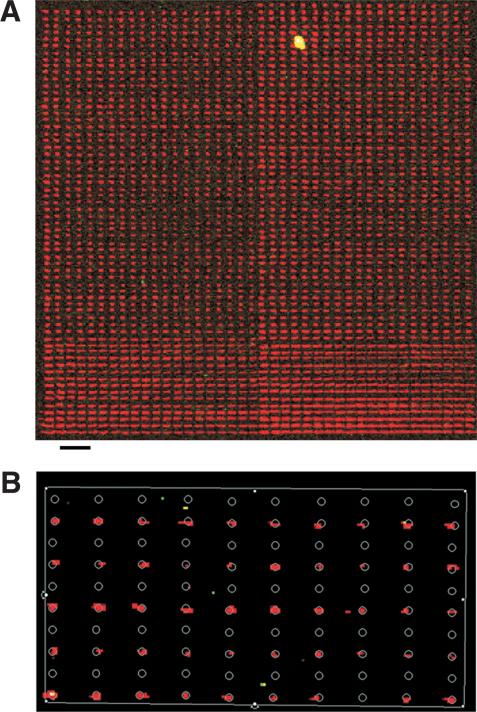
These pens were used to print arrays of fluorescent dye of up to 2500 spots with densities as high as 25,000 spots/cm2. Such an array was produced in a 3.2 × 3.2-mm square (Fig. 3A), suggesting slide capacities of ∼75,000, 150,000, or 225,000 spots when using 16, 32, or 48 pens. (We assume the pens are spaced 4.5 mm apart to load from 384-well microtiter plates, and that each pen prints a block ∼4.4 mm on each side.) Some of the variation in feature size in Figure 3A can be eliminated by printing in a dry environment to limit postprinting spot expansion (data not shown), suggesting that improved uniformity could be achieved if the robotic arrayer is fitted with a humidity-controlled chamber. The highest-density arrays printed with the trench pens had feature sizes of 20 × 40 microns. Lower-density arrays were also produced, with rectangular feature sizes ranging from 20 × 80 to 30 × 140 microns. With careful tip cleaning, we observed negligible carryover when printing spots (Fig. 3B). With a single loading, a pen could print on average 5-20 consecutive spots, depending on spot size and blotting conditions.
Figure 3.
A high-density array of xylene cyanol FF spots. (A) An array of >2500 spots was produced in a 3.2 × 3.2-mm square by a single pen. It has a density of 25,000 spots/cm2. Bar, 250 μm. (B) An array with alternating rows of water and concentrated dye solution was printed to test carryover. With careful pen cleaning, observed carryover is negligible. The white circles superimposed onto the image are 30 μm in diameter, and are spaced 180 and 90 μm apart in the horizontal and vertical directions, respectively.
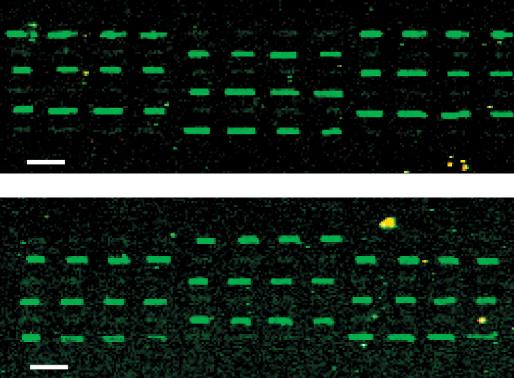
As array densities increase and spot sizes shrink, a concern is having enough material deposited to measure a signal. To prove that the printed arrays could be used to measure DNA hybridization, we spotted down two species of short DNA probes and then hybridized fluorescently labeled complementary oligonucleotides to them. The two different oligonucleotides were printed in blocks of 72 spots with a single microfabricated pen. The blocks were printed with six rows of 12 spots. While printing each row, the pen was loaded prior to each group of four spots, alternating between the two probes on each load. Scans of arrays hybridized with complement A showed successful binding only to probe A. To further illustrate the success of the hybridization, the same slide was washed so as to remove the hybridized target DNA, and a second successful hybridization was performed with complement B (Fig. 4).
Figure 4.
Hybridization results of DNA oligonucleotides to printed probes. Two 10-mer sequences were spotted down with the microfabricated trench pen. (Top) Complement A hybridizes selectively to probe A and does not hybridize to probe B. (Bottom) Complement B hybridizes only to probe B and not to probe A. Bars, 200 μm.
Although the size of the rectangular spots in Figures 3 and 4 appears irregular in comparison with spots printed with conventional pens, in fact, we discovered that they are more regular—the variations are simply more noticeable because the spots are smaller. We analyzed the results of hybridization to several arrays on the same substrate—three independently printed 72-spot arrays made by our stainless steel pens (one shown in Fig. 4, top), and one 72-spot array printed with a conventional pen (data not shown). For the three arrays printed with the new pens, we found the mean spot sizes to be 3500 ± 700, 3750 ± 400, and 3550 ± 550 μm2; the array printed with the conventional pen had a mean spot size of 28,300 ± 4000 μm2. The relative variation in the latter spots is slightly smaller, giving the appearance of more regular spot size, but in absolute terms, the arrays produced by the new pens have a significantly more consistent spot size and quantity of deposited DNA.
We also compared the uniformity of the hybridization intensity to assess accuracy. The three arrays printed with the new pens were found to have mean signals of 122 ± 2, 119 ± 4, and 114 ± 5 fluorescence units (for the 36 spots of each array with sequence matching the hybridization target), showing an accuracy of better than 5% for all arrays. The mean fluorescence intensity for the array printed by the conventional pens was 140 ± 4 units. The higher brightness in this case can probably be attributed to the larger droplet that is deposited by conventional pens. Because larger droplets have more “height” when wet, the spots will have a higher areal density of DNA upon drying. By adjusting the concentration of the spotting solution, one can counteract this effect and achieve similar sensitivity between the two printing methods.
Conclusions
These results demonstrate that microfabricated fountain pens are capable of depositing consistently small features that may be used in DNA hybridization experiments with low amounts of carryover and nonspecific binding. These pens can be mass-produced cheaply because the material is inexpensive and the photolithography process allows parallel production. Higher-resolution lithography will permit the fabrication of pens that print smaller features while storing larger amounts of fluid. This will lead to higher-density DNA arrays, allowing one to measure full genome gene expression of humans and mice with a single array. Finally, increased feature density should improve array sensitivity by reducing the area available for nonspecific binding and by decreasing the surface area a target molecule must diffuse over.
METHODS
Pen Fabrication
Pens were fabricated by using a two-exposure procedure to define a pattern into 12.7-micron-thick 300 series stainless steel shim-stock sheets. During the lithographic exposure, the metal sheet is patterned from both the front and the back surface and is subsequently etched from both sides. Masks for the front and back of the pen were designed with Adobe Photoshop, printed onto transparencies by using a 3386-dpi laser printer, cut out, and individually secured by their edges to glass plates with tape. The masks were designed to be larger than the stainless steel shim-stock sheets from which the pens were etched. Moreover, alignment marks were defined in mask areas that extended beyond the stainless steel sample edges. The stainless steel sheets were spin-coated with thin layers of Microposit S1818 photoresist on both sides, and a soft-bake was performed for 10 min at 90°C on each side. The foil was then cut into smaller pieces, each of which would ultimately become a separate set of pens. These smaller pieces were then attached to a back mask (mask 1) transparency with tape, and exposed with a front mask pattern (mask 2) in a Carl Suss MJB-3 contact mask aligner. The front mask (mask 2) pattern, which is used for the initial exposure, was registered to the back mask pattern (onto which the sample was attached) by using the alignment marks from the back mask that were defined beyond the edges of the stainless steel shim-stock pieces. In the second photolithography step, the sample was turned over and exposed from the rear with the attached back mask (mask 1) pattern. By using this method, the front and rear of the shim-stock could be lithographically patterned with very accurate aligned features.
By performing lithography on both sides of the shim-stock, it was possible to etch through the 12.7-micron-thick steel layer in a single chemical etch, and it was also possible to define slightly different features on the front and back of the shim-stock sample. After both exposures were completed, the sample was developed in a Microposit CD-30 developer, followed by a 140°C hard-bake for 15 min. The photoresist-masked stainless steel shim-stock was subsequently immersed into a mixture of 40% HCl: 40% H2O: 20% HNO3 (v/v/v), which removed the unmasked areas of stainless steel. During the etch, the sample was gently shaken in the solution to avoid gas bubble formation on the steel surface and to ensure a uniformly etched surface. The etch time was typically 8 to 10 min, or until excess steel was completely separated from the base of the pen. The pens were finally cleaned in baths of acetone, isopropyl alcohol, and distilled water. Low-power ultrasonic cleaning was used to completely remove the photoresist mask layer, and the pens were dipped into a thinned polyurethane solution (1 part ebecryl CL 1039 acrylated urethane: 1 part ethyl alcohol: 1% Irgacure 500) and then inverted and exposed in a UV curing oven for 10 min. At this point, the pens were ready for use.
Mechanics of Printing
Arrays were printed using a homemade robotic arrayer constructed according to the design of Schena et al. (1995). Custom control software was written in order to improve precision of the arrayed spots. Average error was reduced from 41.6 to 13.6 microns by introducing a zeroing algorithm to make use of the more accurate positional repeatability of the motors as opposed to the positional accuracy that is used in the Stanford software. The remaining error is due largely to the use of two motor slides for the x- and y-axes, each with comparable errors. The software developed introduced functions that allowed us to better study printing dynamics as well as giving greater flexibility over printing parameters, including independent row/column spacing, introduction of test print algorithms to calibrate slides quickly, easier positional control of multiple block placements done in several prints on a single slide, alternating printing between arbitrary wells, and the replacement of the vacuum station with a heat reservoir. The code was written in Visual Basic 5.0 by using ActiveX controls from Galil Motion Control. Both its source and executable code are available on the Web at http://thebigone.caltech.edu/genomics/arrayer/software.html.
The cleaning process consists of two stations: a sonication wash station and a drying station. The sonicator used was a Koh-I-Noor Ultrasonic Cleaner 25K42. The drying station was converted from the original Stanford vacuum station to a heat reservoir. The heat reservoir was constructed of two nested aluminum sheet-metal boxes separated by an insulating layer of glass wool. The heat was produced by a heat gun on its low setting, delivered through a hole in the side of the reservoir and deflected upward by an internal shield. Pens dip into the reservoir through the top. The reservoir was preheated for 1 min before a print commenced and was reheated during each sonication. The heat reservoir was measured to maintain temperatures of ∼150°C consistently. Sonication and dry times of 6 and 5 sec, respectively, were found sufficient with two cleaning cycles on each reload.
Slide Calibration Protocol
Considering that microscope slides may vary in thickness between slides by as much as 500 microns, and on a single slide itself by 10s of microns, we established tip-slide distance calibration by using a special algorithm written into the robot control software. This allowed the user to specify a maximum contact distance and then incrementally step this distance on each successive print, reducing contact between the pen and slide. Thus, a block could quickly be produced in which spot sizes would vary (data not shown), from which appropriate contact settings could quickly be established. This was typically done with a solution of either fluorescein or xylene cyanol FF. This step will not be necessary for arrayers that measure the distance to the slide surface.
Hybridization Protocol
Oligonucleotide probes were synthesized at the Caltech Biopolymer Synthesis and Analysis Resource Center with the following sequences: 5′-AACCCCACAA-s-a (probe A) and 5′-ACAACCCA AA-s-a (probe B). “s” indicates the C12 Spacer Phosphoramidite, and “a” indicates the C7 Amino Modifier (both from Glen Research). The complementary fluorescent targets had the sequences: 5′-TTGTGGGGTT-Cy3-A (complement A) and 5′-TTTGGGTTGT-Cy3-A (complement B). Probes were printed onto ArrayIt silylated slides in a printing solution consisting of 5× SSC (sodium chloride sodium citrate), 0.001% SDS (sodium dodecyl sulfate), and 50 μM DNA. The slides were then left to dry at room temperature for 24 h, and subsequently washed and blocked according to the slide manufacturer's recommended protocol, which was modified by extending all wash steps to 5-min duration. Before hybridization, the slides were incubated at 37°C with a solution of 5× SSC, 0.1% SDS, and 10 mg/mL bovine serum albumin (BSA) to reduce background due to nonspecific binding. A separate hybridization solution was prepared for each target oligonucleotide because they are labeled with the same fluorophore: 4× SSC, 0.05% SDS, 0.2 mg/mL BSA, and 0.16 μM DNA. Hybridizations were carried out under a cover slip, for 2 h at a temperature of 15°C. Subsequently, slides were washed in a series of four solutions (W1,W2,W3,W4) for 5 min each. W1 consists of 1× SSC and 0.03% SDS at ≈9°C; W2, 0.2× SSC at ≈11°C; W3, 0.05× SSC at ≈13°C; andW4, H2O at ≈15°C. The ramping temperature was achieved by refrigerating plastic test tubes containing 50 mL of each wash solution to ≈9°C, then performing the entire wash sequence with all tubes exposed to room temperature. Washed slides were dried with nitrogen and scanned immediately on a GenePix 4000A microarray scanner.
Acknowledgments
This work was supported by the Whittier Foundation. We are grateful to Barbara Wold, Brian Williams, James Brody, Chris Hart, Cary Gunn, and David Barsic for helpful discussions and technical assistance.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.623903. Article published online before print in September 2003.
References
- Alizadeh, A.A., Eisen, M.B., Davis, R.E., Ma, C., Lossos, I.S., Rosenwald, A., Boldrick, J.C., Sabet, H., Tran, T., Yu, X., et al. 2000. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403: 503-510. [DOI] [PubMed] [Google Scholar]
- Becker, E.W., Ehrfeld, W., Hagmann, P., Maner, A., and Mynchmeyer, D. 1986. Fabrication of microstructures with high aspect ratios and great structural heights by synchrotron radiation lithography, galvanoformung, and plastic moulding (LIGA process). Microelectronic Eng. 4: 35-56. [Google Scholar]
- Chu, S., DeRisi, J., Eisen, M., Mulholland, J., Botstein, D., Brown, P.O., and Herskowitz, I. 1998. The transcriptional program of sporulation in budding yeast. Science 282: 699-705. [DOI] [PubMed] [Google Scholar]
- DeRisi, J.L., Iyer, V.R., and Brown, P.O. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278: 680-686. [DOI] [PubMed] [Google Scholar]
- Dreyer, M., Delgado, A., and Rath, H.-J. 1994. Capillary rise of liquid between parallel plates under microgravity. J. Colloid Interface Sci. 163: 158-168. [Google Scholar]
- Dziurdzia, B., Nowak, S., Ciez, M., and Gregorczyk, W. 2000. Metal foil screens: Manufacturing and techniques of printing. Proceedings of the XXIV Conference of the International Microelectronics and Packaging Society, IMAPS (International Microelectronics and Packaging Society), Washington D.C.
- George, R.A., Woolley, J.P., and Spellman, P.T. 2001. Ceramic capillaries for use in microarray fabrication. Genome Res. 11: 1780-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub, T.R., Slonim, D.K., Tamayo, P., Huard, C., Gaasenbeek, M., Mesirov, J.P., Coller, H., Loh, M.L., Downing, J.R., Caligiuri, M.A., et al. 1999. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science 286: 531-537. [DOI] [PubMed] [Google Scholar]
- Jin, W., Riley, R.M., Wolfinger, R.D., White, K.P., Passador-Gurgel, G., and Gibson, G. 2001. The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat. Genet. 29: 389-395. [DOI] [PubMed] [Google Scholar]
- Lipshutz, R.J., Fodor, S.P., Gingeras, T.R., and Lockhart, D.J. 1999. High density synthetic oligonucleotide arrays. Nat. Genet. 21: 20-24. [DOI] [PubMed] [Google Scholar]
- Lockhart, D.J. and Winzeler, E.A. 2000. Genomics, gene expression and DNA arrays. Nature 405: 827-836. [DOI] [PubMed] [Google Scholar]
- Matson, D.W., Martin, P.M., Stewart, D.C., Tonkovich A., Lee, Y., White, M., Zilka, J.L., Roberts, G.L. 1999. Fabrication of microchannel chemical reactors using a metal lamination process. Proceedings of the Third International Conference on Microreaction Technology, Springer-Verlag, Berlin, Germany.
- Nuwaysir, E.F., Huang, W., Albert, T.J., Singh, J., Nuwaysir, K., Pitas, A., Richmond, T., Gorski, T., Berg, J.P., Ballin, J., et al. 2002. Gene expression analysis using oligonucleotide arrays produced by maskless photolithography. Genome Res. 12: 1749-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, T., Suzuki, T., and Yamamoto, N. 2000. Microarray fabrication with covalent attachment of DNA using bubble jet technology. Nat. Biotech. 18: 438-441. [DOI] [PubMed] [Google Scholar]
- Petersen, K.E. 1982. Silicon as a mechanical material. Proc. IEEE 70: 420-457. [Google Scholar]
- Reese, M. 2001. “Microfluidic fountain pens.” Undergraduate thesis, California Institute of Technology, Pasadena, CA.
- Schena, M., Shalon, D., Davis, R.W., and Brown, P.O. 1995. Quantitative monitoring of gene-expression patterns with a complementary-DNA microarrays. Science 270: 467-470. [DOI] [PubMed] [Google Scholar]
- Shoemaker, D.D., Schadt, E.E., Armour, C.D., He, Y.D., Garrett-Engele, P., McDonagh, P.D., Loerch, P.M., Leonardson, A., Lum, P.Y., Cavet, G., et al. 2001. Experimental annotation of the human genome using microarray technology. Nature 409: 922-927. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.affymetrix.com/support/technical/datasheets/hgu133_datasheet.pdf; GeneChip Human Genome U133 Set.
- http://www.nimblegen.com/products/human.html; NimbleGen Systems, Inc., Homo sapiens 60-mer microarray.
- http://www.majerprecision.com/pins.htm; Majer Precision MicroQuill Array Pins.
- http://arrayit.com/Products/Printing/Stealth/stealth.html; Stealth Micro Spotting Pins and Printheads.
- http://www.biorobotics.com/micspot.html; BioRobotics: Web site of MicroSpot Pins, tungsten pins for microarraying.
- http://thebigone.caltech.edu/genomics/arrayer/software.html; Caltech MicroArrayer, Matthew Reese.