Abstract
One of the most reliable methods for protein function annotation is to transfer experimentally known functions from orthologous proteins in other organisms. Most methods for identifying orthologs operate on a subset of organisms with a completely sequenced genome, and treat proteins as single-domain units. However, it is well known that proteins are often made up of several independent domains, and there is a wealth of protein sequences from genomes that are not completely sequenced. A comprehensive set of protein domain families is found in the Pfam database. We wanted to apply orthology detection to Pfam families, but first some issues needed to be addressed. First, orthology detection becomes impractical and unreliable when too many species are included. Second, shorter domains contain less information. It is therefore important to assess the quality of the orthology assignment and avoid very short domains altogether. We present a database of orthologous protein domains in Pfam called HOPS: Hierarchical grouping of Orthologous and Paralogous Sequences. Orthology is inferred in a hierarchic system of phylogenetic subgroups using ortholog bootstrapping. To avoid the frequent errors stemming from horizontally transferred genes in bacteria, the analysis is presently limited to eukaryotic genes. The results are accessible in the graphical browser NIFAS, a Java tool originally developed for analyzing phylogenetic relations within Pfam families. The method was tested on a set of curated orthologs with experimentally verified function. In comparison to tree reconciliation with a complete species tree, our approach finds significantly more orthologs in the test set. Examples for investigating gene fusions and domain recombination using HOPS are given.
The concepts of orthology and paralogy (Fitch 1970) are widely used. A search in PubMed reveals an increase of the use of the regular expression “ortholog*” in abstracts from 28 in 1990, 68 in 1994, 302 in 1998, to 840 in 2001. An in-depth explanation of orthology and paralogy can be found in recent publications (Fitch 2000; Sonnhammer and Koonin 2002). Numerous applications and analyses rely on the use of orthologous sequences, for instance, transferring functional annotation (Stein 2001), phylogenetic footprinting (Blanchette et al. 2002), and evolutionary and comparative studies (Makalowski et al. 1996; Mushegian et al. 1998; Xie and Ding 2000).
A standard approach for assigning orthology in a phylogenetic tree is tree reconciliation (Goodman et al. 1979; Page 1994). Here a given species tree is compared with a gene tree. This works by postulating the minimum number of duplication and gene-loss events in the gene tree necessary to reconcile it with the species tree. Orthologous assignments can then be made from this reconciled tree. Given a correct species and gene tree, this method can reliably distinguish between orthologs and paralogs. In theory, tree reconciliation is superior to BLAST-based (Altschul et al. 1997) approaches for finding orthologs (Tatusov et al. 1997; Remm et al. 2001). Such methods neither use the information provided by a species tree, nor take unequal rates of evolution into account.
However, one drawback of tree reconciliation is that it uses a given, fixed species tree: For some species the evolutionary history is still controversial, for example, the phylogenetic relationship of Homo sapiens, Caenorhabditis elegans, and Drosophila melanogaster (Mushegian et al. 1998; Xie and Ding 2000; Blair et al. 2002). In addition, a reconstructed phylogenetic tree, especially for short sequences, might not reflect the species tree because of random effects. Simplifications in the phylogenetic model used can also lead to an incorrect sequence tree. For such cases tree reconciliation might not find the correct orthologous sequences.
Here we present an approach to resolve these problems by organizing the sequences into evolutionarily distinct subgroups. Orthology is then inferred between these subgroups using ortholog bootstrapping (Storm and Sonnhammer 2002). The results are saved in a database named HOPS (Hierarchical analysis of Orthologous and Paralogous Sequences). The HOPS data can be analyzed and displayed graphically with a tree in an extended version of the NIFAS browser (Storm and Sonnhammer 2001).
Recent studies indicate a high rate of horizontal transfer for bacteria (Doolittle 1999; Koonin et al. 2001; Snel et al. 2002). The present algorithms for tree reconciliation do not account for horizontal transfer of genes. If a gene has been horizontally transferred, tree reconciliation might fail to find its orthologous genes (Gogarten and Olendzenski 1999). Therefore, bacterial sequences are not included in the analysis.
METHODS
Data
This paper is based on the 3735 protein families in Pfam 7.2 (Bateman et al. 2002). The sequences in each alignment are clustered following a hierarchical scheme derived from the species tree (see Fig. 1). Clades that can be considered equidistant to each other from an evolutionary perspective are assigned to the same level. At present, the grouping scheme consists of two levels: eukaryotes and metazoans. A higher level containing the top level of all three kingdoms could be used as well, but is presently not implemented because of frequent misassignments that would be caused by horizontal transfer in prokaryotes. Therefore, the bacterial and archaeal kingdoms were excluded from HOPS.
Figure 1.
The HOPS hierarchy of species groups. Only the levels marked in black are used for orthology analysis in this paper because of the high levels of horizontal transfer among eubacteria and archaea.
Each level is divided into evolutionarily distinct species groups built around completely sequenced genomes. This minimizes the chance that paralogs could be mistaken for orthologs, because the true orthologous sequence might not be known yet. These species groups, or lineages, are treated as one “pseudospecies” in the orthology analysis, which is carried out only between species groups at the same level. Orthologous relations within a species group are not analyzed.
The eukaryotic level is split up into the clades of Metazoa, Viridiplantae, and Fungi. The metazoan level is divided into Chordata, Nematoda, and Arthropoda. Sequences from species that do not belong to any of those clades are not analyzed. For instance, a sequence from an Echinodermata species (sea urchins, starfish, etc.) would not be taken into account on the metazoan level, because it doesn't belong to any of the species in the Chordata, Nematoda, or Arthropoda groups. But it would be analyzed on the eukaryotic level, because it is part of the metazoan group. Therefore, orthologs to this sequence may be assigned in the Viridiplantae and Fungi groups, but not on the metazoan level.
To improve the quality of ortholog assignments, we can use an outgroup criterion at the metazoan level. All sequences from the eukaryotic groups that are not part of the metazoan group are treated as outgroup sequences. If an outgroup sequence is found between two candidate orthologs in the tree, this means that they are probably not true orthologs but were clustered together because the true ortholog was lost in one of the species. The outgroup species did not lose the ortholog and is hence more closely related than the false ortholog. We have not used the outgroup criterion at the eukaryotic level, however, because of the frequent horizontal transfers from eukaryotes to prokaryotes.
Orthostrapper
Orthology assignments are calculated by a program called Orthostrapper (Storm and Sonnhammer 2002) that performs “ortholog bootstrapping,”, or “orthostrapping.” Orthostrapper analyzes a set of bootstrap trees instead of the optimal tree for orthologs. The algorithm detects orthologous relations between two (groups of) species. The frequency of orthology assignments in the bootstrap trees can be interpreted as a confidence value for the possible orthology of two proteins.
Here Orthostrapper is run on pairs of species groups at the same level. Each Pfam alignment is split according to the phylogenetic groups mentioned above, and six pairwise comparisons are carried out: on the eukaryotic level, Metazoa-Viridiplantae, Metazoa-Fungi, and Viridiplantae-Fungi; on the metazoan level, Chordata-Nematoda, Chordata-Arthropoda, and Nematoda-Arthropoda.
Ortholog groups between more than two species groups are generated by merging the pairwise results. This is only allowed between species groups at the same HOPS level. If there are incongruencies between the pairwise results, this needs to be flagged. Partial sequences/fragments shorter than 50% of the alignment length were removed to improve the tree quality. Ortholog bootstrapping with 200 pseudosamples was then used to analyze the alignments. Alignments with more than 1200 sequences were not analyzed. Because of their arbitrary length, Pfam families of type “repeats” were excluded from the analysis.
Test Set
To evaluate the accuracy of ortholog assignments, we compiled a set of curated orthologs for H. sapiens-Saccharomyces cerevisiae, H. sapiens-C. elegans, and H. sapiens-D. melanogaster.
To avoid any ambiguity, the sequences in the test set should fulfill the following criteria:
Experimentally verified function
Same substrate and activity for putative orthologs
One-to-one orthology
Single-domain proteins
To narrow the set of proteins, all single-domain proteins (according to Pfam-A) in one species were scored with BLAST to all single-domain proteins in the second species. From the alignments, best-best hit pairs (sequences that rank each other as best match when searching both ways) were extracted. To remove many-to-many orthologous relations, each sequence of the best-best hit pairs was then aligned to all sequences within its species. If any sequence within the same species had a lower e-value than the corresponding ortholog, the pair was excluded from the test set. From the remaining pairs, sequences with functional annotation by homology were excluded. The functional descriptions of each sequence pair were then compared. Only if both proteins use the same substrate and show the same activity were they included in the final test set.
This procedure resulted in a set of 102 human-yeast, 19 human-worm, and 47 human-fly putative orthologs, shown in Tables 1, 2, 3. It should be pointed out that the main criteria for the test set are `experimentally verified function' and `same substrate and activity,' which is why the test set is relatively small. The reciprocal BLAST scoring was done to limit the necessary number of (manual) comparisons of sequence annotations.
Table 1.
Results of HOPS and RIO for the Test Set of Putative Orthologs for H. sapiens and D. melanogaster
| H. sapiens | D. melanogaster | Domain | HOPS (%) | RIO (%) |
|---|---|---|---|---|
| 143E_HUMAN | 143E_DROME | 14-3-3 | 100 | 98 |
| ADHX_HUMAN | ADHX_DROME | adh_zinc | 16.5 | 0 |
| APE1_HUMAN | RRP1_DROME | Exo_endo_phos | 100 | 85 |
| ARF1_HUMAN | ARF1_DROME | arf | 89.5 | 92 |
| ARI1_HUMAN | ARI1_DROME | IBR | 100 | 96 |
| ARL1_HUMAN | ARL1_DROME | arf | 100 | 100 |
| ATPG_HUMAN | ATPG_DROME | ATP-synt | 100 | 100 |
| ATPO_HUMAN | ATPO_DROME | OSCP | 100 | 63 |
| CAPB_HUMAN | CAPB_DROME | F_actin_cap_B | 100 | 95 |
| CG1C_HUMAN | CG1C_DROME | Cyclin | 100 | 80 |
| CLAT_HUMAN | CLAT_DROME | Carn_acyltransf | 94 | 89 |
| COXA_HUMAN | COXA_DROME | COX5A | 99 | 78 |
| DYNA_HUMAN | DYNA_DROME | CAP_GLY | 50.5 | 29 |
| ERH_HUMAN | ER_DROME | ER | 100 | 98 |
| FAC2_HUMAN | FAC2_DROME | Abi | 87 | 62 |
| H2AZ_HUMAN | H2AV_DROME | Histone | 76 | 21 |
| H4_HUMAN | H4_DROME | Histone | 62 | 1 |
| IF2A_HUMAN | IF2A_DROME | S1 | 78 | 62 |
| KC2B_HUMAN | KC2B_DROME | CK_II_beta | 18.5 | 36 |
| MCM2_HUMAN | MCM2_DROME | MCM | 100 | 97 |
| MCM4_HUMAN | MCM4_DROME | MCM | 100 | 100 |
| PP1B_HUMAN | PP1B_DROME | Metallophos | 76 | 43 |
| PRS8_HUMAN | PRS8_DROME | AAA | 84.5 | — |
| PSA2_HUMAN | PSA2_DROME | Proteasome | 99.5 | 98 |
| PSA3_HUMAN | PSA3_DROME | Proteasome | 74.5 | 13 |
| PSA4_HUMAN | PSA4_DROME | Proteasome | 100 | 100 |
| PSA5_HUMAN | PSA5_DROME | Proteasome | 79 | 95 |
| PSA6_HUMAN | PSA6_DROME | Proteasome | 77 | 44 |
| PSB1_HUMAN | PSB1_DROME | Proteasome | 50.5 | 37 |
| PSB3_HUMAN | PSB3_DROME | Proteasome | 72.5 | 22 |
| PURA_HUMAN | PURA_DROME | Adenylsucc_synt | 100 | 100 |
| RA51_HUMAN | RA51_DROME | HHH | 0 | 0 |
| RBJK_HUMAN | RBJK_DROME | TIG | 97 | 48 |
| RLA1_HUMAN | RLA1_DROME | 60s_ribosomal | 35 | 12 |
| RLA2_HUMAN | RLA2_DROME | 60s_ribosomal | 45.5 | 21 |
| RPB2_HUMAN | RPB2_DROME | RNA_pol_B | 100 | 96 |
| RPB6_HUMAN | RPB6_DROME | RNA_pol_Rpb6 | 90 | 62 |
| RS18_HUMAN | RS18_DROME | Ribosomal_S13 | 24 | 3 |
| RTC1_HUMAN | RTC1_DROME | RTC | 100 | 6 |
| SSB_HUMAN | SSB_DROME | SSB | 97.5 | 92 |
| T2DA_HUMAN | T2DA_DROME | TFIID_A | 82.5 | 87 |
| T2FB_HUMAN | T2FB_DROME | TFIIF_beta | 100 | 98 |
| TCPA_HUMAN | TCPA_DROME | cpn60_TCP1 | 99.5 | 60 |
| TCPG_HUMAN | TCPG_DROME | cpn60_TCP1 | 97 | 57 |
| UBCI_HUMAN | O62622 | UQ_con | 100 | 96 |
| VATF_HUMAN | VAF1_DROME | ATP-synt_F | 97 | 54 |
| XPA_HUMAN | XPA_DROME | XPA | 100 | 67 |
A “—” indicates no results reported.
Table 2.
Results of HOPS and RIO for the Test Set of Putative Orthologs for H. sapiens and C. elegans
| H. sapiens | C. elegans | Domain | NIFAS (%) | RIO (%) |
|---|---|---|---|---|
| ACBP_HUMAN | ACBP_CAEEL | ACBP | 4.5 | 0 |
| CAPB_HUMAN | CAPB_CAEEL | F_actin_cap_B | 100 | 0 |
| CRTC_HUMAN | CRTC_CAEEL | Calreticulin | 72 | 0 |
| CUL1_HUMAN | CUL1_CAEEL | Cullin | 12.5 | 0 |
| DAD1_HUMAN | DAD1_CAEEL | DAD | 93.5 | 49 |
| EAA2_HUMAN | EAA1_CAEEL | SDF | 62.5 | 70 |
| EF2K_HUMAN | EF2K_CAEEL | MHCK EF2 kinase | 100 | 100 |
| ERH_HUMAN | ERH_CAEEL | ER | 81.5 | 0 |
| GRE1_HUMAN | GRPE_CAEEL | GrpE | 45 | 32 |
| KC2B_HUMAN | KC2B_CAEEL | CK_II_beta | 100 | 20 |
| O43447 | CYPB_CAEEL | pro_isomerase | 8 | 10 |
| O60573 | IFE4_CAEEL | IF4E | 95.5 | 89 |
| OM20_HUMAN | OM20_CAEEL | MAS20 | 86.5 | 51 |
| RNHL_HUMAN | RNHL_CAEEL | RNase_HII | 91 | 22 |
| RPB2_HUMAN | RPB2_CAEEL | RNA_pol_B | 100 | 31 |
| S61G_HUMAN | S61G_CAEEL | SccE | 99.5 | 58 |
| SAHH_HUMAN | SAHH_CAEEL | AdoHcyase | 80.5 | 25 |
| TCPA_HUMAN | TCPA_CAEEL | cpn60_TCP1 | 91.5 | 33 |
| TCPD_HUMAN | TCPD_CAEEL | cpn60_TCP1 | 36 | 2 |
Table 3.
Results of HOPS and RIO for the Test Set of Putative Orthologs for H. sapiens and S. cerevisiase
| H. sapiens | S. cerevisiae | Domain | HOPS (%) | RIO (%) |
|---|---|---|---|---|
| LSM6_HUMAN | LSM6_YEAST | Sm | 0 | 1 |
| SODC_HUMAN | SODC_YEAST | sodcu | 0 | 0 |
| LSM4_HUMAN | LSM4_YEAST | Sm | 0 | 0 |
| SMD1_HUMAN | SMD1_YEAST | Sm | 0 | 0 |
| SMD2_HUMAN | SMD2_YEAST | Sm | 0.5 | 3 |
| LSM5_HUMAN | LSM5_YEAST | Sm | 1 | 0 |
| LSM3_HUMAN | LSM3_YEAST | Sm | 2.5 | 0 |
| ACBP_HUMAN | ACBP_YEAST | ACBP | 4 | 2 |
| UBCJ_HUMAN | UBC7_YEAST | UQ_con | 5 | 4 |
| RUXF_HUMAN | RUXF_YEAST | Sm | 8.5 | 0 |
| H4_HUMAN | H4_YEAST | Histone | 10 | 0 |
| FAC2_HUMAN | RCE1_YEAST | Abi | 12 | 24 |
| RPCX_HUMAN | RPCX_YEAST | DNA_RNApol_7kD | 17 | 65 |
| CAPB_HUMAN | CAPB_YEAST | F_actin_cap_B | 24 | 36 |
| CCHL_HUMAN | CCHL_YEAST | Cyto_heme_lyase | 27 | 80 |
| CDD_HUMAN | CDD_YEAST | dCMP_cyt_deam | 29.5 | 1 |
| UCR6_HUMAN | UCR7_YEAST | UCR_14kD | 31 | 56 |
| PRSA_HUMAN | PRSA_YEAST | AAA | 36.5 | — |
| RS18_HUMAN | RS18_YEAST | Ribosomal_S13 | 37 | 2 |
| NO56_HUMAN | SIK1_YEAST | Nop | 40 | 13 |
| COXG_HUMAN | COXG_YEAST | COX6B | 46.5 | 60 |
| OM20_HUMAN | OM20_YEAST | MAS20 | 47.5 | 24 |
| KIME_HUMAN | KIME_YEAST | GHMP_kinases | 50 | 24 |
| RA51_HUMAN | RA51_YEAST | HHH | 50.5 | 14 |
| NOP5_HUMAN | NOP5_YEAST | Nop | 52 | 1 |
| T2D5_HUMAN | T2D5_YEAST | TAF | 55 | 74 |
| TBCA_HUMAN | TBCA_YEAST | TBCA | 56.5 | 16 |
| SNX8_HUMAN | MVP1_YEAST | PX | 57.5 | 84 |
| CP51_HUMAN | CP51_YEAST | p450 | 59 | — |
| S61G_HUMAN | S61G_YEAST | SecE | 63 | 1 |
| GPT_HUMAN | GPT_YEAST | Glycos_transf_4 | 64.5 | 31 |
| OXA1_HUMAN | OXA1_YEAST | 60KD_IMP | 69 | 30 |
| LSM2_HUMAN | LSM2_YEAST | Sm | 71 | 2 |
| PHB_HUMAN | PHB_YEAST | Band_7 | 72.5 | 11 |
| PSB2_HUMAN | PSB2_YEAST | Proteasome | 73 | 11 |
| RPBX_HUMAN | RPBX_YEAST | RNA_pol_N | 75.5 | 23 |
| MPPA_HUMAN | MPPA_YEAST | Peptidase_M16 | 77.5 | 84 |
| IF2A_HUMAN | IF2A_YEAST | S1 | 77.5 | 49 |
| RPB9_HUMAN | RPB9_YEAST | RNA_POL_M_15KD | 78 | 96 |
| FOLC_HUMAN | FOLE_YEAST | Mur_ligase_C | 78 | 65 |
| PSA1_HUMAN | PSA1_YEAST | Proteasome | 78 | 45 |
| PRS4_HUMAN | PRS4_YEAST | AAA | 78.5 | — |
| PRI1_HUMAN | PRI1_YEAST | DNA_primase_S | 78.5 | 1 |
| DHYS_HUMAN | DHYS_YEAST | DS | 79 | 18 |
| GRE1_HUMAN | GRPE_YEAST | GrpE | 82.5 | 24 |
| FAC1_HUMAN | ST24_YEAST | Peptidase_M48 | 83 | 2 |
| ORN_HUMAN | ORN_YEAST | Exonuclease | 83 | 1 |
| HEMZ_HUMAN | HEMZ_YEAST | Ferrochelatase | 84 | 97 |
| RNHL_HUMAN | RNHL_YEAST | RNase_HII | 84 | 7 |
| PSB1_HUMAN | PSB1_YEAST | Proteasome | 85.5 | 9 |
| GCSP_HUMAN | GCSP_YEAST | GDC-P | 86 | 0 |
| PSA6_HUMAN | PSA6_YEAST | Proteasome | 86.5 | 53 |
| PSA4_HUMAN | PSA4_YEAST | Proteasome | 86.5 | 23 |
| DNL1_HUMAN | DNLI_YEAST | DNA_ligase | 86.5 | 0 |
| IF4E_HUMAN | IF4E_YEAST | IF4E | 87 | 2 |
| CY1_HUMAN | CY1_YEAST | Cytochrome_C1 | 88 | 21 |
| NTF2_HUMAN | NTF2_YEAST | NTF2 | 88.5 | 39 |
| TCPB_HUMAN | TCPB_YEAST | cpn60_TCP1 | 88.5 | 0 |
| PRS8_HUMAN | PRS8_YEAST | AAA | 89 | — |
| PSB3_HUMAN | PSB3_YEAST | Proteasome | 89 | 38 |
| T2EB_HUMAN | T2EB_YEAST | TFIIE_beta | 90 | 100 |
| VATF_HUMAN | VATF_YEAST | ATP-synt_F | 90 | 18 |
| PSA7_HUMAN | PSA7_YEAST | Proteasome | 90.5 | 20 |
| UBCH_HUMAN | UBC8_YEAST | UQ_con | 91 | 19 |
| KAD2_HUMAN | KAD1_YEAST | Adenylatekinase | 91.5 | 80 |
| MD21_HUMAN | MAD2_YEAST | HORMA | 91.5 | 7 |
| T2DB_HUMAN | T2DB_YEAST | TFIID-18 | 92 | 83 |
| RANG_HUMAN | YRB1_YEAST | Ran_BP1 | 92 | 82 |
| MSRA_HUMAN | MSRA_YEAST | PMSR | 92 | 0 |
| UFD1_HUMAN | UFD1_YEAST | UFD1 | 93 | 67 |
| KCY_HUMAN | UMPK_YEAST | Adenylatekinase | 94 | 20 |
| VA0D_HUMAN | VA0D_YEAST | vATP-synt_AC39 | 94.5 | — |
| ATPO_HUMAN | ATPO_YEAST | OSCP | 94.5 | 6 |
| RPB6_HUMAN | RPB6_YEAST | RNA_pol_Rpb6 | 94.5 | 6 |
| XPB_HUMAN | RA25_YEAST | Helicase_C | 95 | — |
| RCL1_HUMAN | RCL1_YEAST | RTC | 96.5 | 17 |
| PRS7_HUMAN | PRS7_YEAST | AAA | 97 | — |
| COXX_HUMAN | COXX_YEAST | UbiA | 97 | 45 |
| PSA3_HUMAN | PSA3_YEAST | Proteasome | 97 | 1 |
| PRS6_HUMAN | PRS6_YEAST | AAA | 98 | — |
| T2EA_HUMAN | T2EA_YEAST | 1FILE_alpha | 98 | 66 |
| TCPA_HUMAN | TCPA_YEAST | cpn60_TCP1 | 98 | 52 |
| RUXG_HUMAN | RUXG_YEAST | Sm | 98 | 41 |
| PSB4_HUMAN | PSB4_YEAST | Proteasome | 98 | 13 |
| PSA5_HUMAN | PSA5_YEAST | Proteasome | 98 | 1 |
| COPB_HUMAN | COPB_YEAST | Adaptin_N | 98.5 | 1 |
| COXA_HUMAN | COX6_YEAST | COX5A | 99 | 40 |
| TCPD_HUMAN | TCPD_YEAST | cpn60_TCP1 | 99 | 29 |
| ATPG_HUMAN | APTG_YEAST | ATP-synt | 99.5 | 96 |
| TCPH_HUMAN | TCPH_YEAST | cpn60_TCP1 | 99.5 | 20 |
| PSA2_HUMAN | PSA2_YEAST | Proteasome | 99.5 | 8 |
| TCPG_HUMAN | TCPG_YEAST | cpn60_TCP1 | 99.5 | 7 |
| T2FB_HUMAN | T2FB_YEAST | TFIIF_beta | 100 | 100 |
| BLMH_HUMAN | BLH1_YEAST | Pept_C1-like | 100 | 100 |
| PURA_HUMAN | PURA_YEAST | Adenylsucc_synt | 100 | 98 |
| XPA_HUMAN | RA14_YEAST | XPA | 100 | 80 |
| NFS1_HUMAN | NFS1_YEAST | Aminotran_5 | 100 | 68 |
| E2BA_HUMAN | E2BA_YEAST | IF-2B | 100 | 61 |
| MCM4_HUMAN | CC54_YEAST | MCM | 100 | 60 |
| MCM2_HUMAN | MCM2_YEAST | MCM | 100 | 53 |
| RPB2_HUMAN | RPB2_YEAST | RNA_pol_B | 100 | 51 |
| TCPQ_HUMAN | TCPQ_YEAST | cpn60_TCP1 | 100 | 4 |
A “—” indicates no results reported.
Phylogenetic Analysis
To analyze why HOPS fails in some cases to find the correct ortholog, we used MrBayes (Huelsenbeck and Ronquist 2001) to reconstruct trees from the sequences. The substitution matrix used was Jones-Taylor-Thornton. The trees were built both with homogeneous and heterogeneous evolution among sides, using a γ distribution in the latter case.
Access to the Data
All data are available at ftp://ftp.cgb.ki.se/pub/HOPS/. The file “manually removed” lists protein pairs that show a reciprocal best BLAST score and have experimentally verified function, but were excluded from the test set based on differences in the catalytic activity or the substrate used.
Comparison With Tree Reconciliation
RIO version 0.3 (Zmasek and Eddy 2002) was chosen for the comparison with a tree reconciliation method. RIO searches Pfam families for orthologs by reconciling the sequences tree with the complete tree of all species. We submitted all human proteins in the test set through the Web interface at http://www.rio.wustl.edu, and the resulting ortholog support values for the corresponding ortholog in the test set were taken for the comparison. If a sequence other than the ortholog from the test set showed a higher orthology score, this was marked.
RESULTS
Accessing the HOPS Database
The HOPS database can be accessed either via the NIFAS browser or a standard Internet browser. NIFAS is a Java applet for viewing phylogenetic trees of domains, connected to schematic graphical representations of the proteins' domain structure (Storm and Sonnhammer 2001).
The increase of computer power over the last years makes it possible to now provide precalculated neighbor-joining trees with bootstrap support for families of up to 250 sequences and up to 1500 sequences with no bootstrap support. Although the information content of phylogenetic trees of this size with no bootstrap support is at best questionable, it allows viewing the ortholog bootstrap values calculated for the larger families within NIFAS.
NIFAS
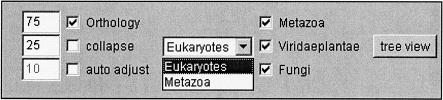
The original functionality of NIFAS remains unchanged. A button is added to the navigation bar to change to orthology mode. If no orthology information for a given family is available (because it contains, e.g., only bacterial sequences), this button is deactivated. The new commands of the orthology mode can be seen in Figure 2.
Figure 2.
New commands in NIFAS for analyzing orthology. The three checkboxes to the left are for applying cutoffs. If the “collapse” checkbox is marked, all proteins that have an orthology bootstrap support below the cutoff are hidden. The “Orthology” checkbox groups orthologs based on an average-linkage algorithm described in Hollich et al. (2002). If the orthologous groups are ambiguous, “auto adjust” tries to minimize the number of overlapping orthologous groups by going through all cluster cutoffs within the range of the corresponding value, with a mean of the original cutoff. In the box to the middle, the user can choose the HOPS level for which ortholog assignments are shown. Presently these are the eukaryotic and metazoan levels. The checkboxes to the right allow choosing what species of the present level should be included in the clustering.
While the orthology navigation panel is shown, a click with the left mouse button on any domain schematic will open a new window. In this window, all proteins are displayed that contain at least one domain with an orthology score above the Orthology value. A right click on a domain schematic will show the same information in the browser window in text format.
The species group of a sequence is shown by color-coding the background of the species name next to the sequence identifier. Colored boxes around the sequence identifier highlight orthologous relations. Each group of orthologs and paralogs has a different color assigned (Fig. 3). Sometimes the results of the clustering can be ambiguous, especially if clustering sequences from more than two species. In these cases, the boxes for sequences that are grouped in more than one cluster are drawn with multiple colors, one for each cluster to which the sequence is assigned.
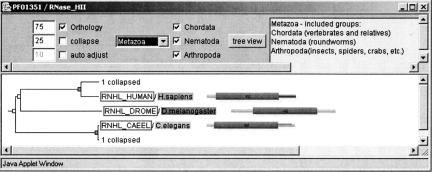
Figure 3.
Orthologs for RNHL_HUMAN in HOPS, based on the domain RNase_HII. The orthology between RNHL_HUMAN and RNHL_CAEEL is not found by RIO, because the tree topology for RNHL_HUMAN, RNHL_DROME, and RNHL_CAEEL does not reflect the Ecdysozoa hypothesis (see text for further explanation).
Access Through an Internet Browser
An html page is available at http://pfam.cgb.ki.se/HOPS/. Here one can enter a SWISS-PROT/TrEMBL identifier and get a list of its HOPS orthologs. This page shows the same information as right clicking on a domain schematic in NIFAS does, but also includes domain diagrams and annotation.
Accuracy of the Test Set
For some genes, both methods fail to find the correct orthologous sequence. An explanation would be that the sequences are not true orthologs. However, for none of these sequences does either HOPS or RIO find a sequence with a higher ortholog bootstrap support, indicating that the test set is correct but contains some very difficult cases.
Comparing HOPS With RIO
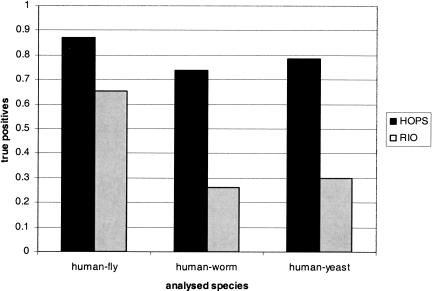
Figure 4 presents a summary of the HOPS-RIO comparison. A detailed list of the results is shown in Tables 1, 2, 3. For some families, RIO does not provide orthology scores. These families are not taken into account for calculating the percentage of true positives for RIO in Figure 4.
Figure 4.
Performance of HOPS and RIO on the putative orthologs from our test set. A 50% cutoff was used to assign orthology. Shown are the fractions of orthologs from the test set found with HOPS, respectively, RIO. See text for further information.
Results for the H. sapiens-D. melanogaster Test Set
The human-fly orthology inference gave the most similar result between HOPS and RIO. Applying a 50% orthology support cutoff for assigning orthology, HOPS found 87.5% of the orthologs in the test set. RIO assigned 65% of the orthologs correctly. In all cases when only HOPS found the ortholog, RIO gave a score >0. As indicated in Figure 5A, most of these orthologs were given a lower bootstrap support by RIO because more sequences were included or because of other differences in the tree-building method. In general, larger trees tend to have lower bootstrap support than smaller trees (Zharkikh and Li 1995).
Figure 5.
Detailed analysis for the cases in which RIO fails to assign the correct ortholog contrary to HOPS. The labels show the reason, which is usually a sequence from a particular species that prevents RIO from assigning orthology correctly. The cases that could not be traced back to a sequence preventing correct orthology assignment are due to differences in the tree-building method or the inclusion of more sequences by RIO. (A) A total of 10 potential orthologous relations are not found by RIO in the human-fly test set, but are assigned correctly by HOPS. (B) Nine orthologous pairs in the human-worm test set assigned correctly by HOPS are not found by RIO. In all cases, the sequences reflect the Coelomata, not the Ecdysozoa model of evolution (see text for further explanation). (C) Results of the human-yeast comparison, in which 51 orthologous relations found with HOPS are missed by RIO. The majority of the cases (35) are not found by RIO because the human sequence is an outgroup to a plant/yeast clade. In three cases, bacterial sequences break the assignment of orthology by RIO, indicating possible horizontal gene transfers. Four of the orthologous relations are missed by RIO because of Plasmodium sp. sequences on the Chordata branch.
Results for the H. sapiens-C. elegans Test Set
The outcome for the human-worm analysis was quite different. Based on a 50% orthology support cutoff, HOPS assigns 74 % of the orthologs correctly. RIO, on the other hand, found 26% of the orthologs in the test set. In 26% of the cases, RIO gave a score of 0 to a sequence pair that was orthologous by HOPS.
An analysis of the corresponding sequence trees shows why orthology inference with a complete species tree as done by RIO fails to find most of the orthologs (Fig. 5B). In these cases, the trees do not follow the “Ecdysozoa” phylogeny in the species tree used by RIO, which places C. elegans in a clade with arthropods. The fly sequences in these trees are in the same clade as the H. sapiens sequences, and the nematode sequences are basal to both—representing the “Coelomata” hypothesis (Fig. 3).
In this combination of gene and species tree, the tree reconciliation algorithm will assign the human and the worm sequence to be paralogous. In HOPS, the fly sequences are not included for the human-worm analysis; therefore, the orthologous sequence is correctly assigned.
Results for the H. sapiens-S. cerevisiae Test Set
In the human-yeast analysis, HOPS assigned 78% of the orthologs correctly, RIO 31%. The reasons for this difference are summarized in Figure 5C. In 68% of these cases, the sequence trees did not reflect the species tree used by RIO, which places plants as an outgroup to a fungi/metazoa clade. For the human-yeast analysis, horizontal transfer of genes into intracellular parasites prevents tree reconciliation with a complete species tree from finding the correct ortholog. In 14% of the missed orthologous relations, bacterial or Plasmodium sp. sequences were on a branch with either yeast or human, thus breaking the orthology assignment.
False Negatives in HOPS
Figure 6 summarizes the cases in which HOPS failed to find the correct ortholog from the test set (all of these orthologous relations were also not found by RIO). In nearly one-fifth of the cases, this was caused by rooting problems. This can happen in small domain families with varying rates of evolution among the lineages. Then the mid-tree rooting method used might place the root in the wrong branch.
Figure 6.
Analysis of the 30 orthologous relations in the test set HOPS fails to find. The labels describe the reason for the misassignment. Of the 13 cases in which the HOPS clustering scheme is not reflected, eight go back to a single-domain family, the SM domain. Here the S. cerevisiae orthologs evolved much faster than the corresponding S. pombe ortholog. This leads to a tree in which only the S. pombe sequence is assigned as orthologous, but not the S. cerevisiae sequence. Other reasons are wrong rooting (five cases) of the tree and general low bootstrap support for the whole sequence family.
Nearly all (12 out of 13) of the cases in which the HOPS clustering scheme is not reflected by the gene tree come from a misplaced Schizosaccharomyces pombe sequence. More than 60% (eight out of 13) of these cases are observed in a single domain family (the SM domain). For instance, in the phylogenetic tree for the SM family, the S. cerevisiae LSM6_YEAST sequence appears as an outgroup to an S. pombe + metazoan clade. Therefore, both methods assign orthology between the S. pombe and the human LSM6 but not between S. cerevisiae and human. Trees reconstructed with MrBayes (data not shown) give the same results. This indicates that this behavior does not originate from an error specific to the tree reconstruction method used in HOPS. For the SM family, the S. cerevisiae sequences show an elevated rate of evolution. In combination with the relative shortness of the alignment (111 residues), this might explain why none of the tree methods reconstructs the correct tree: a monophyletic S. pombe and S. cerevisiae branch basal to the metazoan clade.
In the remaining cases, orthology was not assigned because of generally low bootstrap support for the whole family. The alignment simply does not contain enough phylogenetic information to reconstruct a reliable phylogenetic tree. In other words, the ratio (informative sites)/(sequence number) is too small. This results in low ortholog bootstrap values.
Investigating Gene Fusion
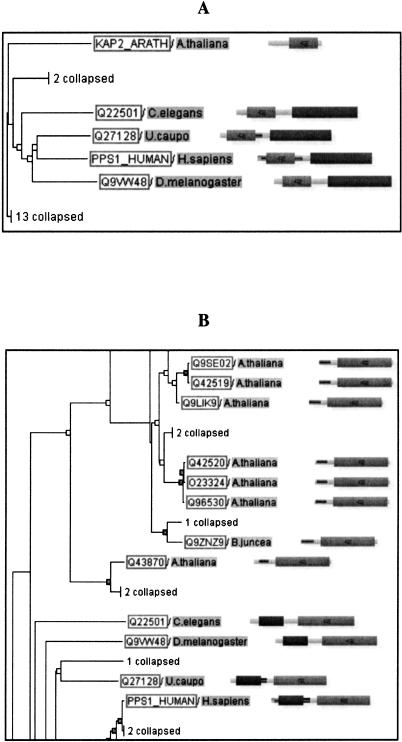
The HOPS database can be used to analyze evolution of domain structure between orthologs. Figure 7 shows an example of genes with different domain structures in plants and metazoa. The metazoan proteins consist of two domains and are bifunctional, with APS-kinase (Pfam: PF01583) and ATP-sulfurylase (Pfam: PF01747) activity. Both domains have orthologous single-domain proteins in Arabidopsis thaliana. This indicates that the metazoan proteins were created by a gene-fusion event. This happened most likely in an early metazoan ancestor, as all metazoan homologs have both domains, and no nonmetazoan proteins are known with this particular dual-domain architecture. However, a set of fungal proteins has the same domains in the inverted order. Only one domain (ATP-sulfurylase) is orthologous to the metazoan proteins in HOPS, however, indicating two independent domain accretion events in the metazoan and fungal lineages. Oddly, a bacterial protein from Aquifex aeolicus (SWISSPROT: O67174/SATC_AQUAE) has the same domain structure as the fungal proteins, and both domains are grouped with fungi in the NIFAS tree (data not shown). This implies a horizontal transfer from fungus to bacteria.
Figure 7.
Detecting gene fusion. (A) Shown are selected members of the APS_kinase family (seed alignment) with orthologous assignments between the Viridiplantae and the Metazoa group. The proteins of the two clades are orthologous to each other. The A. thaliana protein (KAP2_ARATH) consists of one domain, APS_KINASE. The proteins in the metazoan clade all have an additional domain, ATP_Sulfurylase. (B) The same metazoan proteins are shown as in A, but this time with the orthologous assignments displayed for the ATP_Surylase domain (full alignment, here the ATP_Surylase domain has a tree icon). Because both domains in the metazoan proteins have orthologous single-domain proteins in A. thaliana, this indicates a gene fusion in an early metazoan ancestor that lead to the bifunctional metazoan proteins.
Orthology Between Multidomain Proteins
Figure 8 shows an example of domain orthology between multidomain proteins. These plant and human molybdopterin biosynthesis proteins have the same domains, but in different order. Despite this, all domains are orthologous. A molybdopterin binding domain (Pfam: PF00994) is found N-terminally in chordates, whereas it is C-terminal in plants. The same domain is found N-terminally in arthropods as well, whereas in nematodes the four-domain combination does not appear. Instead, a single-domain nematode protein is the closest relative to the additional PF00994 domain (see Fig. 9). It thus appears that this domain was inserted early in the metazoan lineage. What is unusual here is that the additional PF00994 domain in plants and chordates appears orthologous (ortholog bootstrap support = 95%), but the arthropod domain is placed as an outgroup and is thus not orthologous to either the plant or chordate domain.
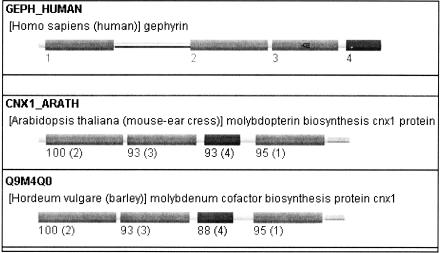
Figure 8.
Analyzing orthology for multidomain proteins. Displayed are the two highest scoring orthologs for GEPH_HUMAN: CNX1_ARATH and Q9M4Q0. The numbers below the domains are the ortholog bootstrap values (%). The number in brackets after the score indicates the corresponding orthologous domain in GEPH_HUMAN. Although the order of the domains is different, all domains show a high orthology score to the corresponding domain in GEPH_HUMAN.
Figure 9.
Orthologous domains in multidomain proteins. Most molybdopterin biosynthesis proteins in metazoa and plants contain four domains (two molybdopterin-binding domains, PF00994 [green], and one PF03453 [blue] and PF03453 [red] domain). This domain arrangement appears to have been constructed by joining a common cassette of PF00994, PF03453, and PF03453 to another copy of PF00994. The three-domain cassette is intact, and all domains are orthologous in the same order. (A) Metazoa-plant orthologs of the central domain of the cassette. (B) Orthology of the additional PF00994 domain. Oddly, here only the chordate lineage of metazoa appears orthologous to the plant domain. The Drosophila protein CIN_DROME cannot be considered orthologous to GEPH_HUMAN in this domain despite having the same domain architecture. This indicates that the N-terminal domain in the arthropod and metazoan proteins derive from paralogous domains, which were added to the cassette independently in each lineage. The additional PF00994 domain was also added independently in the plant protein CNX1_ARATH, but here the orthologous domain was used.
There could be several reasons for this observation. Assuming that horizontal transfer between chordates and plants is ruled out, it can be explained by independent recombination events of paralogous copies of the PF00994 domain in each of the arthropod, chordate, and plant lineages. Given that the plant protein has a different domain order, it must have happened independently in the plant lineage. A scenario in which the arthropod recombination selected a paralogous copy of the PF00994 domain present early in the metazoan lineage, whereas the plant and chordate recombinations both selected the same (orthologous) copy, and the other copy was lost in all lineages, would be consistent with the present tree. On the other hand, if horizontal transfer of a single domain from chordates to plants were biologically feasible, this would be an alternative explanation. It would be interesting to know if the chordate and plant proteins have evolved into identical functions despite the different domain architecture.
DISCUSSION
Conclusion
The HOPS/RIO comparison shows that the simplified HOPS approach has a higher sensitivity for finding orthologs than classical tree reconciliation with a complete species tree.
Orthology assignments are done on the domain level by HOPS. The orthologous relations can be viewed graphically in NIFAS. This combination provides a comprehensive and user-friendly analysis not only of orthologous relations but also of gene fusion and domain rearrangements. The two examples of orthology between genes with different domain architecture (Figs. 7, 8, 9) demonstrate the potential of HOPS to study the mechanism of domain rearrangements.
Additionally, these examples show that with increasing evolutionary distance, domain rearrangements and gene fusion can become an issue for assigning orthology. Any approach not taking into account the modular architecture of the proteins would fail to extract all orthologous information in the given examples.
The results of this study indicate that there is a limit to how much phylogenetic information can be included sensibly for finding orthologs. Up to a certain point, including additional information will improve the results. For instance, if one only analyzes sequences from two species for orthology, this can lead to a situation in which a paralog is incorrectly taken for an ortholog. This will happen if the true ortholog was lost or is not sequenced yet. Including sequences from additional species in the analysis would increase the chance that at least one true ortholog is present in the same clade. Tree-based methods like HOPS and RIO will then assign orthology and paralogy correctly.
But the lower success rate of RIO for finding orthologs between H. sapiens and C. elegans shows that the inclusion of additional sequences can have the opposite effect. Here the inclusion of all species in the analysis, especially arthropod sequences, prevents the tree-reconciling algorithm from finding the correct orthologs.
Even if the Coelomata hypothesis were used instead of the Ecdysozoa hypothesis in combination with tree reconciliation, this would not solve the problem. The investigated sequences are rather short, compared with full-length proteins. Therefore a tree reconstructed from them is more prone to “statistical fluxes” in evolution. Assigning orthology based on speciation events that followed as close in time as for D. melanogaster, H. sapiens, and C. elegans is bound to fail for a high fraction of sequences. The only viable approach is to say that the exact grouping of these three species is unknown.
For the assignment of orthology in HOPS, we try to find the right balance between including too little and too much phylogenetic information. This is not always possible. If there is only one sequence for each lineage present in a family and no sequence from an outgroup species is available, HOPS would assign these sequences as orthologous. However, it is possible that these sequences are not orthologous, but paralogous. This would be the case if the true orthologs were lost.
We would like to point out that although RIO was used for the comparison with tree reconciliation, the program was not specifically designed to solve the problem of finding all orthologs between two species. Rather, the idea of RIO is to find orthologs in any species to a given query sequence. In most of the examples in which RIO fails to find the ortholog pair on which we have focused, it typically does report an ortholog in some other species. We expect RIO to have a lower rate of false positives than HOPS. This expectation is based on RIO's stringent application of all available phylogenetic information from the species tree. However, estimating the false-positive rate is very hard, if not impossible. In the absence of the true ortholog, one cannot reliably say that two sequences that appear orthologous in a tree are not. In cases in which the true ortholog exists, both RIO and HOPS have very low false-positive rates, especially compared with cases in which many different species are clustered together (Remm et al. 2001), as in, for example, COGs (Tatusov et al. 1997).
Applying more advanced phylogenetic methods will allow including additional phylogenetic information in the analysis. The HOPS clustering scheme is set up in a way that it can handle ambiguity in the species tree and some errors encountered in neighbor-joining tree reconstruction. But the clustering scheme could easily be adjusted to include more phylogenetic information from the species tree in case a more advanced method for the tree reconstruction would be used.
At the moment, a large-scale analysis using, for instance, trees constructed in a Bayesian framework would not be practical. Calculating the tree for only a small protein family can easily take more than a day with MrBayes. In contrast, the computation of all trees and inference of orthology for all 3735 families done in this paper took less than a week on a 6 CPU UltraSPARC III system. With the increase of computational processing power, especially the increase due to parallel systems like Beowulf clusters, it will be possible to use advanced algorithms and methods for finding orthologs in the near future.
Acknowledgments
We thank Lars Arvestad for helpful comments on this work and Maido Remm, Jennifer Lee, and Alistair Chalk for their suggestions on the clustering scheme of HOPS.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
[The NIFAS viewer is integrated in the Stockholm Pfam site (http://Pfam.cgb.ki.se) and the test set of putative orthologs and detailed results are available at ftp://ftp.cgb.ki.se/pub/HOPS/.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr1305203.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S.R., Griffiths-Jones, S., Howe, K.L., Marshall, M., and Sonnhammer, E.L.L. 2002. The Pfam protein families database. Nucleic Acids Res. 30: 276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair, J.E., Ikeo, K., Gojobori, T., and Hedges, S.B. 2002. The evolutionary position of nematodes. BMC Evol. Biol. 2: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette, M., Schwikowski, B., and Tompa, M. 2002. Algorithms for phylogenetic footprinting. J. Comput. Biol. 9: 211-223. [DOI] [PubMed] [Google Scholar]
- Doolittle, W.F. 1999. Lateral genomics. Trends Cell Biol. 9: M5-M8. [PubMed] [Google Scholar]
- Fitch, W.M. 1970. Distinguishing homologous from analogous proteins. Syst. Zool. 19: 99-113. [PubMed] [Google Scholar]
- ____. 2000. Homology: A personal view on some of the problems. Trends Genet 16: 227-231. [DOI] [PubMed] [Google Scholar]
- Gogarten, J.P. and Olendzenski, L. 1999. Orthologs, paralogs and genome comparisons. Curr. Opin. Genet. Dev. 9: 630-636. [DOI] [PubMed] [Google Scholar]
- Goodman, M., Czelusniak, J., Moore, G.W., Romero-Herrera, A.E., and Matsuda, G. 1979. Fitting the gene lineage into its species lineage: A parsimony strategy illustrated by cladograms constructed from globin sequences. System. Zool. 28: 132-168. [Google Scholar]
- Hollich, V., Storm, C.E., and Sonnhammer, E.L.L. 2002. OrthoGUI: Graphical presentation of Orthostrapper results. Bioinformatics 18: 1272-1273. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck, J.P. and Ronquist, F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754-755. [DOI] [PubMed] [Google Scholar]
- Koonin, E.V., Makarova, K.S., and Aravind, L. 2001. Horizontal gene transfer in prokaryotes: Quantification and classification. Annu. Rev. Microbiol. 55: 709-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makalowski, W., Zhang, J., and Boguski, M.S. 1996. Comparative analysis of 1196 orthologous mouse and human full-length mRNA and protein sequences. Genome Res. 6: 846-857. [DOI] [PubMed] [Google Scholar]
- Mushegian, A.R., Garey, J.R., Martin, J., and Liu, L.X. 1998. Large-scale taxonomic profiling of eukaryotic model organisms: A comparison of orthologous proteins encoded by the human, fly, nematode, and yeast genomes. Genome Res. 8: 590-598. [DOI] [PubMed] [Google Scholar]
- Page, R.D.M. 1994. Maps between trees and cladistic analysis of historical associations among genes, organisms, and areas. System. Biol. 43: 58-77. [Google Scholar]
- Remm, M., Storm, C.E., and Sonnhammer, E.L.L. 2001. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 314: 1041-1052. [DOI] [PubMed] [Google Scholar]
- Snel, B., Bork, P., and Huynen, M.A. 2002. Genomes in flux: The evolution of archaeal and proteobacterial gene content. Genome Res. 12: 17-25. [DOI] [PubMed] [Google Scholar]
- Sonnhammer, E.L.L. and Koonin, E.V. 2002. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 18: 619-620. [DOI] [PubMed] [Google Scholar]
- Stein, L. 2001. Genome annotation: From sequence to biology. Nat. Rev. Genet. 2: 493-503. [DOI] [PubMed] [Google Scholar]
- Storm, C.E. and Sonnhammer, E.L.L. 2001. NIFAS: Visual analysis of domain evolution in proteins. Bioinformatics 17: 343-348. [DOI] [PubMed] [Google Scholar]
- ____. 2002. Automated ortholog inference from phylogenetic trees and calculation of orthology reliability. Bioinformatics 18: 92-99. [DOI] [PubMed] [Google Scholar]
- Tatusov, R.L., Koonin, E.V., and Lipman, D.J. 1997. A genomic perspective on protein families. Science 278: 631-637. [DOI] [PubMed] [Google Scholar]
- Xie, T. and Ding, D. 2000. Investigating 42 candidate orthologous protein groups by molecular evolutionary analysis on genome scale. Gene 261: 305-310. [DOI] [PubMed] [Google Scholar]
- Zharkikh, A. and Li, W.H. 1995. Estimation of confidence in phylogeny: The complete-and-partial bootstrap technique. Mol. Phylogenet. Evol. 4: 44-63. [DOI] [PubMed] [Google Scholar]
- Zmasek, C.M. and Eddy, S.R. 2002. RIO: Analyzing proteomes by automated phylogenomics using resampled inference of orthologs. BMC Bioinformatics 3: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- ftp://ftp.cgb.ki.se/pub/HOPS/; HOPS.
- http://Pfam.cgb.ki.se; Stockholm Pfam site.
- http://www.rio.wustl.edu; RIO.