Abstract
Generating knockout mice is still an expensive and highly time-consuming process. Target construct generation, the first labor-intensive step in this process, requires the manipulation of large fragments of DNA and numerous, and often cumbersome, cloning steps. Here we show the development of a rapid approach for generating targeting constructs that capitalizes on efficient homologous recombination between linear DNA fragments and circular plasmids in Escherichia coli (“recombineering”), the availability of bacterial artificial chromosomes (BACs), and the accessibility of the sequence of the mouse genome. Employing recombineering, we demonstrate with only 1–2 template plasmids, short homologies (40–50bp) between donor and target DNA, and one subcloning step that we can efficiently manipulate BACs in situ to generate a complicated targeting vector. This procedure avoids the need to construct or screen genomic libraries and permits the generation of most standard, conditional, or knock-in targeting vectors, often within two weeks.
The generation of knockout mice through gene targeting by homologous recombination in embryonic stem (ES) cells is still an expensive and highly time-consuming process. Knockout construct assembly is the first step in a laborious process that can take up to a year even in the most technically skilled laboratories. Typically, the process requires screening a genomic library to obtain the gene of interest, followed by restriction mapping and numerous cloning steps.
One strategy that can avoid time-consuming subcloning steps and may offer a more efficient approach for generating targeting constructs is the use of homologous recombination in Escherichia coli (Eggleston and West 1996). The basis of the technology is to utilize strains of E. coli that can efficiently carry out homologous recombination between short terminal homology regions on a linear PCR-derived DNA fragment and sequences on a recipient plasmid. This strategy has been referred to as either recombineering—for recombinogenic engineering (Copeland et al. 2001; Court et al. 2002)—or recombination cloning (Zhang et al. 2002). This approach has been utilized to alter various DNA targets, including the E. coli chromosome, high copy number plasmids, and bacterial artificial chromosomes (BACs; Zhang et al. 1998, 2002; Angrand et al. 1999; Muyrers et al. 1999; Datsenko and Wanner 2000; Yu et al. 2000; Copeland et al. 2001; Lee et al. 2001; Liu et al. 2003).
While two groups have employed this strategy for the construction of gene targeting vectors, simplifying some of the time-consuming steps for targeting vector construction, these approaches still required gene isolation or the construction of genomic libraries and/or numerous complex gene manipulations (Angrand et al. 1999; Zhang et al. 2002). We desired a construction strategy that would preclude the need to screen for and subsequently map genomic sequences and, most importantly, would avoid all of the cumbersome cloning steps. To simplify the process, we reasoned that one could identify the genomic sequence of the gene of interest from publicly available databases and then purchase an E. coli clone containing a BAC encompassing these sequences rather than employing standard genomic library screening. With the appropriately constructed template vectors (described below) and the essential recombination machinery, we could then use homologous recombination in E. coli to manipulate directly the gene of interest in the BAC. Finally, in one step, we could remove the manipulated sequence within the BAC by identifying flanking restriction sites from the published sequence, and under appropriate antibiotic selection, generate a targeting construct. Overall, this approach would obviate the need for the numerous time-consuming cloning steps typically required for generating complicated targeting constructs.
RESULTS AND DISCUSSION
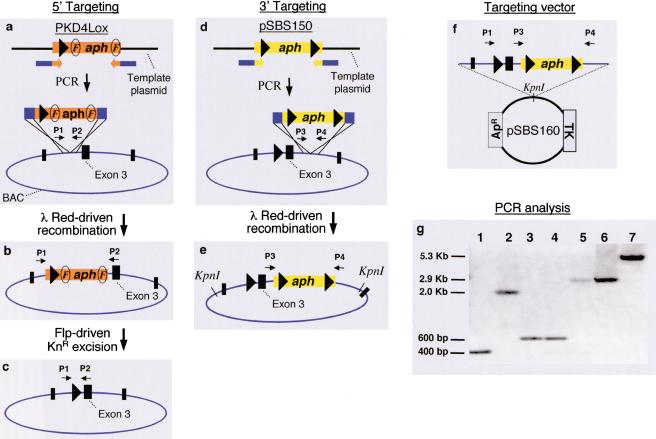
We sought to generate a variety of constructs for the conditional targeting of the NOD2/CARD15 gene (the murine homolog of a novel Crohn's disease gene; Hugot et al. 2001; Ogura et al. 2001). From available sequence databases, we identified the NOD2 gene and purchased a BAC clone containing all 11 coding exons. The overall strategy for generating a conditional targeting vector was to employ two successive recombination steps to introduce a loxP site 5′ to exon 3 and an aminoglycoside phosphotransferase (aph) gene cassette (mediating kanamycin resistance or neomycin resistance when expressed in prokaryotes or eukaryotes, respectively) flanked by loxP sites (“floxed”) 3′ to exon 3 in the BAC clone (Fig. 1). To mediate each homologous recombination step, we introduced a plasmid containing the arabinose-inducible Red recombination system from bacteriophage λ (Murphy 1998; Datsenko and Wanner 2000) into the BAC containing strain.
Figure 1.
Conditional knockout vector construction. (a–f) Sequential steps leading to a NOD2 exon 3 recombinant BAC and targeting vector (see text for details). (g) Gel electrophoresis depicting PCR products confirming the recombinants schematized in diagrams: (a) lane 1 (WT-BAC), (b) lane 2 (BACloxP/frt/aph/frt), and (c) lane 3 (BACloxP), generated with primers 1 (P1) and 2 (P2); (d) lane 4 (BACloxP) and (e) lane 5 (BACloxP-loxP/aph/loxP), generated with P3 and P4; and (f) lanes 6,7 (targeting construct) using respectively P3 and P4, and P1 and P4. Circled “F”, frt site; large arrowhead, loxP site.
In the first recombination step, intronic BAC sequences 5′ to exon 3 were targeted with PCR products containing a loxP site and an frt-flanked aph gene (Fig. 1a). These PCR products were amplified with 60-bp primer pairs consisting of 40 bp of sequence homologous to the intronic region flanking the targeted exon, and 20 bp complementary to the template plasmid containing the DNA to be inserted (Fig. 1a). The template plasmid, pKD4Lox, was derived from pKD4 (Datsenko and Wanner 2000), a conditional replicon (oriRγ) that requires the pir gene product (absent in most E. coli strains that carry BACs) for replication. Transformation with the loxP/aph-containing PCR fragment resulted in several kanamycin-resistant (KnR) recombinants (Fig. 1b). Following transformation with a temperature-sensitive replicon that expresses the flp recombinase (Cherepanov and Wackernagel 1995), which recognizes frt sites, the aph gene was excised by site specific recombination (Fig. 1c).
The second step was to manipulate BAC intronic sequence 3′ to exon 3. Kanamycin-sensitive strains isolated above (Fig. 1c) were targeted with a PCR product containing a floxed aph cassette generated from pSBS150. This pKD4-based template contains the aph gene under the control of both prokaryotic and eukaryotic promoters to allow selection in bacterial and ES cells (Fig. 1d). From the genomic sequence, we identified a restriction enzyme site (KpnI) flanking the manipulated exon that would yield a fragment of sufficient length for homologous recombination in ES cells (Fig. 1e). To isolate the construct, the recombinant BAC was digested with KpnI, and the fragments were subcloned into a multicopy cloning vector (e.g., pBluescript or a thymidine kinase [TK]-containing plasmid; Fig. 1f), and selected for ampicillin resistant (ApR) and KnR colonies. The presence of the aph gene within the region of the BAC of interest allowed, upon kanamycin selection, enrichment of clones containing only the desired DNA fragments. PCR (Fig. 1g), restriction enzyme, and sequencing analyses confirmed the desired products in all steps.
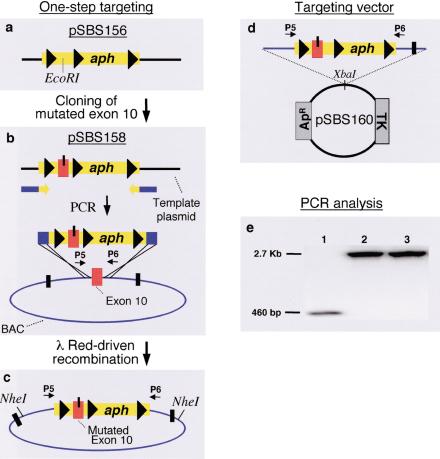
We also developed a similar method for a “one-step” targeting strategy that would be applicable for either conditional knockout targeting construct generation or for the “knock-in” of a mutated exon. We first generated pSBS156, another conditional replicon. This plasmid contained a unique EcoRI site to allow the cloning of a wild-type or “mutated” exon of interest flanked by a loxP sequence and a floxed aph gene cassette (Fig. 2a). Subsequently, we developed a NOD2 “knock-in” template by introducing into pSBS156 a mutated exon 10 that corresponded to the mutant genotype in CD (Fig. 2b, pSBS158; Hugot et al. 2001; Ogura et al. 2001). Homologous recombination between linear PCR fragments and the BAC generated KnR recombinants containing the mutated exon and a 3′ floxed aph gene, but not the 5′ loxP. To circumvent this recombination that occurred within the exon (which would be incompatible with a conditional targeting construct), we increased the homology arm in the sense primer to 50 bp, and selected a target sequence located 100 bp upstream of the mutated exon as the 5′ region for homologous recombination. Appropriate recombinants were identified and confirmed (Fig. 2c,e). Restriction analyses of the recombinant BACs revealed the absence of unwanted internal rearrangements (data not shown). A targeting construct was generated by digesting the recombinant BAC with NheI, followed by subcloning into a TK-containing vector and selection on ampicillin/kanamycin plates (Fig. 2d).
Figure 2.
“Knock-in” vector construction. (a–d) Sequential steps leading to a mutated NOD2 exon 10 “knock-in” BAC and targeting construct (see text for details); (e) Gel electrophoresis depicting PCR products confirming the recombinants schematized in diagrams: (c) lane 1 (WT-BAC), (d) lane 2 (BACloxP/exon 10m/loxP/aph/loxP), and (e) lane 3 (the targeting vector), generated with P5 and P6.
Although not described here, construction of simple targeting vectors that aim to replace a portion of a gene with an aph gene cassette can be generated using pSBS150 as template. PCR primers homologous to the sequence flanking the deleted regions can be used to generate a linear fragment for homologous recombination with the BAC. It was previously shown that primers designed in this way efficiently loop out intervening sequence with replacement by the selectable marker (Zhang et al. 1998; Angrand et al. 1999; Datsenko and Wanner 2000; Yu et al. 2000).
Two groups previously described methods for the construction of targeting vectors utilizing recombineering (Angrand et al. 1999; Zhang et al. 2002). Both of these approaches required screening genomic libraries and did not utilize the availability of BACs. Here we describe a simple and rapid method of generating complex gene knockout targeting constructs using recombination engineering requiring only one or two plasmid templates (Figs. 1, 2). This approach exploits the commercial availability of mouse genomic BAC clones spanning known sequences in the mouse genome. Since they can accommodate 5–10-fold larger inserts than bacteriophage, the use of BACs ensures that sufficient sequence is available to generate large regions of homology required for efficient homologous recombination in ES cells. Furthermore, this strategy avoids library construction and most subcloning steps that are characteristic of conditional targeting construct generation, and can be applied to the rapid generation of most standard, conditional, or knock-in targeting vectors.
While our work was being finalized for publication, another recombineering-based approach that manipulated BACs for the generation of targeting vectors was reported (Liu et al. 2003). Those authors employ gap-repair for the sub-cloning of a BAC fragment into a recipient plasmid, followed by further targeting of this plasmid to construct a conditional targeting vector. Although both approaches utilize recombineering and BAC DNA sequences for vector construction, there are unique and notable distinctions. Because we did not use Cre recombinase in the BAC targeting process, the presence of loxP sites in the BAC vectors has not posed any constraints to the insertion of further loxP sites for conditional targeting vector construction. On the other hand, the presence of three loxP sites leads to three possibilities of recombinants when the Cre recombinase is expressed in ES cells; the utilization of additional sequences (e.g., frt sites) which orient site-specific deletion can further simplify this process (Liu et al. 2003). In addition, in our approach, the requirement for unique restriction sites in the BAC that are located outside the regions of homology is one potential obstacle. Although we have been able to generate three different targeting constructs using this approach without difficulty, one can circumvent this obstacle by subcloning via gap repair (Liu et al. 2003).
Most importantly, in our studies, we manipulated BACs in situ in the original host, which avoids numerous subcloning steps and permits additional uses of this technology. Furthermore, we utilized a plasmid, rather a prophage, to encode the lambda Red recombination system and achieved efficient recombination utilizing only two templates for all targetings and very short terminal homologies (40–50 bp). Short homologies did not pose any constraint to our targetings. We achieved a specific targeting efficiency of 65%±30% for eight unique targetings within four different genes (average total number of colonies tested per targeting: 13±9; average number of correct colonies identified per targeting: 7±3). Aberrant recombination in the alternative approach employing gap repair may have resulted from short homologies between the donor sequences and repetitive sequences in the target DNA (Liu et al. 2003). It is likely that this potential problem can be avoided by surveying the BAC sequence prior to designing primers, to avoid using repetitive sequences for the site of recombination. Finally, manipulating BACs directly can permit the use of the entire BAC as a targeting construct in ES cells—avoiding constraints on the size of the homology arms.
METHODS
Construction of Template Plasmids
The template plasmids pKD4Lox, pSBS150, and pSBS158 were derived from pKD4, and each maintained in pir+ bacteria (Datsenko and Wanner 2000). The use of pKD4 as the backbone was essential to prevent replication of the template plasmid in transformed strains.
pKD4Lox was constructed by cloning a single loxP site upstream of the frt-flanked TN5-aph segment in pKD4. pSBS150 was constructed by cloning a floxed aph gene (driven by both TN5 and PGK promoters) in the pKD4 vector double-digested with BamHI/NcoI. To allow single targeting of BACs (see below), pSBS156 was generated from pSBS150 by cloning a loxP site upstream of the floxed aph gene. pSBS158 was constructed by cloning the mutated CARD15 exon (see text) into a unique EcoRI site located between the loxP sequence and the floxed aph gene cassette in pSBS156 (see Fig. 2b).
Generation of BAC Targeting Constructs
PCR products for BAC targeting were generated with 60–74-bp primer pairs using Advantage 2 polymerase (Clontech). Within each primer, the first 40–54 bp (homology arm) was homologous to the BAC sequence, and the next 20 bp was complementary to the template plasmid of interest. The 60-bp sequences to prime pKD4Lox, pSBS150, and pSBS158 (and further BAC targeting) were, respectively: CCTTGTTTAACCATATCAAAGTTCCCG GCTGTAT TCCTGCTCTTGAGCGATTGTGTAGGC (forward) and GGACCCAGGGGCTTCC TAAACCTGGTTCTTCCCCTG CAA ACATATGAATATCCTCCTTAG (reverse), ATT CCACCTCTGC CACCCTTCATACAACACCAGCCAGACCCCGATCATA TTCAATA ACCC (forward) and GCTATAGTTATTTGTTCT GAAACTAAGGCCCAGAAGAGGG CTATGCTACTCCGT CAATAA (reverse), AGCCTGGCAATGAGCTGTGGCTCCT CAGCCCTTCCTCCCTTCCCAGTGTGGGTACCGGGGG ATCCGGAACCCTT (forward) and ATACCAAATGTTG GTCA AAACTGATCTTCAGAATTCCACCCTATG CTACTCCGTCAATAA (reverse). Each PCR product (1.6-, 2.2-, and 2.5-Kb in length for pKD4Lox, pSBS150, and pSBS158, respectively) was purified (QIAquick PCR purification kit, QIAGEN), digested with DpnI (New England Biolabs), ethanol-precipitated, and resuspended in Tris-Cl buffer pH 8.5.
Two-Step BAC Gene Targeting
Bacteria carrying the target BAC and pKD46 (λ Red recombination system) were grown in 100-mL LB cultures with chloramphenicol (12.5 μg/mL), ampicillin (100 μg/mL), and L-arabinose (150 μg/mL, Sigma) at 30°C to an OD600 of ∼0.6. Electrocompetent bacteria were transformed with 100 ng of the pKD4Lox-amplified and aph gene-containing PCR construct. Chloramphenicol-resistant (CmR)/KnR transformants were first replated to confirm adequate resistance, and then screened by PCR (using P1 and P2 primers, AGCCCTGCCCCCTT CTATTT and TCACAGCGGGACCTACACAG, respectively; see Fig. 1), and the PCR products were sequenced to confirm the desired targeting. Positive colonies were grown overnight at 42°C to cure pKD46 (confirmation was performed by testing ampicillin sensitivity). In order to remove the aph gene, a single CmR/KnR and ampicillin-sensitive colony was transformed with pCP20 (an ApR and temperature-sensitive replicon containing the thermally-induced flp recombinase gene; Cherepanov and Wackernagel 1995) and grown overnight at 30°C on chloramphenicol/ampicillin plates. Some colonies were selected again in chloramphenicol plates at 43°C, and then tested for kanamycin sensitivity (i.e., loss of the aph gene) and loss of ampicillin resistance (i.e., the curing of pCP20).
A single CmR and kanamycin-sensitive colony was transformed with pKD46. Chloramphenicol/ampicillin selected transformants were then transformed with 100 ng of the pSBS150-amplified (containing the floxed aph gene) PCR product. As detailed above, to confirm the desired targeting, CmR/KnR transformants were first screened by PCR and sequencing (using P3 and P4 primers, TGGGCCTGTGACAAATGCTACTG and GTTGTGGGGATGGAT GGGGTTATT, respectively; see Fig. 1), and then cured of pKD46.
Mutation of the Target Exon
PCR amplification of mNOD2/CARD15 exon 10 was performed in the BAC clone RP23–447G4, and the product was subcloned in PCR4-TOPO (TOPO TA cloning kit, Invitrogen). A mutation analogous to the 3020incC frameshifit mutation in NOD2/CARD15 found in Crohn's disease patients (Hugot et al. 2001; Ogura et al. 2001) was introduced into this subclone, using site-directed mutagenesis (QuickChange site-directed mutagenesis kit, Stratagene).
One-Step BAC Gene Targeting
pSBS158 was used as the template for the one-step BAC targeting in E. coli. This targeting used the same BAC clone and was performed as described above for the two-step targeting protocol. Confirmation of the desired recombinant BAC was performed by PCR screening and sequencing (using primers P5 and P6, TGGGCCTGTGACAAATGCTACTG and GTTGTGGGGATGGATGGGGTTATT, respectively; see Fig. 2).
Generation of the Targeting Vector
In both targeting strategies, the desired recombinant BAC DNA (∼50 μg) was digested with a suitable restriction enzyme in order to achieve homology arms with appropriate size for further homologous recombination in ES cells. Following digestion, BAC DNA was shotgun-cloned into a linearized pBluescript-derived TK-containing vector, and the desired transformants were selected by growth on ampicillin/kanamycin plates.
Acknowledgments
We gratefully acknowledge members of the Snapper laboratory for their support and the critical reading of the manuscript by Drs. Daniel Podolsky, Vijay Yajnik, Antonio Oliveira-dos-Santos, Sheila Thomas and Anna Lyubimova. These studies were supported by the National Institutes of Health (AI50950 and HL59561—S.B.S.) and by CAPES (Fundação Coordenação de Aperfeiçoamento de Pessoal de Nível Superior/Brazil—V.C.-A.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1356503. Article published online before print in August 2003.
References
- Angrand, P.O., Daigle, N., van der Hoeven, F., Scholer, H.R., and Stewart, A.F. 1999. Simplified generation of targeting constructs using ET recombination. Nucleic Acids Res. 27: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov, P.P. and Wackernagel, W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158: 9–14. [DOI] [PubMed] [Google Scholar]
- Copeland, N.G., Jenkins, N.A., and Court, D.L. 2001. Recombineering: A powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2: 769–779. [DOI] [PubMed] [Google Scholar]
- Court, D.L., Sawitzke, J.A., and Thomason, L.C. 2002. Genetic engineering using homologous recombination. Annu. Rev. Genet. 36: 361–388. [DOI] [PubMed] [Google Scholar]
- Datsenko, K.A. and Wanner, B.L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggleston, A.K. and West, S.C. 1996. Exchanging partners: Recombination in E. coli. Trends Genet. 12: 20–26. [DOI] [PubMed] [Google Scholar]
- Hugot, J.P., Chamaillard, M., Zouali, H., Lesage, S., Cezard, J.P., Belaiche, J., Almer, S., Tysk, C., O'Morain, C.A., Gassull, M., et al. 2001. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 411: 599–603. [DOI] [PubMed] [Google Scholar]
- Lee, E.C., Yu, D., Martinez de Velasco, J., Tessarollo, L., Swing, D.A., Court, D.L., Jenkins, N.A., and Copeland, N.G. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73: 56–65. [DOI] [PubMed] [Google Scholar]
- Liu, P., Jenkins, N.A., and Copeland, N.G. 2003. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 13: 476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, K.C. 1998. Use of bacteriophage λ recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 180: 2063–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyrers, J.P., Zhang, Y., Testa, G., and Stewart, A.F. 1999. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 27: 1555–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura, Y., Bonen, D.K., Inohara, N., Nicolae, D.L., Chen, F.F., Ramos, R., Britton, H., Moran, T., Karaliuskas, R., Duerr, R.H., et al. 2001. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 411: 603–606. [DOI] [PubMed] [Google Scholar]
- Yu, D., Ellis, H.M., Lee, E.C., Jenkins, N.A., Copeland, N.G., and Court, D.L. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. 97: 5978–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P., Li, M.Z., and Elledge, S.J. 2002. Towards genetic genome projects: Genomic library screening and gene-targeting vector construction in a single step. Nat. Genet. 30: 31–39. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Buchholz, F., Muyrers, J.P., and Stewart, A.F. 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20: 123–128. [DOI] [PubMed] [Google Scholar]