Abstract
GANESH is a software package designed to support the genetic analysis of regions of human and other genomes. It provides a set of components that may be assembled to construct a self-updating database of DNA sequence, mapping data, and annotations of possible genome features. Once one or more remote sources of data for the target region have been identified, all sequences for that region are downloaded, assimilated, and subjected to a (configurable) set of standard database-searching and genome-analysis packages. The results are stored in compressed form in a relational database, and are updated automatically on a regular schedule so that they are always immediately available in their most up-to-date versions. A Java front-end, executed as a stand alone application or web applet, provides a graphical interface for navigating the database and for viewing the annotations. There are facilities for importing and exporting data in the format of the Distributed Annotation System (DAS), enabling a GANESH database to be used as a component of a DAS configuration. The system has been used to construct databases for about a dozen regions of human chromosomes and for three regions of mouse chromosomes.
One of the underpinning aims of the Human Genome Project is to provide the resources to support genetic analysis of human conditions and disorders. This aim is incomplete in part because neither the finished DNA sequence of the whole of the human genome has yet been established, nor has the task of identifying all of the genes within even the available DNA sequence been completed (Lander et al. 2001). This is an inevitable consequence of the continuing nature of the HGP. But even if the genome were complete, software tools in the public domain are not optimally configured for gene discovery efforts in circumscribed genomic regions.
Presently, there are several partially independent sources of annotated human genomic sequences that include Ensembl (Hubbard et al. 2002), UCSC (Kent et al. 2002), and Celera (Venter et al. 2001). Each of these aims at producing an accurate assembly of the genomic sequence and using available databases of expressed sequences, combined with in silico gene prediction, to identify the location and structure of human genes with the greatest possible accuracy. Necessarily, these databases increase their coverage incrementally as both the finished DNA sequence and further computational resources become available, but, in turn, this means that the annotation processes lag behind relevant data. Explicitly, these databases are databases of record, and as a consequence, they tend to sacrifice completeness for the sake of accuracy and certainty. Evidence for the existence or location of a gene may be rejected if the appropriate level of confidence is lacking. Furthermore, we note that Hogenesch et al. (2001) have argued that the concordance between Ensembl and Celera's data is partial, suggesting that neither database can be considered as definitive.
In contrast to the requirements of databases of record, genetic research places rather different constraints upon the annotation and analysis of genomic data. Modern genetic research generally takes a positional cloning approach, frequently using whole-genome scans for linkage in appropriately constructed pedigree collections to identify regions likely to contain a variant gene(s) that predisposes to the condition or disorder under study. Such approaches generally identify regions ranging from a few megabases (in the case of monogenic disorders) to tens of megabases (in the case of multifactorial traits). The challenge for the genetic researcher is then to identify the disease-susceptibility variant or variants within this region.
Genetic analysis software and gene identification software as represented by databases of record might, in time, become congruent. At present, this is not the case. There are three main distinguishing features. First, although gene identification is a key element of both types of software, the ultimate goal of genetic analysis software is to derive an exhaustive list of genes and gene-like objects within a specified region, so that these can be subjected to experimental analysis to identify sequence variants that might be correlated with disorder or condition state. Accuracy of gene prediction, especially the elimination of false positives, is of lesser importance because experimental analyses can be deployed readily to validate the in silico predictions (Shoemaker et al. 2001). Secondly, genetic analysis requires as exhaustive an identification of potential genes in the target region as possible, and so must be able to exploit any additional data sources that may be relevant to the target region. In contrast, for databases of record, the sheer scope of the target region necessarily limits the sources of data that can be accommodated. Thirdly, genetic analysis requires comparative analysis of all of the genes within the target region, to allow candidate prioritization. This is not a necessary feature of databases of record, which require classification tools that have a global scope. It has been said that the annotations served by the reference databases of record are broad, but shallow, whereas those required by genetic researchers tend to be narrow, but deep (Stein 2001).
These three design differences make the databases of record cumbersome and restricted in their utility for specifically genetic analysis. To overcome these limitations, we have developed a specialist software package (named GANESH), designed in explicit recognition of the differing goals and requirements of geneticist and genomicist. GANESH is a set of software components that may be assembled to construct a newself-updating database providing annotation for a specified region of human (or other) genomic sequence. Sequence and other relevant data for the target region are gathered from various distributed data sources, assimilated, and subjected to a range of database-searching and genome-analysis programs. The results are stored in the database in compressed form and updated on a regular schedule, so that they are always available immediately in their most up-to-date form. A front-end in Java, executed as a stand-alone application or as a web applet, provides a graphical interface for navigating the database and visualization of the genome features detected. There are utilities for importing and exporting data in a variety of formats, including those of DAS, the Distributed Annotation System (Dowell et al. 2001).
At the time of writing, GANESH has been used to construct databases for about a dozen regions of human chromosomes and for three regions of mouse chromosomes. Example databases and further details can be found at http://zebrafish.doc.ic.ac.uk. In this work, we describe the structure of GANESH and its components, and demonstrate its role in the genetic analysis of several regions of the human genome.
RESULTS
GANESH: Overview
A GANESH application has the following main components:
the assimilation module (including downloading scripts, sequence analysis packages, and sequence database searching tools);
the database;
the updating module;
the graphical front-end;
visualization software; optional analysis tools; utilities.
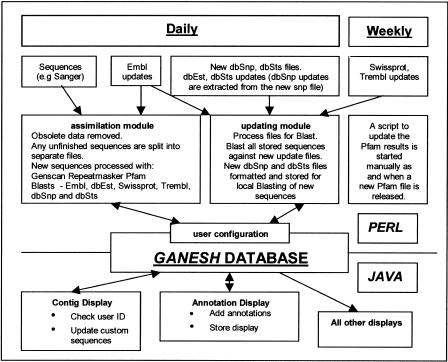
Figure 1 provides an overview of the system structure.
Figure 1.
GANESH Overview.
For construction of a new application, GANESH is focused upon a region of the genome (human or other) by first identifying DNA markers or genomic positions that flank the region of genetic interest. These markers, in turn, are used to define a set of DNA clones that span the interval and that have been, or are being, sequenced. Several databases can be used to select these clones including the UCSC golden path (Kent and Hausler 2001), Ensembl, or the total human fingerprint database (McPherson et al. 2001). The next step is to specify one or more sources of DNA sequences for these clones. In principle, any site is acceptable, provided the files have a fixed location and there is a well-defined update file structure for sequences still to be collected. We have successfully used the Sanger Institute and EMBL. For regions being sequenced by the Sanger Institute, we download from the relevant ftp archive (ftp.sanger.ac.uk/pub and human/sequences or mouse/sequences) and check daily for updates. For other regions, the sequences are retrieved initially from EMBL and updates extracted from the EMBL daily update files available from the European Bioinformatics Institute (EBI) ftp site (ftp.ebi.ac.uk/pub/databases/embl/new).
All sequence data from the target region is thereafter downloaded by the assimilation module and processed using a range of (standard) genome analysis tools. The standard configuration presently includes RepeatMasker (http://ftp.genome.washington.edu/RM/RepeatMasker.html), Genscan (Burge and Karlin 1997) for exon predication, poly(A) addition site and promoter finding, and Pfam analysis (Bateman et al. 2002) using the Wise/halfwise package available from the Sanger Institute (http://www.sanger.ac.uk/Software/Wise2). We have chosen this set of programs to provide maximal flexibility for the analysis of any region of any genome; there are, of course, a number of sources of precomputed data, for example, repeats in human DNAs, that we could have used, but this would necessarily have significant implications on the range of DNAs that could be studied with GANESH.
BLAST searching (Altschul et al. 1990) is performed for all of the sequences against EMBL, dbEST, SWISS-PROT, and TrEMBL for homology, and against dbSNP and dbSTS for identity. These searches are all performed on local copies of the databases. Parameters for the different kinds of BLAST searches, and for the other analysis packages, can all be specified by the user though the standard configuration defaults to settings that we have determined to be most useful. Similarly, the configuration of components can also be adjusted. The system is designed to be modular and user-configurable; newcomponents—newgenome analysis tools, other forms of sequence database searches, in-house annotations (e.g., from in-house experiments)—are added easily.
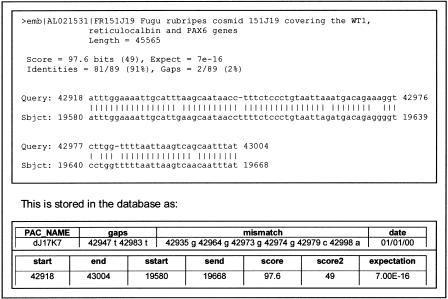
The results of all of these computations are compressed and stored in a standard relational database (specifically, the MySQL system (http://www.mysql.com), which we chose because it is used widely, and is freely available in the public domain). Scripts are provided to reconstruct the original form from the compressed form whenever required. For illustration, Figure 2 shows the compressed form of results of BLAST searches. Links to less frequently used data sources, or data sources requiring little computation (such as publication databases, OMIM, some gene expression collections) are provided as standard WWW links.
Figure 2.
BLAST storage. Storing the BLAST results. Shown is the standard BLAST output of a hit for the sequence of PAC dJ17K7 (Query). In the Subject description, emb signifies this is from the EMBL database; AL021531 is the accession number, the unique identifier for this sequence; Score is a measure of the quality of the hit; Expect is the probability of this hit occurring by chance in a database of this size; Identities is the number of matching bases; Gaps is the number of gaps introduced into the sequence (query) in order to produce the alignment. Following this is the alignment itself.
Once a GANESH database is set up, a set of procedures in the updating module scan the remote data sources periodically and download, process, and assimilate any new sequences from the target region as they are deposited. New sequences are passed to the assimilation module for processing in the usual way. The results for all stored sequences already processed are also updated. BLAST searches are repeated for every stored sequence as new versions of the archival databases are released—daily for the incremental releases of EMBL-nr (nonredundant EMBL that does not contain ESTs or STSs), EMBLnew (everything that has been added to EMBL since the last release), dbEST, TrEMBLnew (everything added to TrEMBL since the last release), dbSNP, and dbSTS, and weekly for other databases such as SWISSNEW (everything added to SWISS-PROT since the last release). Computationally intensive tasks, such as bulk searches against the EMBL, EST, and protein sequence databases, and the execution of the various DNA sequence analysis programs (e.g., repeat sequence detection and gene/exon prediction) are performed automatically, usually overnight. For our GANESH installation at Imperial College, we make use of a general purpose system, Disperse (Clifford and Mackey 2000; http://www.doc.ic.ac.uk/-rc5/Disperse), which distributes the updating computations to any available (PC) machine left free in the Department of Computing (and elsewhere) overnight. Large computational facilities, therefore, are not necessarily required for maintaining the databases.
It is also possible for users to add their own annotations, as discussed further below. The system reports any new results automatically as they are discovered for regions that have been registered by the user as being of particular interest.
To make EST data more meaningful, and at the same time to reduce the amount of storage required, matches between ESTs and the region of interest are stored in the database according to various stringency criteria. The standard configuration uses the criteria identified in Bailey et al. (1998). EST hits not satisfying these criteria are not stored, but are still available, in that they can be reconstructed whenever required by triggering a BLAST search of the relevant database.
We also provide an optional component that attempts to predict the presence of genes/exons by comparing the output from several of the annotation tools. This has been of particular interest to the user groups working on identification of candidate disease genes. It is described below separately.
The first application of GANESH, and the application used to drive its development, was a reconstruction of the 11Db database, implemented previously in ACeDB (R. Durbin and J. Thierry-Mieg, unpubl.; http://www.acedb.org), maintained at the Department of Biochemistry, Imperial College. The 11Db contains the sequence and annotation for the WAGR region of human chromosome 11. At the time of writing, the GANESH version of the 11Db database has been operational (and publicly accessible) for about 2 yr. Similar GANESH databases have nowbeen set up for regions of human chromosomes 1, 2, 3, 5, 6, 11, 12, 14, 16, and 20, and regions of mouse chromosomes 2, 4, and 12. We also constructed a GANESH database of the complete human chromosome 21 to test whether that system can cope with this volume of data, and to provide comparisons with annotations produced elsewhere. Further details of current applications may be found at http://zebrafish.doc.ic.ac.uk.
The initial analysis of a new region ∼10 Mb in size typically takes ∼24 h to complete, depending, of course, on the finished state of the sequences and the processing power available. The daily update of a 10-Mb region typically takes ∼1.5 h, again depending on the size of the BLAST database updates and the number of updated sequences. It should be noted, however, that the time to process and update several GANESH databases does not necessarily increase, as the assimilation and updating processes are inherently parallel. As already mentioned, we use the Disperse system (Clifford and Mackey 2000) to distribute the tasks to any number of available machines.
Sequence and Annotation Display
The display of sequence and annotation data is of central importance. The challenge here is to allowthe ultimate synthesis of a list of genes or potential genes within the region. The decision about what to include in this list, in contrast to the criteria used within the databases of record, is entirely within the user's control.
We have chosen to use a nested series of displays organized at the first level around individual DNA clones (but, see below for the display of larger sequenced fragments). The display software is written in Java, and can be used both as a Web-based applet and an application. It uses a library of Java display utilities that were developed independently following trials with the Neomorphic Genome Software Development Kit (http://www.neomorphic.com), with which it shares some look and feel. Interactive access to GANESH, allowing individual user annotations, is controlled by a user login.
Figure 3 shows the opening screen of the display, in this case a human chromosome region, 11p13, that is deleted in WAGR syndrome patients. The display can be scrolled, and zoomed in and out to focus on areas of interest. Selection of a clone in this screen (by clicking) accesses information about it and (by double-clicking) brings up the annotation display shown in Figure 4. The content of this display is configurable. The user can also add or remove displayed features as required during viewing. As with all displays, it is also possible to zoom in and out; increased detail (names, markers, etc.) appears automatically as display resolution allows.
Figure 3.
Contig display. The initial display shows the minimal tiling path for the region (in this case the chromosome 11 WAGR region) with markers above to assist in navigation. The markers are linked to the associated sequence clones and vice versa. The display can be zoomed in and out. From it, the Annotation Display can be launched. Window image is clipped for presentation purposes.
Figure 4.
Annotation Display. (A)The Annotation display provides an overview of the features of the sequence. For the application version, a configuration file can set the features to be displayed. The display can be scrolled and zoomed. From this display, the details of an individual feature, including a BLAST hit or all of the BLAST hits, can be viewed. There are also links to several external data sources and options to view the sequence (or partial sequence) in fasta format or a six-frame translation. Window image is clipped for presentation purposes. (B) Annotation Display. Zoomed view, at small-medium resolution.
Clicking on any of the individual features brings up details of the annotation, depending on the nature of the probe. There is a separate graphical display for viewing BLAST hits. Clicking on a BLAST feature brings up the BLAST window shown in Figure 5. This can be scrolled and zoomed as usual. The BLAST significance level for display is controlled with a slider (shown to the left of the screen in Fig. 5), which is used to specify the stringency threshold for displayed BLAST hits. Clicking on a BLAST hit brings up a detailed view of the BLAST alignment, as shown in Figure 6. Links are created automatically to access source data, via the WWW, from GenBank, Entrez, dbSNP, UniGene, and SRS. Because of security restrictions, it is not possible to copy and paste from an applet. Some output is therefore displayed in a separate browser window via a CGI Perl script to enable the user to cut and paste information into other software. Additional windows have also been created to display the complete FPC clone map of the region (Marra et al. 1997), YAC-based maps, and several radiation hybrid maps (James et al. 1994; Deloukas et al. 1998). As with all displays, these can be scrolled and zoomed to access display details at varying levels of resolution.
Figure 5.
BLAST Display. The BLAST Display shows a summary of all the BLAST hits. The display can be scrolled and zoomed. An index of all of the hits can be displayed optionally—as shown—which is linked to the main display. The slider on the left of the display is used to screen the BLAST hits on minimum score value. Window images are clipped for presentation purposes.
Figure 6.
BLAST Alignment Display. The BLAST Alignment Display shows the detail of an individual BLAST hit with the gaps/mismatches highlighted. The display can be scrolled and zoomed. The standard BLAST alignment and details are also provided in a text window from which they can be copied. Window image is clipped for presentation purposes.
A key feature superimposed on these displays is the automatic notification of updated information from the daily/weekly BLAST analyses or resulting from updates to the sequence itself. The last viewed version can be retrieved for comparison to the latest information. There is a further option to viewjust the information added since the user last accessed the system. This feature is particularly useful when combined with the tools provided for users to add their own annotations. These are keyed to individual operators and are intended to be the primary tool for recording acceptance or rejection of features within the displays—genes, gene-like objects, SNPs, regulatory regions, etc. It is important to stress that these annotations do not necessarily have to represent definitive genome features as in the databases of record, and can take the form of a simple statement of the form `gene X is located at position Y' in the sequence. The purpose of GANESH is primarily to facilitate the identification of as complete a list as possible of genes and other genome features of interest within the target region, and only secondarily to provide definitive descriptions of their structures.
Unfinished Sequences
A complicating factor in both the processing and display of a sequence is the presence of unfinished DNA sequences. These are treated by GANESH as a series of smaller sequences contained within a large BAC clone; in order not to mislead, the display has a prominent warning that these fragments are being located in the display in arbitrary order. As the appropriate genome center further finishes these sequences, GANESH updates its displays so that the number of subsequences for the BAC decreases and their length increases. Eventually, they are displayed as a continuous finished DNA sequence.
All annotations, and user annotations in particular, have to be updated as each newrelease of the sequence is assimilated. We deal with this by first performing a Smith-Waterman comparison of the new sequence with the previously stored version to identify the parts that have changed. We use crossmatch (http://www.phrap.org/phrap.docs/general.html) for this purpose. Typically, large fragments of the sequence will have remained the same. Only those parts that are neware reprocessed (subject to the sequence length requirements discussed below), thus reducing the computational effort, and more importantly, preserving user annotations as far as possible. As already mentioned in the description of the display components, users are able to compare updated sequences with the previously stored and analyzed versions.
Clearly, the problems of dealing with unfinished sequence for the human genome will reduce as more and more genome sequence is finished, but the need to deal with unfinished sequence will not disappear. Although GANESH was developed originally to support the genetic analysis of human genomic regions, it is being used for other model organisms as well (at present, for mouse), and it is expected that it will be used increasingly for other organisms, including organisms whose genomes will remain in largely unfinished draft form.
Long Sequences
In some cases, the regions that are displayed within a GANESH application are not organized around (BAC) clones, but are contained within long, continuous, merged DNA sequences. This is particularly true of those chromosomes and regions that have been the subject of concentrated finishing analyses, such as human chromosomes 21 and 22, and now increasingly more and more regions of the other chromosomes. Most of the analysis packages deployed within GANESH work optimally on sequences up to ∼250 Kb, and thus, initially the sequences are cut into 300-Kb lengths with a 50-Kb overlap. Any feature that starts within the first 270 Kb of a fragment is stored, and those that start after this position are ignored, as the next subsequence will record them. For Genscan, sequences are broken into 3-Mb fragments with 300-Kb overlaps, as this software requires a longer sequence context to operate optimally. GANESH automatically detects large sequences and displays a sliding selection box to allow the operator to select a region of the sequence for viewing in the annotation display. The position of the selection box within the sequence is displayed in the text box.
These large genomic sequences also cause some problems of focus, because not all regions are of similar interest to all users. Provision is therefore made within GANESH for the creation of custom sets of sequences. Users can pick out and register subregions and sets of subsequences as being of special interest. For the purposes of the annotation display and update notification, these custom sets are treated in the same way as normal (BAC-indexed) sequences. The user is notified of any new annotation data within their range as the parent sequences are updated, as usual. The difference is that displays of custom sets of sequence are only available to their registered users. Different users may create their own custom sets, and these regions may overlap, but they are only displayed when the registered user logs in, and then only their own custom sets are displayed.
Gene Identification Tools
An optional GANESH module is provided, which can be used to predict the presence of genes/exons. Predictions can be made by comparing evidence from the following three sources: (1) similarity to known expressed sequences, (2) in silico prediction programs such as Genscan, and (3) similarity to genomic regions of closely related organisms. The strategy adopted is to consider all lines of evidence in parallel and retain all gene/exon predictions, however low the likelihood. This is in contrast to the strategies adopted by databases of record, in which genes are only predicted if several lines of evidence are available. Our reason for adopting this approach is that in any positional cloning strategy, particularly once localization is down to the submegabase level, researchers will wish to be aware of every sequence within that region with evidence supporting transcriptional or regulatory function, less the disease-susceptibility gene be overlooked. We therefore categorize the gene predictions on the basis of the strength of evidence available, enabling the researcher to focus on the stronger candidates initially, but without eliminating other weaker candidates.
The gene predictions are subdivided into:
category 1 predictions (Ganesh-1): match a known Ensembl gene;
category 2 predictions (Ganesh-2): evidence from all three main sources of evidence: (1) similarity to known expressed sequences, (2) in silico prediction programs such as Genscan, and (3) similarity to genomic regions of closely related organisms;
category 3 predictions (Ganesh-3): evidence from two of the three lines of evidence;
category 4 predictions (Ganesh-4): evidence from a single line of evidence.
We make our gene predictions available to our collaborators by utilizing the Distributed Annotation System (DAS; Dowell et al. 2001; http://www.biodas.org). A DAS server (zebrafish.doc.ic.ac.uk:8080) was set up using the Dazzle server (http://www.biojava.org/dazzle), enabling our collaborators to view the predictions using the standard Ensembl Web-based browser with the relevant GANESH database selected as an additional DAS source. Figure 7 shows the Ensembl display of chromosome 17: 12–12.3 Mbp, highlighting the Ensembl-predicted transcripts and GANESH category 1, 2, 3, and 4 gene predictions. They are summarized in Table 1 for the first 100 Kbp.
Figure 7.
Ensembl DAS display. Ensembl display of the 12–12.3-Mbp region of human chromosome 17 highlighting the Ensembl predicted transcripts and GANESH category 1, 2, 3, and 4 gene predictions.
Table 1.
GANESH Gene Predictions for the First 100 Kbp of Human Chromosome 17:12-12.3
| Ensembl transcripts | Ganesh category 1 | Ganesh category 2 | Ganesh category 3 | Ganesh category 4 |
|---|---|---|---|---|
| ENST00000226207 | SQ:chr17 - :8844 | SQ:chr17 + :43979 | SQ:chr17 + :41289 | |
| ENST00000245503 | SQ:chr17 - :37683 | SQ:chr17 + :27635 | SQ:chr17 + :41756 | |
| SQ:chr17 + :8826 | SQ:chr17 + :55977 | |||
| SQ:chr17 + :32398 | SQ:chr17 + :50033 | |||
| SQ:chr17 + :24862 | SQ:chr17 + :57127 | |||
| SQ:chr17 + :58939 | SQ:chr17 + :43131 | |||
| SQ:chr17 + :44749 | SQ:chr17 - :42224 | |||
| SQ:chr17 + :17368 | SQ:chr17 + :54196 | |||
| SQ:chr17 + :15128 | SQ:chr17 + :51808 | |||
| SQ:chr17 + :61779 | SQ:chr17 + :48052 | |||
| SQ:chr17 + :20406 | SQ:chr17 + :65951 | |||
| SQ:chr17 + :63852 | SQ:chr17 + :56461 | |||
| SQ:chr17 + :49818 | ||||
| SQ:chr17 - :32446 | ||||
| SQ:chr17 - :20850 | ||||
| SQ:chr17 + :31403 | ||||
| SQ:chr17 + :51606 | ||||
| SQ:chr17 - :32444 | ||||
| SQ:chr17 + :48088 | ||||
| SQ:chr17 - :9840 | ||||
| SQ:chr17 - :9411 | ||||
| SQ:chr17 + :55738 | ||||
| SQ:chr17 + :2773 | ||||
| SQ:chr17 + :68464 | ||||
| SQ:chr17 + :65763 | ||||
| SQ:chr17 + :62328 | ||||
| SQ:chr17 + :66994 | ||||
| SQ:chr17 + :23916 | ||||
| SQ:chr17 + :38183 | ||||
| SQ:chr17 + :34809 | ||||
| SQ:chr17 + :72196 | ||||
| SQ:chr17 + :71100 | ||||
| SQ:chr17 + :35753 | ||||
| SQ:chr17 + :99468 |
At the time of writing, we have not completed experimental work on the predicted genes in this table beyond some preliminary microarray analysis to confirm/discard some of the gene predictions. It seems likely that a proportion of the low-category predictions represent genuine transcripts. According to a recent microarray-based validation of predicted genes on chromosome 22q (Shoemaker et al. 2001), as many as 57% of sequences predicted to be transcribed on the basis of in silico evidence alone had their expression confirmed experimentally.
Portability and Requirements
We maintain a GANESH system at Imperial College for our own users and collaborators annotating the regions detailed. The GANESH system itself is freely available for other researchers to install to annotate their own regions, and GANESH can be deployed to any region of the human genome, or to any region of another species once the parameters for the various genome analysis packages have been adjusted accordingly. The default version of GANESH requires the installation of the analysis programs and other software detailed, all of which are either open source or free to academic users. If any of the default analysis programs are not required, the system requires minimal modification to accommodate the changes. GANESH requires access to the EMBL, SWISS-PROT, and TrEMBL databases, and if these are not available locally to the installer, a Perl script is provided to build the required databases, and GANESH automatically maintains the updates. Also required are the installation of Perl, including DBD, DBI, and FTP modules, and Java 1.3. It uses the MySQL database system (although it does not rely on anything specific to that system and can be adapted easily for use with any relational database system). GANESH can be installed and run satisfactorily on a single processor linux machine, either as a standalone or a client-server system. The Java viewer can be run on any operating system that has access to the machine hosting the GANESH system.
The applet requires Netscape 6(+), or for other browsers, the installation of the Java Runtime Environment 1.3.1_02(+) and can be installed on any web server that can access the GANESH installation database.
GANESH is designed to be as straightforward to install and maintain as possible, but some knowledge of the unix/linux operating system is required. Furthermore, some knowledge of Perl would be beneficial in order to modify the scripts if required, particularly if newanalysis programs are to be added.
We have also developed a portable version of the system that allows databases to be stored and queried off-line. A laptop or notebook is sufficient to run this version. These stand-alone databases can be synchronized with the central database over the internet.
The GANESH software is available under an Open Source license. Details may be found at http://zebrafish.doc.ic.ac.uk.
DISCUSSION
Our main objective in developing GANESH was to provide a local tool for genetic analysis, but clearly, such studies can contribute to whole-genome annotation efforts. GANESH shares many features with other systems for the automated annotation of genomic sequence. The main distinguishing features of GANESH as we see them are as follows:
GANESH provides a means to set up and tailor a sequence database and annotation system for small groups with limited computational resources, or groups dealing with unusual model organisms. We see the latter point as being a significant contribution, as the major annotation systems such as Ensembl focus on a few organisms; GANESH, in contrast, provides the opportunity to annotate any organism of choice.
GANESH is designed to support the detailed analysis of relatively small, often <10–20 cM (∼10–20 Mb), regions of the genome, and to incorporate varied, and sometimes speculative, tools, external data sources, and in-house annotations and experimental data, that could not be included in the archival databases of record.
GANESH provides update mechanisms to ensure that data and annotations for the target region are as up-to-date as possible.
We have recently added facilities for importing and exporting data in the format of the Distributed Annotation System (DAS), enabling a GANESH database to be used as a component of a DAS configuration. A GANESH database of annotated human, and now also mouse, sequence can thus be accessed and viewed in two different ways as follows: (1) via the Ensembl site, using the Ensembl Web-based browser with the GANESH database selected as an additional DAS source; or (2) using the GANESH front end, either as a standalone application or as a web applet, with relevant data extracted from Ensembl and stored in GANESH as desired. The advantage of the first method is that the Ensembl browser is used widely and is increasingly familiar to researchers, and is likely to be the subject of sustained further development. It also provides an easy mechanism for disseminating results to our collaborators (as there is less for us to maintain). The advantages of the second method are that the GANESH front end is (comparatively) easy to adapt and customize, and can also be used as a stand-alone application, which some users seem to prefer. Only the second method is applicable, of course, to the analysis of genomes not presently stored in Ensembl.
Acknowledgments
The development of GANESH was supported by BBSRC/EPSRC Bioinformatics Initiative grant BIF28/10483. D.S. was supported by the EU Framework V project QLRT-CT-1999-00546. The library of Java graphics utilities used in the GANESH front-end was designed and implemented by Manuel Cardoso and Chris Iannou as part of their MSc projects in the Department of Computing, Imperial College. We thank Win Hide for many useful discussions and for his continuing support of this project.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.698103.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Bailey Jr., L.C., Searls, D.B., and Overton, G.C. 1998. Analysis of EST-driven gene annotation in human genomic sequence. Genome Res. 8: 362–376. [DOI] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S.R., Griffiths-Jones, S., Howe, K.L., Marshall, M., and Sonnhammer, E.L.L. 2002. The Pfam protein families database. Nucleic Acids Res. 30: 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge, C. and Karlin, S. 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78–94. [DOI] [PubMed] [Google Scholar]
- Clifford, R.J. and Mackey, A.J. 2000. Disperse: A simple and efficient approach to parallel database searching. Bioinformatics 16: 564–565. [DOI] [PubMed] [Google Scholar]
- Deloukas, P., Schuler, G.D., Gyapay, G., Beasley, E.M., Soderlund, C., Rodriguez-Tome, P., Hui, L., Matise, T.C., McKusick, K.B., Beckmann, J.S., et al. 1998. A physical map of 30,000 human genes. Science 282: 744–746. [DOI] [PubMed] [Google Scholar]
- Dowell, R.D., Jokerst, R.M., Day, A., Eddy, S.R., and Stein, L. 2001. The distributed annotation system. Bioinformatics 2: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenesch, J.B., Ching, K.A., Batalov, S., Su, A.I., Walker, J.R., Zhou, Y., Kay, S.A., Schultz, P.G., and Cooke, M.P. 2001. A comparison of the Celera and Ensembl predicted gene sets reveals little overlap in novel genes. Cell 106: 413–415. [DOI] [PubMed] [Google Scholar]
- Hubbard, T., Barker, D., Birney, E., Cameron, G., Chen, Y., Clark, L., Cox, T., Cuff, J., Curwen, V., Down, T., et al. 2002. The Ensembl genome database project. Nucleic Acids Res. 30: 38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, M.R., Richard III, C.W., Schott, J.J., Yousry, C., Clark, K., Bell, J., Terwilliger, J.D., Hazan, J., Dubay, C., Vignal, A., et al. 1994. A radiation hybrid map of 506 STS markers spanning human chromosome 11. Nat. Genet. 8: 70–76. [DOI] [PubMed] [Google Scholar]
- Kent, W.J. and Haussler, D. 2001. Assembly of the working draft of the human genome with gigassembler. Genome Res. 11: 1541–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., and Haussler, A.D. 2002. The Human genome browser at UCSC. Genome Res. 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., and FitzHugh, W. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Marra, M.A., Kucaba, T.A., Dietrich, N.L., Green, E.D., Brownstein, B., Wilson, R.K., McDonald, K.M., Hillier, L.W., McPherson, J.D., and Waterston, R.H. 1997. High throughput fingerprint analysis of large-insert clones. Genome Res. 7: 1072–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson, J.D., Marra, M., Hillier, L., Waterston, R.H., Chinwalla, A., Wallis, J., Sekhon, M., Wylie, K., Mardis, E.R., Wilson, R.K., et al. 2001. A physical map of the human genome. Nature 409: 934–941. [DOI] [PubMed] [Google Scholar]
- Shoemaker, D.D., Schadt, E.E., Armour, C.D., He, Y.D., Garrett-Engele, P., McDonagh, P.D., Loerch, P.M., Leonardson, A., Lum, P.Y., Cavet, G., et al. 2001. Experimental annotation of the human genome using microarray technology. Nature 409: 922–927. [DOI] [PubMed] [Google Scholar]
- Stein, L. 2001. Genome annotation: From sequence to biology. Nat. Rev. Genet. 2: 493–503. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304–1351. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.sanger.ac.uk/Software/Wise2; Birney, E., Wise2.
- http://www.doc.ic.ac.uk/∼rc5/Disperse; Clifford, R.J., Mackey, A.J., Disperse.
- http://www.biodas.org; Distributed Annotation System (DAS) Home page.
- http://www.biojava.org/dazzle; Down, T. The Dazzle server.
- http://www.ensembl.org; Ensembl Home page.
- ftp.ebi.ac.uk/pub/databases/embl/new; European Bioinformatics Institute. EMBL daily update files.
- http://zebrafish.doc.ic.ac.uk; GANESH Home page.
- http://www.phrap.org/phrap.docs/general.html; Green, P. Crossmatch documentation.
- http://www.mysql.com; MySQL Home page.
- http://www.neomorphic.com; Neomorphic Home page.
- ftp.sanger.ac.uk/pub/human/sequences and ftp.sanger.ac.uk/pub/mouse/sequences; Sanger Institute ftp archives.
- http://ftp.genome.washington.edu/RM/RepeatMasker.html; Smit, A.F.A., Green, P. RepeatMasker documentation.
- http://www.acedb.org; AceDB home page.