Abstract
The identification of orthologous groups is useful for genome annotation, studies on gene/protein evolution, comparative genomics, and the identification of taxonomically restricted sequences. Methods successfully exploited for prokaryotic genome analysis have proved difficult to apply to eukaryotes, however, as larger genomes may contain multiple paralogous genes, and sequence information is often incomplete. OrthoMCL provides a scalable method for constructing orthologous groups across multiple eukaryotic taxa, using a Markov Cluster algorithm to group (putative) orthologs and paralogs. This method performs similarly to the INPARANOID algorithm when applied to two genomes, but can be extended to cluster orthologs from multiple species. OrthoMCL clusters are coherent with groups identified by EGO, but improved recognition of “recent” paralogs permits overlapping EGO groups representing the same gene to be merged. Comparison with previously assigned EC annotations suggests a high degree of reliability, implying utility for automated eukaryotic genome annotation. OrthoMCL has been applied to the proteome data set from seven publicly available genomes (human, fly, worm, yeast, Arabidopsis, the malaria parasite Plasmodium falciparum, and Escherichia coli). A Web interface allows queries based on individual genes or user-defined phylogenetic patterns (http://www.cbil.upenn.edu/gene-family). Analysis of clusters incorporating P. falciparum genes identifies numerous enzymes that were incompletely annotated in first-pass annotation of the parasite genome.
With the progress of large-scale sequencing efforts, comparative genomic approaches have increasingly been employed to facilitate both evolutionary and functional analyses: Conserved sequences can be used to infer evolutionary history, and to the extent that homology implies conserved biochemical function, this information may be used to facilitate genome annotation. The concepts of orthology and paralogy originated from the field of molecular systematics (Fitch 1970), and have recently been applied to functional characterizations and classifications on the scale of whole-genome comparisons (Tatusov et al. 1997, 2000, 2001; Chervitz et al. 1998; Mushegian et al. 1998; Wheelan et al. 1999; Rubin et al. 2000). Orthologs and paralogs constitute two major types of homologs: The first evolved from a common ancestor by speciation, and the latter are related by duplication events (Fitch 1970, 2000). Although we can assume that paralogs arising from ancient duplication events are likely to have diverged in function (as in the case of α- and β-tubulins), true orthologs (e.g., α-tubulin from yeast and flies) are likely to retain identical function over evolutionary time, making ortholog identification a valuable tool for gene annotation. In comparative genomics, the clustering of orthologous genes provides a frame-work for integrating information from multiple genomes, high-lighting the divergence and conservation of gene families and biological processes. For pathogens such as the human malaria parasite of Plasmodium falciparum (Gardner et al. 2002; Kissinger et al. 2002; Bahl et al. 2003), orthologous groupings can facilitate the identification of candidates for drug and/or vaccine development.
The identification of orthologous groups in prokaryotic genomes has permitted cross-referencing of genes from multiple species, facilitating genome annotation, protein family classification, studies on bacterial evolution, and the identification of candidates for antibacterial drug development (Tatusov et al. 1997; Galperin and Koonin 1999; Natale et al. 2000a,b; Forterre 2002). The Clusters of Orthologous Groups (COG) database (http://www.ncbi.nlm.nih.gov/COG/) is constructed based on all-against-all BLAST searches of complete proteomes (Tatusov et al. 2000, 2001). Sequences from distinct genomes that are reciprocal best hits (i.e., the first sequence finds the second sequence as its best hit in the second species, and vice versa) are identified as a pair of orthologs, and “COGs” recognizing relationships among at least three distinct lineages (triangles) have been identified across distant phylogenetic lineages.
Although Saccharomyces cerevisiae is included in the COG database, general application of this approach in the construction of orthologous groups for other eukaryotic genomes has proved problematic (even for complete prokaryotic genomes, extensive manual inspection of COGs is often required to correct false-positives and split mega-clusters). Complications associated with ortholog group construction for eukaryotic genomes include extensive gene duplication and functional redundancy, the multidomain structure of many proteins, and the predominance of incomplete eukaryotic genome sequencing (Doolittle 1995; Henikoff et al. 1997). These challenges demand an approach able to distinguish between “recent” paralogs (i.e., gene duplications occurring subsequent to speciation, such as the multiple β-tubulins found in the human genome), and “ancient” paralogs likely to exhibit different function(s). Recent paralogs (which are equally related to orthologs in other species) are likely to retain similar function, and should be grouped with true orthologs, It is also important to assess global relationships among orthologs, without being misled by local relationships coming from complicated domain structures or incorrect ortholog assignments. Unfortunately, the computational costs of multiple sequence alignments and phylogenetic tree construction, and the difficulty in interpreting such alignments and trees, preclude a phylogenetic approach for whole-genome comparisons in eukaryotes.
The INPARANOID algorithm (Remm et al. 2001) exploits a BLAST-based strategy to identify orthologs as reciprocal best hits between two species, while applying additional rules to accommodate paralogs arising from duplication after speciation (a.k.a. in-paralogs). Note that the resulting ortholog groups include paralogs derived by recent gene duplication, as each of these proteins is orthologous to a protein in another species. This algorithm performs well in the identification of ortholog groups compared to a curated set of yeast versus mammalian orthologs defined by phylogenetic methods, providing evidence that the strategy based on reciprocal best hits works well in separating orthologs from “ancient” paralogs. Unfortunately, the rule-based approach used by INPARANOID assumes that pairwise comparison is limited to comparisons between two species. EGO (previously named TOGA; Lee et al. 2002) applies a COG-based approach to the TIGR gene indices (Quackenbush et al. 2000, 2001), and this method is applicable to multiple species. In the absence of a rigorously curated data set of orthologs from multiple eukaryotic species, it is difficult to assess performance, but EGO is easily misled by the functional redundancy of multiple paralogs, and by the absence of true orthologs within incomplete genome data sets.
Motivated by these challenges, we developed OrthoMCL as an alternative approach for automated eukaryotic ortholog group identification. To distinguish functional redundancy from divergence, this method identifies “recent” paralogs to be included in ortholog groups as within-species BLAST hits that are reciprocally better than between-species hits. This approach is similar to INPARANOID, but differs primarily in the requirement that recent paralogs must be more similar to each other than to any sequence from other species. To resolve the many-to-many orthologous relationships inherent in comparisons across multiple genomes, OrthoMCL applies the Markov Cluster algorithm (MCL; Van Dongen 2000; http://micans.org/mcl/), which is based on probability and graph flow theory and allows simultaneous classification of global relationships in a similarity space. MCL simulates random walks on a graph using Markov matrices to determine the transition probabilities among nodes of the graph. The MCL algorithm has previously been exploited for clustering a large set of protein sequences, where it was found to be very fast and reliable in dealing with complicated domain structures (Enright et al. 2002). OrthoMCL generates clusters of proteins where each cluster consists of orthologs or “recent” paralogs from at least two species. We have now employed OrthoMCL to examine the proteomes from several other genomes, including Homo sapiens (human), Drosophila melanogaster (fruit fly), Caenhorhabditis elegans (nematode worm), Saccharomyces cerevisiae (yeast), the flowering plant Arabidopsis thaliana, the protozoan malaria parasite Plasmodium falciparum, and the bacterium Escherichia coli; results can be examined online (http://www.cbil.upenn.edu/gene-family). The underlying object-based relational storage model GUS (Genomic Unified Schema; Davidson et al. 2001) also hosts the human–mouse DoTS gene index (http://www.allgenes.org) and the Plasmodium Genome Database PlasmoDB (http://PlasmoDB.org; Kissinger et al. 2002; Bahl et al. 2003), permitting these results to be integrated with various organismal data types to facilitate comprehensive data mining.
RESULTS
Identification of Orthologous Groups by OrthoMCL
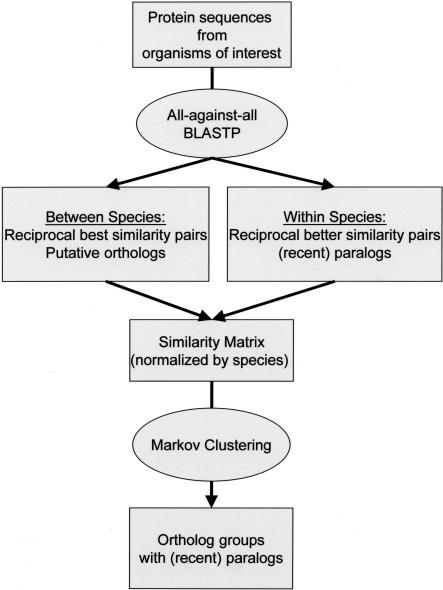
The OrthoMCL procedure starts with all-against-all BLASTP comparisons of a set of protein sequences from genomes of interest (Fig. 1). Putative orthologous relationships are identified between pairs of genomes by reciprocal best similarity pairs. For each putative ortholog, probable “recent” paralogs are identified as sequences within the same genome that are (reciprocally) more similar to each other than either is to any sequence from another genome. A P-value cut-off of 1e-5 was chosen for putative orthologs or paralogs, based on empirical studies.
Figure 1.
Flow chart of the OrthoMCL algorithmfor clustering orthologous proteins.
Next, putative orthologous and paralogous relationships are converted into a graph in which the nodes represent protein sequences, and the weighted edges represent their relationships. As shown in Figure 2, weights are initially computed as the average –log10 (P-value) of BLAST results for each pair of sequences. Because the high similarity of “recent” paralogs relative to orthologs can bias the clustering process, edge weights are then normalized to reflect the average weight for all ortholog pairs in these two species (or “recent” paralogs when comparing within species). Although more sophisticated weighting schemes can be envisioned, this simple method for adjusting the systematic bias between edges connecting sequences within the same genome and edges connecting sequences from different genomes seems to generate satisfactory results, judging from the comparison with INPARANOID, the EGO database, and EC annotations (see below). The resulting graph is represented by a symmetric similarity matrix to which the MCL algorithm (Enright et al. 2002) is applied. MCL uses flow simulation and considers all the relationships in the graph globally and simultaneously during clustering, providing a robust method for separating diverged paralogs, distant orthologs mistakenly assigned based on (weak) reciprocal best hits, and sequences with different domain structures. An important parameter in the MCL algorithm is the inflation value, regulating the cluster tightness (granularity); increasing the inflation value increases cluster tightness (see below). Clusters containing sequences from at least two species form the final output of this procedure: clustered groups of orthologs and “recent” paralogs.
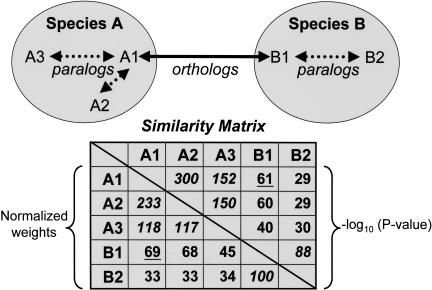
Figure 2.
Illustration of sequence relationships and similarity matrix construction. Dotted arrows represent “recent” paralogy (duplication subsequent to speciation); solid arrows represent orthology. The upper right half of the matrix contains initial weights calculated as average –log10 (P-value) frompairwise WU-BLASTP similarities. The lower left half contains corrected weights supplied to the MCL algorithm; the edge weight connecting each pair of sequences wij is divided by Wij/W, where W represents the average weight among all ortholog (underlined) and “recent” paralog (italicized) pairs, and Wij represents the average edge weight among all ortholog pairs from species i and j. The net result of this normalization is to correct for systematic differences in comparisons between two species (e.g., differences attributable to nucleotide composition bias), and when i = j, to minimize the impact of “recent” paralogs (duplication within a given species) on the clustering of cross-species orthologs.
OrthoMCL Performance on a Pairwise Comparison of Worm and Fly Proteomes
In order to evaluate the performance of OrthoMCL on pairwise comparisons between two species, both OrthoMCL and INPARANOID were applied to the complete set of protein predictions for the fly and worm. Because OrthoMCL uses WU-BLAST results for sequence similarities and INPARANOID uses NCBI-BLAST, INPARANOID was adapted to use the –log10 (P-value) from WU-BLAST as a similarity measure, rather than the NCBI-BLAST bit score (see Methods). INPARANOID identified slightly more orthologous sequences than previously reported (Remm et al. 2001), due to a lower stringency in filtering BLAST results (see Methods). The computational time required for the application of either method is primarily attributable to BLAST analysis and postprocessing of these results. Given processed BLAST results, OrthoMCL required ∼35 min on a Linux i686 computer to cluster the worm and fly protein data sets (including database transactions), whereas INPARANOID required 15 min (note that OrthoMCL is implemented as a pipeline on a relational database whereas INPARANOID operates on flat files). Clusters obtained using the two methods were compared by determining the number of groups that are identical, and those that are coherent—that is, where the sequences in a group generated by one method are a subset of sequences in a group generated by the other (note that identical groups are a subset of coherent groups).
As shown in Table 1, from a total of 33,062 proteins (13,288 fly; 19,774 worm), OrthoMCL clustered 10,849 sequences (33% of the total data set) into 4061 groups, whereas INPARANOID clustered 11,357 sequences (34%) into 4135 groups. We found that 10,597 sequences (32% of the total data set) were recognized by both OrthoMCL and INPARANOID. Thus, 98% of the proteins grouped by OrthoMCL were also grouped by INPARANOID, whereas 93% of the proteins grouped by INPARANOID were also grouped by OrthoMCL. In addition, 8629 proteins (81% of the total number grouped by both algorithms) were grouped into 3735 identical groups, representing 92% of the total number of orthologous groups identified by OrthoMCL, and 90% of the INPARANOID groups. It was revealed that 10,229 proteins (97%) formed coherent groups; 3888 OrthoMCL groups (96%) were a subset of an INPARANOID group, and 3912 INPARANOID groups (95%) were a subset of an OrthoMCL group. These results demonstrate that when employed for the comparison of two genomes, OrthoMCL and INPARANOID exhibit very similar performances.
Table 1.
Comparison of Ortholog Groups Identified by OrthoMCL vs. INPARANOID
| Total | OrthoMCLa | INPARANOID | Grouped by both (∩)b | Identical groups | Coherent groups | |
|---|---|---|---|---|---|---|
| # Protein sequences | 33,062 | 10,849 (33%) | 11,357 (34%) | 10,597 (98/93%) | 8,629 (81%)c | 10,229 (97%)c |
| Fly data set | 13,288 | 5,133 (39%) | 5,550 (42%) | 5,006 (98/90%) | 4,058 (81%) | 4,820 (96%) |
| Worm data set | 19,774 | 5,716 (29%) | 5,807 (29%) | 5,591 (98/96%) | 4,571 (82%) | 5,409 (97%) |
| # Groups | 4,061 | 4,135 | 3,735 (92/90%)d | 3,888/3,912e (96/95%)d |
Using inflation index I = 1.5 (see text).
Percentages indicate percent of sequences grouped by either OrthoMCL (left) or INPARANOID (right).
Percent of sequences grouped by both OrthoMCL and INPARANOID.
Percent of OrthoMCL groups (left); percent of INPARANOID groups (right).
OrthoMCL groups entirely contained within INPARANOID groups (left); INPARANOID groups entirely contained within OrthoMCL groups (right).
OrthoMCL Performance on a Three-Species Data Set (Yeast, Worm, Fly)
A serious limitation to the general application of INPARANOID for comparative genomics applications is that this algorithm can only be employed to compare two sets of proteins, as noted above. In contrast, OrthoMCL can be applied to all-against-all comparisons of multiple genomes. As a test case, we applied OrthoMCL to the complete proteomes of yeast, fly, and worm; results are summarized in Table 2. From the data set of 39,420 proteins (6358 from yeast, 13,288 fly, and 19,774 worm), OrthoMCL placed 13,851 proteins (2531 yeast, 5409 fly, 5911 worm) into 4425 groups (using I = 2.5). The bottom row of Table 2 shows that 1748 (40%) of these groups contain sequences from all three species.
Table 2.
Comparison of Ortholog Groups Identified by OrthoMCL vs. EGO
| Total | OrthoMCLa | EGOb | Grouped by both (∩) | Identical groups | EGO extends OrthoMCLe | OrthoMCL extends EGOf | Coherent groups | |
|---|---|---|---|---|---|---|---|---|
| # Protein sequences | 39,420 | 13,851 (35%) | 5,286 (13%) | 4,959 (36/94%)c | 2,432 (49%)d | 158 (3%)d | 3,004 (61%)d | 4,716 (95%)d |
| Yeast data set | 6,358 | 2,531 (40%) | 923 (15%) | 882 (35/96%) | 452 (51%) | 38 (4%) | 617 (70%) | 827 (94%) |
| Fly data set | 13,288 | 5,409 (41%) | 2,138 (17%) | 2,018 (37/94%) | 987 (49%) | 66 (3%) | 1,174 (58%) | 1,928 (96%) |
| Worm data set | 19,774 | 5,911 (30%) | 2,225 (11%) | 2,059 (35/93%) | 993 (48%) | 54 (3%) | 1,213 (59%) | 1,961 (95%) |
| # Groups | 4,425 | 3,620 | not applicable | 989 (22/27%)g | 70 (2%)i | 2,038 (56%)j | 1,059/3,027h (24/84%)g | |
| Yeast, fly not worm | sequences | 586 (4%)k | 816 (17%)l | 56 (10/7%) | 40 (71%) | 2 (4%) | 9 (16%) | 51 (92%) |
| groups | 215 (5%)i | 440 (12%)j | not applicable | 20 (9/5%) | 1 (0.5%) | 5 (1%) | 21/25 (10/6%) | |
| Yeast, worm not fly | sequences | 470 (3%) | 911 (17%) | 62 (13/7%) | 28 (45%) | 0 (0%) | 30 (48%) | 58 (94%) |
| groups | 155 (4%) | 492 (14%) | not applicable | 14 (9/3%) | 0 (0%) | 19 (4%) | 14/33 (9/7%) | |
| Fly, worm not yeast | sequences | 6,337 (46%) | 3,568 (67%) | 1,614 (25/45%) | 1,161 (72%) | 18 (1%) | 390 (24%) | 1,535 (95%) |
| groups | 2,307 (52%) | 1,874 (52%) | not applicable | 571 (25/30%) | 9 (0.4%) | 208 (11%) | 580/779 (25/42%) | |
| Fly, worm and yeast | sequences | 6,458 (47%) | 2,105 (40%) | 1,868 (29/89%) | 1,203 (64%) | 48 (3%) | 654 (35%) | 1,763 (94%) |
| groups | 1,748 (40%) | 814 (22%) | not applicable | 384 (22/47%) | 16 (1%) | 263 (32%) | 400/647 (23/79%) |
Using inflation index I = 2.5 (see text).
See text for description of pruning method used to identify groups containing sequences from at least two of the three species under consideration (yeast, fly, and worm).
Percent of sequences grouped by OrthoMCL (left) or EGO (right).
Percent of sequences grouped by both OrthoMCL and EGO.
Coherent (but not identical) groups extended by EGO (EGO ⊃ OrthoMCL).
Coherent (but not identical) groups extended by OrthoMCL (OrthoMCL ⊃ EGO).
Percent of OrthoMCL groups (left); percent of EGO groups (right).
OrthoMCL groups entirely contained within EGO groups (left); EGO groups entirely contained within OrthoMCL groups (right).
Percent of OrthoMCL groups (regardless of species distribution).
Percent of EGO groups (regardless of species distribution).
Percent of all sequences grouped by OrthoMCL (regardless of species distribution).
Percent of all sequences grouped by EGO (regardless of species distribution).
EGO is based on a clustering of transcribed sequences, and provides the most comprehensive published resource of eukaryotic ortholog groups (Lee et al. 2002). These groups are defined by first clustering and assembling ESTs and transcripts into consensus sequences that are represented in the TIGR Gene Indices (Quackenbush et al. 2000, 2001). Gene indices are then used to construct ortholog groups by pairwise comparisons, using an approach similar to that employed in the COG database (Tatusov et al. 2000, 2001); in EGO, reciprocal best match pairs that link at least three species are clustered into groups. Finally, paralogs are identified as one-way best hits, and groups are further combined by merging extensively overlapping groups.
To compare the OrthoMCL results for yeast, fly, and worm with the EGO database (including genes from many species), we first compared the gene indices for these species to their proteomes using BLASTP. We then extracted those EGO groups that contain sequences from at least two of the three species, and discarded sequences derived from other species. This yields a nonredundant set of 5286 proteins (923 from yeast, 2138 fly, 2225 worm), corresponding to 6106 gene index sequences (note that because the EST database used to construct gene indices contains partial cDNA sequences and alternatively spliced isoforms, multiple gene index sequences sometimes map to a single protein). A total of 3620 EGO groups were identified, of which 814 (22%) contain sequences from all three species. In the analysis below, groups derived from the EGO database are referred to as the “EGO subset”.
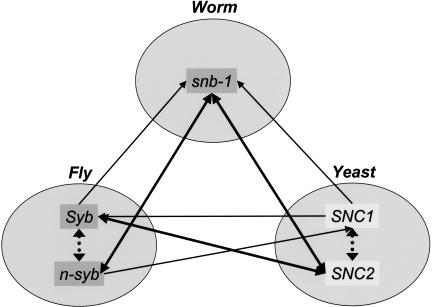
Far more sequences were grouped by OrthoMCL (13,851) than the EGO subset (5286); this accounts for 35% versus 13% of all protein sequences. In some cases, the greater inclusiveness of OrthoMCL is attributable to the recognition of “recent” paralogs that were missed by EGO because they were not best hits. In other cases this reflects the inclusion of ortholog groups containing only two species in OrthoMCL (EGO, like COG, requires a `triangle' of reciprocal best hits joining sequences in three species). Figure 3 provides an illustrative example, showing OrthoMCL group #379767, containing five synaptobrevin genes from worm, yeast, and fly (including “recent paralogs” in the latter two species). Only Syb (fly), n-syb (fly), and snb-1 (worm) — mapping to gene indices TC134828, TC140251 from Drosophila, and TC72314 from C. elegans, respectively—were identified by the EGO subset (Syb and snb-1 were contained in two EGO groups — TOG273790 and TOG272289—whereas n-syb and Snb-1 were contained in TOG257010). The EGO subset failed to include any synaptobrevin genes from yeast, because they did not form a triangle of reciprocal best matches due to independent recent gene duplications in yeast and fly, producing “recent” paralogs. Syb, n-syb, and snb-1 were included in the EGO subset only because they formed triangles of reciprocal best hits with sequences from other species not analyzed here (based on BLASTN searches; note that the BLASTP analysis employed by OrthoMCL identifies n-syb as the best hit when querying with snb-1 against the fly proteome).
Figure 3.
Example of a group from the EGO subset that is extended by OrthoMCL. Five synaptobrevin genes were clustered together by OrthoMCL (GroupID #379767), including yeast SNC1 and SNC2, fly Syb and n-syb, and worm snb-1. Thick solid arrows represent orthology identified by reciprocal best matches, dotted arrows represent “recent” paralogs, and thin solid arrows represent one-way best matches indicating the direction from query to subject (based on BLASTP comparisons). Only snb-1, n-syb, and Syb (dark gray) were identified by the EGO subset (groups TOG257010, TOG272289, TOG273790), and these genes were only grouped because their gene index sequences (TC72314, TC140251, TC134828) formed `triangles' of reciprocal best matches based on BLASTN comparisons with other species not shown in this analysis.
Virtually all of the 5286 proteins grouped in the EGO subset were also grouped by OrthoMCL, (4959 = 94%; Table 2). Of the 327 sequences not represented in OrthoMCL, many represent cases where EGO groupings were dependent on sequences from other species in the complete EGO database, which would presumably be recognized by a larger-scale application of OrthoMCL. Other differences are attributable to the addition of sequences by EGO on the basis of one-way best hits, inappropriately grouping functionally diverged (i.e., “ancient”) paralogs where true orthologs have been lost. In still other cases, reciprocal best hits were separated by OrthoMCL during the clustering step because they exhibit much lower similarity than other sequences in the group. Finally, some differences are attributable to the use of BLASTN in constructing the EGO database, whereas OrthoMCL uses BLASTP.
Of the 4959 sequences grouped by both methods, 2432 (49%) were included in 989 identical groups. To assess the coherence of nonidentical groups, we examined cases where groups identified by one method were extended by the alternative method, that is, cases where all sequences contained in an OrthoMCL group are included in a larger EGO group, or vice-versa. Only 70 OrthoMCL groups were extended by EGO, but OrthoMCL extends 2038 EGO groups. Combining all of these categories ([EGO = OrthoMCL] + [EGO OrthoMCL] + [OrthoMCL EGO]) yields a total of 4716 sequences. Thus 95% of the total number of sequences identified by both algorithms were represented in coherent groups (this number is smaller than the sum of sequences in all three subsets, because many sequences appear in multiple EGO groups, despite a final step in which initial EGO groups are merged).
As shown in Table 3, OrthoMCL successfully clusters multiple overlapping groups identified by EGO. Because the algorithm only clusters reciprocal best hits, EGO places eight glyceraldehyde 3-phosphate dehydrogenase (GAPDH) genes from yeast, fly, and worm into 14 overlapping groups, the largest of which contains seven genes (some of these groups are identical, and some are a subset of others, because the EGO database grouped these sequences with genes from other species not analyzed here). In contrast, OrthoMCL clusters a total of nine GAPDH genes into a single group (#380487; Table 3); the recognition of “recent” paralogs causes multiple sequences to be clustered together by OrthoMCL, reducing the redundant groups presented by EGO.
Table 3.
EGO Groups Combined by OrthoMCL Group 380487 (GAPDH)a
|
Yeast
|
Fly
|
Worm
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene: Protein: | TDH1 YJL052W | TDH2 YJR009C | TDH3 YGR192C | Gapdh1 Fbgn 0001092 | Gapdh2 Fbgn 0001091 | Gpd-1 CE02343 | gpd-2 CE07371 | gpd-3 CE07370 | Gpd-4 CE01568 |
| EGO group | |||||||||
| 248594: | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| 248635: | ✓ | ✓ | ✓ | ✓ | ✓ | ||||
| 248718: | ✓ | ✓ | ✓ | ✓ | ✓ | ||||
| 248791: | ✓ | ✓ | ✓ | ✓ | |||||
| 249074: | ✓ | ✓ | ✓ | ||||||
| 249666: | ✓ | ✓ | ✓ | ||||||
| 249879: | ✓ | ✓ | ✓ | ||||||
| 250009: | ✓ | ✓ | ✓ | ||||||
| 253481: | ✓ | ✓ | ✓ | ||||||
| 250748: | ✓ | ✓ | |||||||
| 251654: | ✓ | ✓ | |||||||
| 256270: | ✓ | ✓ | |||||||
| 252346: | ✓ | ✓ | |||||||
| 255159: | ✓ | ✓ | |||||||
| Gene Index (TC#): | 11801 | 10521 | 8903 | 139089 | 128588 | 70553 | 70631 | 70524 | none |
OrthoMCL group 380487 includes nine glyceraldehyde 3-phosphate dehydrogenase (GAPDH) genes: yeast TDH1, -2, and -3; fly Gapdh1 and -2; and worm gpd-1, -2, -3, and -4. Eight of these genes are included in 14 distinct EGO groups. Correlation of proteins with Gene Index TC numbers was carried out by comparing TCs with the organism proteomes using BLASTP to identify best matches with P values <1e-5.
We also examined OrthoMCL and EGO groups exhibiting distinct phylogenetic patterns, as shown in the bottom half of Table 2. Many groups from both analyses contain sequences from all three species (bottom line of Table 2: 1748 = 40% of OrthoMCL groups, 814 = 22% of EGO groups), and comparisons between OrthoMCL and the EGO subset reveal a high degree of coherence: 79% of EGO groups were subgroups of OrthoMCL groups. The majority of groups identified by both analyses contain sequences from fly and worm, but not yeast (2307 = 52% of OrthoMCL groups, 1874 = 52% of EGO groups), reflecting the many shared derived characters associated with metazoa. Forty-two percent of EGO groups containing fly+worm but not yeast sequences were subgroups of OrthoMCL groups exhibiting the same species distribution. For such phylogenetically restricted groups, the coherence between OrthoMCL and EGO is somewhat lower than for unrestricted groups, because OrthoMCL extends many EGO groups to include sequences from the excluded species. The grouping of individual sequences recognized by both algorithms is highly coherent, however (>90%). Far fewer groups were identified with other phylogenetic distribution patterns (215 OrthoMCL groups contain yeast and fly but not worm sequences; 155 OrthoMCL groups contain yeast and worm but not fly sequences), and the coherence between OrthoMCL and EGO was lower for these groups.
In sum, the OrthoMCL and EGO algorithms exhibit highly consistent ortholog groupings. By distinguishing “recent” paralogs from “ancient” paralogs, and clustering “recent” paralogs together with orthologs, OrthoMCL improves the accuracy of the ortholog group assignments, increases data coverage, and decreases the redundancy of data representation.
Application of OrthoMCL to P. falciparum, Human, and Other Eukaryotic Genomes
Based on the success of these analyses, we applied OrthoMCL to a data set of seven proteomes, totaling 101,047 proteins, including A. thaliana (25,009 proteins), C. elegans (19,774), D. melanogaster (13,288), Homo sapiens (27,049), P. falciparum (5279), S. cerevisiae (6358), and E. coli (4290). As shown in Table 4, application of OrthoMCL at an inflation index of 2.5 to the complete proteomes for these seven species identified 7681 groups, representing a total of 45,473 protein sequences (see below for further discussion of the inflation index). In comparison to the 4425 groups identified in the three species data set from Table 2, 74% more groups were identified, representing an increase of 228% in the number of sequences for which putative orthologs could be identified. Thus, most new sequences added to the data set could be accommodated within previously defined groups (the average number sequences/group rose from 3.1 for the three-taxon data set to 5.9 for the seven-taxon data set).
Table 4.
Consistency of OrthoMCL Groups with EC Assignments
| Total data set
|
Groups with ≥1 protein for which complete EC annotation is available
|
Groups with ≥2 proteins for which complete EC annotations are available
|
Consistent EC assignmentsc
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Inflation (cluster tightness) | Groups | Proteins (% of proteome)a | Groups | Proteins | EC-annotated (% of total)b | Groups | Proteins | EC-annotated (% of total) | Groups (% poss.) | Proteins | EC-annotated (% possible) |
| 1.1 | 6,249 | 50,771 (50) | 999 | 12,032 | 2,921 (82) | 664 | 9,561 | 2,586 (73) | 528 (80) | 5,476 | 1,958 (76) |
| 1.5 | 7,265 | 47,668 (47) | 1,117 | 8,730 | 2,877 (81) | 696 | 6,318 | 2,456 (69) | 596 (86) | 4,768 | 2,067 (84) |
| 2.0 | 7,569 | 46,245 (46) | 1,148 | 8,343 | 2,849 (80) | 701 | 5,916 | 2,402 (67) | 611 (87) | 4,610 | 2,073 (86) |
| 2.5 | 7,681 | 45,473 (45) | 1,160 | 8,171 | 2,840 (80) | 705 | 5,789 | 2,385 (67) | 617 (88) | 4,556 | 2,062 (86) |
| 3.0 | 7,786 | 44,729 (44) | 1,172 | 7,975 | 2,831 (79) | 706 | 5,553 | 2,365 (66) | 621 (88) | 4,450 | 2,059 (87) |
| 3.5 | 7,857 | 44,263 (44) | 1,180 | 7,889 | 2,821 (79) | 707 | 5,506 | 2,348 (66) | 624 (88) | 4,444 | 2,057 (88) |
| 4.0 | 7,896 | 43,900 (43) | 1,186 | 7,784 | 2,811 (79) | 704 | 5,414 | 2,329 (65) | 623 (88) | 4,372 | 2,048 (88) |
Total proteome size = 101,047 (Arabidopsis thaliana, 25,009 sequences; Caenorhabditis elegans, 19,774; Drosophila melanogaster, 13,288; Homo sapiens, 27,049; Plasmodium flaciparum, 5279; Saccharomyces cerevisiae, 6358; Escherichia coli, 4290).
A total of 3562 EC-annotated proteins were obtained from the ENZYME database (A. thaliana, 370; C. elegans, 269; D. melanogaster, 210; H. sapiens, 1160; S. cerevisiae, 778; E. coli, 775).
All EC-annotated sequences in the group were assigned the same EC number. Percentages indicate fraction of ortholog groups containing at least two complete EC assignments (the only data set for which consistency can be assessed), or percentage of EC-annotated sequences properly identified.
One important application of orthologous group identification lies in the functional characterization of proteins. In order to assess the utility of OrthoMCL results for protein functional analysis, we examined the consistency of these groups with respect to enzyme commission (EC) numbers assigned by the ENZYME database (http://us.expasy.org/enzyme), reasoning that EC numbers are probably among the most reliable functional assignments that have been widely applied during genome annotation. The complete data set includes a total of 3562 sequences for which a complete EC number has been assigned (described as `EC-annotated' in the analysis below). At an inflation index of 2.5, 2840 of these (80%) were included in OrthoMCL groups. As expected, changing the inflation index affects cluster tightness: Lower `I' values result in the inclusion of more sequences in fewer groups (50,771 sequences in 6,249 groups at I = 1.1), whereas increasing the inflation index fragments clusters and reduces the number of sequences included (43,900 sequences in 7896 groups at I = 4). Recognition of sequences associated with EC numbers parallels this trend: Increasing the inflation value from 1.1 to 4 reduced the number of EC-annotated sequences that were clustered from 2921 to 2811, and increased the number of associated groups from 999 to 1186. It is worth noting that sequences annotated with EC numbers were relatively insensitive to cluster tightness (this range of `I' values affects the inclusion of 7% of total sequences in OrthoMCL groups, but only 3% of EC-annotated sequences), presumably due to the fact that conserved sequences are more likely well annotated.
Only ∼6% of all sequences in the combined proteome are associated with EC annotations (2840/45,473 at I = 2.5), but their distribution is decidedly nonrandom: Only 1160/7681 groups contain any EC-annotated sequences at all, but 705 of these 1160 groups contain more than one EC-annotated sequence. Groups containing two or more EC-annotated sequences can be used to assess the consistency with which OrthoMCL groups these enzymes, by examining the percentage of groups within which all EC annotations are identical. Of the 705 OrthoMCL groups containing at least two EC-annotated sequences (at I = 2.5), all EC annotations were identical within 617 (88%) of these groups, representing 2062 EC-annotated sequences (4556 total sequences). Of the few cases where enzymes from the same ortholog group were associated with different EC numbers, most show inconsistency only at the fourth level of EC hierarchy, suggesting inconsistency in annotation rather than incorrect orthologous group assignment (data not shown). In some cases such inconsistency may also be attributable to cases where multiple EC numbers are assigned to a single (multifunctional) enzyme.
The percentage of groups that are consistent with EC assignments increases from 80% to 88% with increasing cluster tightness. These differences are most pronounced when the inflation value increases from 1.1 to 1.5; the number of EC-annotated sequences in consistent groups was maximal at I = 2.0 (2073 sequences). Tight clustering tends to prevent sequences with different functions from being clustered together, but may also separate true orthologs (e.g., EF-G genes were broken into two clusters when the inflation index was increased from 1.5 to 2.0). An inflation index of I = 1.5 (where 86% of all groups are EC-consistent) appears to balance sensitivity and selectivity: exhibiting consistency close to the maximum observed value, while excluding a minimum number of sequences.
Clustering the entire data set at I = 1.5 yields 7265 groups from a total of 47,668 sequences. We found that 195 groups contained sequences from all seven species analyzed, representing genes that are shared between eukaryotes and bacteria, including DNA helicase, DNA mismatch repair proteins, ribosomal proteins, thymidylate synthase, tRNA synthetases, enolase, ACP synthase, pyruvate kinase, glyceraldehyde-3-phosphate dehydrogenase, etc. (see Supplemental Table 1; http://www.cbil.upenn.edu/gene-family/SupTable1.htm). In addition, 856 groups contain sequences from all six eukaryotic species but not E. coli, representing genes that are conserved among (and may also be restricted to) eukaryotic lineages. The largest groups in this category include calcium-dependent protein kinase, histone proteins (H2B, H3, H4), MAP kinase, cyclophilin-type peptidylprolyl isomerase, myosin, hexokinase, high-mobility group proteins, etc. The distribution of orthologous groups having distinct phylogenetic patterns of gene presence/absence of specific species has been compiled as Supplemental Table 2 (http://www.cbil.upenn.edu/gene-family/SupTable2.htm).
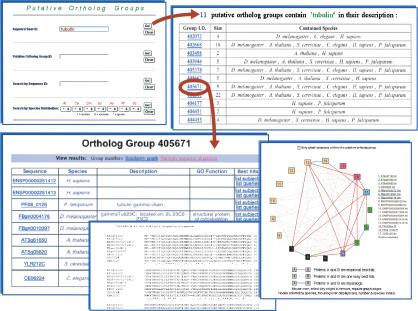
OrthoMCL groups have been stored in the GUS relational database (Davidson et al. 2001) and may be queried through keyword searches based on sequence name or description, or by user-defined patterns of species distribution (http://www.cbil.upenn.edu/gene-family), as indicated in Figure 4. The report for each selected ortholog group provides an interactive graphical display of the similarity relationships among related sequences, a table of information related to each individual member of the group (including, e.g., functional assignments based on Gene Ontologies; Ashburner et al. 2000; The Gene Ontology Consortium 2001; Schug et al. 2002), and a multiple sequence alignment of the group members generated using CLUSTALW (Thompson et al. 1994). The graphical representation can be customized to selectively show two-way best matches, one-way best matches, and “recent” paralog relationships among group members or all of the related sequences (see Fig. 4).
Figure 4.
Screenshots of the Web interface. A keyword search (top left) identifies 11 ortholog groups containing sequences with the word “tubulin” in sequence name or description (top right). Clicking the group ID pulls up a page describing sequences in the group (bottom left), a graphical display of relationships among these sequences (bottom right), and a CLUSTALW multiple sequence alignment (bottom center).
Mining the P. falciparum Proteome: Insights From Ortholog Groupings
The clusters of orthologous genes produced by OrthoMCL can be used to address a variety of biological questions from the perspective of comparative genomics, focusing on shared and divergent biochemical functions. A complete genome sequence for the protozoan parasite Plasmodium falciparum (responsible for the most severe form of human malaria) was recently published (Gardner et al. 2002), and we were interested in exploring whether orthologous group analysis might be useful for functional assignment of predicted proteins by inferring function from other genomes. Where the orthologs have known function in other species, it might be possible to assign putative function in P. falciparum. Cases in which orthologs can be defined for which no function has been assigned may be interesting for evolutionary and functional study. Plasmodium proteins for which no orthologs are identifiable may represent targets for parasite-specific chemotherapy and/or vaccine development. Considering the complete P. falciparum proteome of 5279 sequences, 2191 OrthoMCL groups incorporate a total of 3069 parasite sequences.
Complete EC annotations are available for 349 Plasmodium proteins, and 315 (90%) of these are included in 302 OrthoMCL groups. Of these 302 groups, 270 (89%) also include a total of 931 other EC-annotated proteins, of which 833 (89%) are identical to the P. falciparum EC annotations. We found that 175 P. falciparum sequences without a complete EC number are included in 143 groups containing at least one EC-annotated sequence from another species; 84 of these groups contain at least two EC-annotated sequences. Some of these cases represent properly annotated P. falciparum genes for which EC assignments are missing or incomplete from the annotation, whereas others are annotated as hypothetical proteins. Based on the high level of consistency between OrthoMCL groups and EC assignments (Table 4), the complete EC numbers associated with these groups provide a presumptive functional assignment for these P. falciparum orthologs. Manual curation has confirmed at least 137 of these assignments as valid annotations that were missed during first-pass annotation.
Extending this analysis beyond EC annotations, a total of 1297 P. falciparum sequences annotated as “hypothetical proteins” are included in OrthoMCL groups. Where available, annotations associated with the entire group may prove relevant to the orthologous P. falciparum sequences, providing more reliable transitive annotations than the results from a simple BLAST similarity search. For example, P. falciparum PFD0450c, annotated as “hypothetical protein, conserved”, was clustered in ortholog group #405550 with six other sequences (human ENSP00000263436, fly FBgn0036487, Arabidopsis AT1g60170 and AT3g60610, worm CE24126, and yeast YGR091W). “Unknown protein” AT1g60170 and “putative protein” AT3g60610 were described as similar to a splicing factor, whereas the other four members were all annotated as having pre-mRNA splicing function. Multiple sequence alignment of these sequences provides further support to this putative functional assignment.
One primary goal of pathogen genome projects is to accelerate the search for new drug and vaccine targets. Phylogenetically restricted genes in the parasites that have diverged from (or are absent in) animals are likely to be associated with biological processes that distinguish them from their animal hosts. Searching based on species distribution identifies 447 groups containing sequences from P. falciparum but no human orthologs, and 273 groups containing sequences from P. falciparum but not human, fly, or worm. Proteins with this restricted species distribution include known drug targets such as dihydropteroate synthetase (Group 407390), an ancient gene (shared with E. coli) that has been lost in the animal lineage. Putative orthologs of the chloroquine resistance transporter (Group 403748) are identified only in A. thaliana, although a weaker (but reciprocal) best hit is identified in C. elegans as a chemoreceptor. Many groups containing P. falciparum sequences but not animal sequences include sequences from A. thaliana. Some of these may derive from the secondary endosymbiosis of a eukaryotic alga (Kohler et al. 1997; Roos 1999), giving rise to the apicoplast—a plastid remnant that is essential for parasite survival (Fichera and Roos 1997; He et al. 2001). Known apicoplast proteins include enoyl-acyl carrier reductase (FABI; Group 403190) and 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DOXPR; Group 402142), both of which are under study as potential drug targets. Phylogenetically restricted genes that are conserved in related parasite species provide candidates for broad-spectrum antibiotic development; orthologs of all the genes noted above have also been identified in Plasmodium yoelii yoelli (Carlton et al. 2002).
DISCUSSION
Challenges for Comparative Eukaryotic Genomics
Compared to prokaryotes, eukaryotic genomes tend to exhibit a much higher rate of duplicative gene family expansion. The dynamic fate of these paralogs makes it important—and difficult—to distinguish functional redundancy from functional divergence. Genes that have evolved from relatively “ancient” duplication events (i.e., duplication before speciation) may have diverged to acquire new functions, and these homologs should not be clustered with true orthologs. In contrast, relatively “recent” duplication events (i.e., duplication after speciation) may produce multiple copies of similar or identical genes compared to their orthologs in other species. As noted above, we define these genes as “recent” paralogs, and have devised methods to cluster such genes along with orthologs from other genomes in a many-to-many relationship. Thus OrthoMCL groups six “recent” α-tubulin paralogs in humans with the α-tubulin genes from other eukaryotic species (OrthoMCL group 412325), but not with the “ancient” β- and γ-tubulin paralogs (OrthoMCL groups 412877 and 410694, respectively; see Fig. 4). Incorporating “recent” paralogs into ortholog groups also avoids problems associated with inaccurate or incomplete assembly of eukaryotic genomes—a common problem when microsatellite sequences are abundant.
A second challenge in clustering orthologous groups in eukaryotes comes from the complicated domain architecture of many proteins. In constructing ortholog groups, clustered proteins should have very similar if not identical domain structure. Otherwise, proteins with markedly different functions may be mistakenly clustered into a single group because they share similarities with distinct regions of a multidomain protein, or because they share domains present in many families. For example, the presence of bifunctional dihydrofolate reductase-thymidylate synthase genes in protists and plants should not lead to the inclusion of monofunctional dihydrofolate reductase and thymidylate synthase genes from bacteria, fungi, and animals within a single group. In the COG approach, `triangles' of mutually consistent, genome-specific best hits are merged if they share a common side, to form larger orthologous groups. This straightforward clustering procedure based on transitivity is limited in dealing with complicated domain structures of proteins, as it only considers the local relationship among sequences belonging to the triangles to be merged, while ignoring the global relationship among all proteins to be clustered in the same group. As a result, the original COG method inappropriately merges unrelated proteins based on similarities to different regions of multidomain proteins; further refinement of the COG database requires manual inspection on a case-by-case basis.
A third challenge in identifying eukaryotic ortholog groups derives from the “incompleteness” of genome sequence data. The economics of genome sequencing means that extensive `shotgun' sequencing is (or soon will be) available for many eukaryotes, long before the genome has been sequenced to completion. For example, extensive genome sequence information is now available for at least 10 species of apicomplexan parasites (and extensive EST data sets are available for several additional species), but only the P. falciparum genome has been completely sequenced. This means that true orthologs may be missing, and reciprocal best hits may identify inappropriate substitutes, such as divergent (ancient) paralogs. OrthoMCL evaluates the global pattern of sequence similarities among provisionally grouped sequences during clustering (Fig. 2), minimizing errors attributable to missing genes, because diverged paralogs are likely to exhibit lower similarity to each other when compared with the similarities between true orthologs.
Identification of Eukaryotic Ortholog Groups by OrthoMCL
This report describes a novel approach for the construction of orthologous groups from eukaryotic genomes—identifying both orthologs between species and functionally redundant (“recent”) paralogs within species, and grouping these together using the MCL algorithm to cluster graph vertices by flow simulation. The OrthoMCL approach can be viewed as a two-step process: A graph representation of sequence relationships is generated, and then divided into subgraphs using the MCL algorithm. The first step involves applying rules based on biological knowledge of the problem to determine which sequences should be included, how sequence nodes are connected, and how to weight edges in order to quantify the relationships between sequences. The second step is a graph clustering problem where computational theories and techniques come into play.
The MCL graph clustering algorithm includes a parameter regulating cluster granularity—the inflation index. Table 4 shows that the inflation parameter has relatively little impact on the resultant clusters, however: Rather coarse-grained clustering (I = 1.5) provides sufficient tightness for identifying coherent EC groups, for example. This is even more obvious when a smaller number of genomes is analyzed, as observed from comparisons with INPARANOID and the EGO database using yeast, worm, and fly proteins (data not shown). Overall, the simple structured graph itself seems to capture sequence relationships quite well.
OrthoMCL produces results similar to INPARANOID when applied to two genomes (Table 1), while offering the opportunity to compare multiple genomes. Orthologous group identification across multiple genomes can be very useful for genome annotation, revealing the phylogenetic patterns of proteins from distinct lineages, and providing evolutionary insights into the conservation and diversity of cellular functions in different species. OrthoMCL groupings were coherent with groups produced by EGO (Table 2), with most differences attributable to extending the EGO groups through the addition of “recent” paralogs, and combining EGO groups linked via these paralogs (Fig. 3; Table 3).
OrthoMCL differs from the EGO strategy (and the COG algorithm commonly applied to prokaryotic genomes; Tatusov et al. 2000, 2001) in several ways. OrthoMCL does not insist on a minimum of three species to form a “triangle” of reciprocal best hits, a requirement which poses problems when genome sequence information may be incomplete. Moreover, even when complete genomes are compared, groups representing orthologs in only two species can be very informative, as from the comparison of P. falciparum and A. thaliana cited above. OrthoMCL also differs from EGO by incorporating “recent” paralogs, and by considering these sequences together with orthologs during the clustering process, rather than appending paralogs as one-way best hits after clustering reciprocal best hits. This strategy is particularly useful for the analysis of eukaryotic genomes, where gene duplications may prevent the formation of reciprocal best hits (cf. Fig. 3).
The implementation of OrthoMCL outlined above provides a successful proof of concept, although further improvements may be possible by focusing on individual components of the process. BLASTP comparisons might be modified to provide a more sophisticated weighting scheme for capturing sequence similarities and domain architecture, providing greater robustness in dealing with protein fusions. Modifying the normalization of inter-versus intra-species weights might improve the handling of lineage-specific expansions. Finally, alternative clustering algorithms might be applied (Shi and Malik 1997; Abascal and Valencia 2002).
Mining Comparative Genome Databases
Identification of orthologous groups across multiple eukaryotic taxa provides a valuable resource for automated genome annotation, as noted above. BLAST-based clustering assumes equal evolutionary rates among paralogs, however, masking cases where “recent” paralogs may have evolved to acquire new function, or where distant orthologs differ significantly (e.g., in the substrate specificities of enzymes). The ability to adjust clustering granularity makes OrthoMCL scalable for the identification of functionally conserved orthologs and paralogs. The consistency of EC assignments with OrthoMCL groups justifies the use of this tool in automated annotation.
Distinctive biological features revealed by the phylogenetic patterns from orthologous grouping are particularly useful for analyzing pathogen genomes, offering great potential for biological investigation of pathogen evolution and drug or vaccine targets. In the future, we plan to expand the Web-based database of orthologous groups illustrated in Table 4 and Figure 4 to include other species, and to integrate these results into genome databases such as the malaria parasite genome database PlasmoDB (Kissinger et al. 2002; Bahl et al. 2003), to facilitate data mining by the research community.
METHODS
Data Sources
Data examined in this manuscript were downloaded from the following sources (duplicate entries removed).
Arabidopsis thaliana data (25,009 predicted proteins): The Institute for Genome Research (ATH1_pep.06132001) http://www.tigr.org/tdb/e2k1/ath1/ath1.shtml.
Caenorhabditis elegans (19,774 predicted proteins): Wormpep (release wormpep54) http://www.sanger.ac.uk/Projects/C_elegans/wormpep/wormpep_download.shtml.
Drosophila melanogaster (13,288 predicted proteins): FlyBase (translated polypeptide for every predicted transcript from Release 2) http://www.fruitfly.org/sequence/download.html.
Homo sapiens (27,049 predicted proteins): Ensembl (release 7.29) ftp://ftp.ensembl.org/pub/human-7.29/.
Plasmodium falciparum data (5279 predicted proteins): PlasmoDB (release 4.0, excluding 55 pseudogenes) www.plasmodb.org/restricted/data/P_falciparum/.
Saccharomyces cerevisiae (6358 predicted proteins): SGD (translated yeast ORFs) http://genome-www.stanford.edu/Saccharomyces/DownloadContents.shtml.
Escherichia coli (4290 predicted proteins): The E. coli Genome Project (E. coli K-12 sequence and annotations) www.genome.wisc.edu/sequencing/k12.htm.
TIGR Gene Indices obtained from http://www.tigr.org/tdb/tgi/.
EGO database obtained from http://www.tigr.org/tdb/tgi/ego/index.shtml.
EC associations for SWISS-PROT proteins obtained from the ENZYME database ftp://us.expasy.org/databases/enzyme.
Software
BLAST searches were carried out using WU-BLAST 2.0 http://blast.wustl.edu/. INPARANOID software was obtained from http://kisac.cgb.ki.se/cgb/inparanoid/. MCL software was obtained from http://micans.org/mcl/mcl-02-063/.
Identification of Ortholog Groups Using INPARANOID
The source code of INPARANOID was modified so that it could be applied to WU-BLAST results using –log10(P-value) as the similarity score. Based on the distribution of P-values from BLAST results, a score of 350 was used when the P-value was returned as `0', and the similarity score between a sequence and itself was defined as 350. A cutoff P-value of 1e-5 was used, and no match length coverage is required. The cutoff confidence value for inparalogs (“recent” paralogs) was set at 0.01.
Construction of the “EGO Subset”
Orthologous groups containing gene index sequences (TCs) from yeast, worm, or fly were extracted from the EGO database, and TCs from other Gene Indices in these groups were discarded. Each TC from these groups was then substituted with its best match in the proteome, determined by comparing sequences from yeast, worm, and fly Gene Indices with their respective proteomes using BLASTP (V = 1, B = 1, cutoff P-value of 1e-5). Redundant sequences (arising when more than one TC was associated with a protein) were removed so that each transformed group contained a distinct set of protein sequences, and redundant groups were removed when identical. From the resultant groups, those containing protein sequences from at least two of the three species were used to construct the “EGO subset”.
Evaluation of Consistency With EC Assignments
SWISS-PROT proteins associated with EC number annotations were first cross-referenced with sequences from the relevant proteome using BLASTP (>98% identity over >98% of query sequence length). EC numbers associated with SWISS-PROT proteins were then assigned to those sequences from the proteomes which they had been cross-referenced (only complete EC assignments were considered in this study). Only orthologous groups containing at least two sequences with EC assignments were examined, and a group was considered consistent with EC annotation only if all EC-annotated sequences in this group were assigned exactly the same EC number.
Acknowledgments
We thank Drs. Warren Ewens, Junhyong Kim, and members of the Roos and Stoeckert groups for helpful comments, discussion, and critical reading of the manuscript; Philip Labo for the development of the graphical tool for the Web interface; and Matt Berriman (Sanger Inst.) and Alan Fairlamb (Univ. Dundee) for helpful comments regarding the application of EC annotations to P. falciparum genome sequence data. We thank all those responsible for generating the data, annotation, and software referenced herein. This work was supported by research grants from the NIH and the Burroughs Wellcome Fund. D.S.R. is an Ellison Foundation Senior Scholar in Global Infectious Diseases.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1224503.
References
- Abascal, F. and Valencia, A. 2002. Clustering of proximal sequence space for the identification of protein families. Bioinformatics 18: 908–921. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., Ball, C.A., Blake, J.A., Botstein, D., Butler, H., Cherry, J.M., Davis, A.P., Dolinski, K., Dwight, S.S., Eppig, J.T., et al. 2000. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl, A., Brunk, B., Crabtree, J., Fraunholz, M.J., Gajria, B., Grant, G.R., Ginsburg, H., Gupta, D., Kissinger, J.C., Labo, P., et al. 2003. PlasmoDB: The Plasmodium Genome Resource. A database integrating experimental and computational data. Nucleic Acids Res. 31: 212–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton, J.M., Angiuoli, S.V., Suh, B.B., Kooij, T.W., Pertea, M., Silva, J.C., Ermolaeva, M.D., Allen, J.E., Selengut, J.D., Koo, H.L., et al. 2002. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature 419: 512–519. [DOI] [PubMed] [Google Scholar]
- Chervitz, S.A., Aravind, L., Sherlock, G., Ball, C.A., Koonin, E.V., Dwight, S.S., Harris, M.A., Dolinski, K., Mohr, S., Smith, T., et al. 1998. Comparison of the complete protein sets of worm and yeast: Orthology and divergence. Science 282: 2022–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, S. B., Crabtree, J., Brunk, B.P., Schug, J., Tannen, V., Overton, G.C., and Stoekert, C.J. 2001. K2/Kleisi and GUS: Experiments in integrated access to genomic data sources. IBM Systems J. 40: 512–531. [Google Scholar]
- Doolittle, R.F. 1995. The multiplicity of domains in proteins. Annu. Rev. Biochem. 64: 287–314. [DOI] [PubMed] [Google Scholar]
- Enright, A.J., Van Dongen, S., and Ouzounis, C.A. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30: 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichera, M.E. and Roos, D.S. 1997. A plastid organelle as a drug target in apicomplexan parasites. Nature 390: 407–409. [DOI] [PubMed] [Google Scholar]
- Fitch, W.M. 1970. Distinguishing homologous from analogous proteins. Syst. Zool. 19: 99–113. [PubMed] [Google Scholar]
- Fitch, W.M. 2000. Homology, a personal view on some of the problems. Trends Genet. 16: 227–231. [DOI] [PubMed] [Google Scholar]
- Forterre, P. 2002. A hot story from comparative genomics: Reverse gyrase is the only hyperthermophile-specific protein. Trends Genet. 18: 236–237. [DOI] [PubMed] [Google Scholar]
- Galperin, M.Y. and Koonin, E.V. 1999. Searching for drug targets in microbial genomes. Curr. Opin. Biotechnol. 10: 571–578. [DOI] [PubMed] [Google Scholar]
- Gardner, M.J., Hall, N., Fung, E., White, O., Berriman, M., Hyman, R.W., Carlton, J.M., Pain, A., Nelson, K.E., Bowman, S., et al. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419: 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. 2001. Creating the gene ontology resource: Design and implementation. Genome Res. 11: 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, C.Y., Shaw, M.K., Pletcher, C.H., Striepen, B., Tilney, L.G., and Roos, D.S. 2001. A plastid segregation defect in the protozoan parasite Toxoplasma gondii. EMBO J. 20: 330–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., Greene, E.A., Pietrokovski, S., Bork, P., Attwood, T.K., and Hood, L. 1997. Gene families: The taxonomy of protein paralogs and chimeras. Science 278: 609–614. [DOI] [PubMed] [Google Scholar]
- Kissinger, J.C., Brunk, B.P., Crabtree, J., Fraunholz, M.J., Gajria, B., Milgram, A.J., Pearson, D.S., Schug, J., Bahl, A., Diskin, S.J., et al. 2002. The Plasmodium genome database. Nature 419: 490–492. [DOI] [PubMed] [Google Scholar]
- Kohler, S., Delwiche, C.F., Denny, P.W., Tilney, L.G., Webster, P., Wilson, R.J., Palmer, J.D., and Roos, D.S. 1997. A plastid of probable green algal origin in Apicomplexan parasites. Science 275: 1485–1489. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Sultana, R., Pertea, G., Cho, J., Karamycheva, S., Tsai, J., Parvizi, B., Cheung, F., Antonescu, V., White, J., et al. 2002. Cross-referencing eukaryotic genomes: TIGR Orthologous Gene Alignments (TOGA). Genome Res. 12: 493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushegian, A.R., Garey, J.R., Martin, J., and Liu, L.X. 1998. Large-scale taxonomic profiling of eukaryotic model organisms: A comparison of orthologous proteins encoded by the human, fly, nematode, and yeast genomes. Genome Res. 8: 590–598. [DOI] [PubMed] [Google Scholar]
- Natale, D.A., Galperin, M.Y., Tatusov, R.L., and Koonin, E.V. 2000a. Using the COG database to improve gene recognition in complete genomes. Genetica 108: 9–17. [DOI] [PubMed] [Google Scholar]
- Natale, D.A., Shankavaram, U.T., Galperin, M.Y., Wolf, Y.I., Aravind, L., and Koonin, E.V. 2000b. Towards understanding the first genome sequence of a crenarchaeon by genome annotation using clusters of orthologous groups of proteins (COGs). Genome Biol. 1: research0009.1–0009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush, J., Liang, F., Holt, I., Pertea, G., and Upton, J. 2000. The TIGR gene indices: Reconstruction and representation of expressed gene sequences. Nucleic Acids Res. 28: 141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush, J., Cho, J., Lee, D., Liang, F., Holt, I., Karamycheva, S., Parvizi, B., Pertea, G., Sultana, R., and White, J. 2001. The TIGR Gene Indices: Analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res. 29: 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remm, M., Storm, C.E., and Sonnhammer, E.L. 2001. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 314: 1041–1052. [DOI] [PubMed] [Google Scholar]
- Roos, D.S. 1999. The apicoplast as a potential therapeutic target in Toxoplasma and other apicomplexan parasites: Some additional thoughts. Parasitol Today 15: 41. [DOI] [PubMed] [Google Scholar]
- Rubin, G.M., Yandell, M.D., Wortman, J.R., Gabor Miklos, G.L., Nelson, C.R., Hariharan, I.K., Fortini, M.E., Li, P.W., Apweiler, R., Fleischmann, W., et al. 2000. Comparative genomics of the eukaryotes. Science 287: 2204–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug, J., Diskin, S., Mazzarelli, J., Brunk, B.P., and Stoeckert Jr., C.J. 2002. Predicting gene ontology functions from ProDom and CDD protein domains. Genome Res. 12: 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J. and Malik, J. 1997. Normalized cuts and image segmentation. Proc. IEEE Conf. Comp. Vision Pattern Recognit. 731–737.
- Tatusov, R.L., Koonin, E.V., and Lipman, D.J. 1997. A genomic perspective on protein families. Science 278: 631–637. [DOI] [PubMed] [Google Scholar]
- Tatusov, R.L., Galperin, M.Y., Natale, D.A., and Koonin, E.V. 2000. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28: 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov, R.L., Natale, D.A., Garkavtsev, I.V., Tatusova, T.A., Shankavaram, U.T., Rao, B.S., Kiryutin, B., Galperin, M.Y., Fedorova, N.D., and Koonin, E.V. 2001. The COG database: New developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29: 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dongen, S. 2000. “Graph clustering by flow simulation.” Ph.D thesis, University of Utrecht, The Netherlands.
- Wheelan, S.J., Boguski, M.S., Duret, L., and Makalowski, W. 1999. Human and nematode orthologs—Lessons from the analysis of 1800 human genes and the proteome of Caenorhabditis elegans. Gene 238: 163–170. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.cbil.upenn.edu/gene-family; Putative ortholog groups generated by OrthoMCL, University of Pennsylvania.
- http://www.ncbi.nlm.nih.gov/COG/; The Clusters of Orthologous Groups (COG) database, NCBI.
- http://www.allgenes.org; The human and mouse gene index, University of Pennsylvania.
- http://www.tigr.org/tdb/tgi/; TIGR Gene Indices.
- http://www.tigr.org/tdb/tgi/ego/index.shtml; Eukaryotic Gene Orthologs (EGO), TIGR.
- http://us.expasy.org/enzyme; The ENZYME database, Bairoch A.
- http://blast.wustl.edu/; BLAST2, Washington University.
- http://www.ebi.ac.uk/clustalw/; CLUSTALW alignment, EBI.
- http://micans.org/mcl/; Markov Cluster Algorithm, Stijn van Dongen.
- http://www.cgb.ki.se/inparanoid/; INPARANOID program.
- http://www.plasmodb.org/, The Plasmodium Genome Database, University of Pennsylvania.
- http://www.fruitfly.org; The Berkeley Drosophila Genome Project (BDGP).
- http://genome-www.stanford.edu/Saccharomyces/; The Saccharomyces Genome Database (SGD).
- http://www.sanger.ac.uk/Projects/C_elegans/; The C. elegans Genome Project.
- http://www.genome.wisc.edu/; Escherichia coli Genome Project, University of Wisconsin.
- http://www.ensembl.org/; Ensembl, Sanger.
- http://www.tigr.org/tdb/e2k1/ath1/; TIGR, Arabidopsis thaliana Database.