Abstract
Purpose
Identification of single nucleotide polymorphisms (SNPs) associated with development of advanced colorectal adenomas.
Experimental Design
Discovery Phase: 1,406 Caucasian patients (139 advanced adenoma cases and 1,267 controls) from the Adenoma Prevention with Celecoxib (APC) trial were included in a genome-wide association study (GWAS) to identify variants associated with post-polypectomy disease recurrence. Genome-wide significance was defined as false discovery rate < 0.05, unadjusted p=7.4×10−7. Validation Phase: Results were further evaluated using 4,175 familial colorectal adenoma or CRC cases and 5,036 controls from patients of European ancestry (COloRectal Gene Identification consortium, Scotland, Australia and VQ58).
Results
Our study identified eight SNPs associated with advanced adenoma risk in the APC trial (rs2837156, rs7278863, rs2837237, rs2837241, rs2837254, rs741864 at 21q22.2, and rs1381392 and rs17651822 at 3p24.1, at p<10–7 level with odds ratio – OR>2). Five variants in strong pairwise linkage disequilbrium (rs7278863, rs2837237, rs741864, rs741864 and rs2837241, r2=0.8–1) are in or near the coding region for the tight junction adhesion protein, IGSF5. An additional variant associated with advanced adenomas, rs1535989 (minor allele frequency 0.11; OR 2.09; 95% confidence interval 1.50–2.91), also predicted CRC development in a validation analysis (p=0.019) using a series of adenoma cases or CRC (CORGI study) and 3 sets of CRC cases and controls (Scotland, VQ58 and Australia, N=9,211).
Conclusions
Our results suggest that common polymorphisms contribute to the risk of developing advanced adenomas and might also contribute to the risk of developing CRC. The variant at rs1535989 may identify patients whose risk for neoplasia warrants increased colonoscopic surveillance.
Keywords: Colorectal adenomas, colorectal cancer screening, genetic predisposition
INTRODUCTION
Colorectal cancer (CRC) is a common malignancy, with a prevalence in developed nations of 40–50 cases per 100,000 individuals (1). Approximately one third of those diagnosed with CRC will die of their disease due to diagnosis at a stage not curable by locoregional therapy. Most CRC cases arise from premalignant adenomas that require years or even decades to progress to invasive disease. Colonoscopy to identify and remove precursor adenomas has been recommended for more than 25 years for patients at high CRC risk, and recently completed long-term analyses of screened cohorts confirmed the utility of adenoma removal for preventing deaths due to CRC (2).
Our goal is to understand the biology of CRC in order to develop effective prevention and therapy, and also to characterize individual risk in a manner that will identify patients most likely to benefit from colonoscopy to detect and remove premalignant adenomas. Identification of germline variants conveying an increased risk of CRC could be used to promote adherence to colonoscopy and polypectomy for patients at highest risk, improving utilization and cost-benefit of this life-saving procedure. In addition, more accurate characterization of high risk individuals would facilitate participation in prevention clinical trials. Finally, therapy to treat or prevent CRC would be advanced if the biological consequences of germline susceptibility variants were further characterized to uncover the molecular basis of CRC.
The adenoma-carcinoma sequence in the colorectum represents a disease spectrum. Adenomas with a low risk of cancer development are small (< 0.6 cm diameter) and lack histological features associated with progression, such as the presence of villous features or high grade dysplasia. The identification of an advanced adenoma (size ≥ 1 cm, villous or tubulovillous histology, high grade dysplasia) indicates that a patient has a higher risk of future adenoma and CRC development (3). Advanced adenomas, therefore, are the most important lesions to target for CRC prevention. In this study, we used a large cohort of adenoma patients from a prospective randomized clinical trial to identify SNPs associated with increased risk of developing advanced adenomas. These variants were then further tested using large genotyped cohorts of patients and controls with advanced adenomas and CRC. In doing so, we identified variants associated with both advanced pre-malignant lesions and CRC.
METHODS
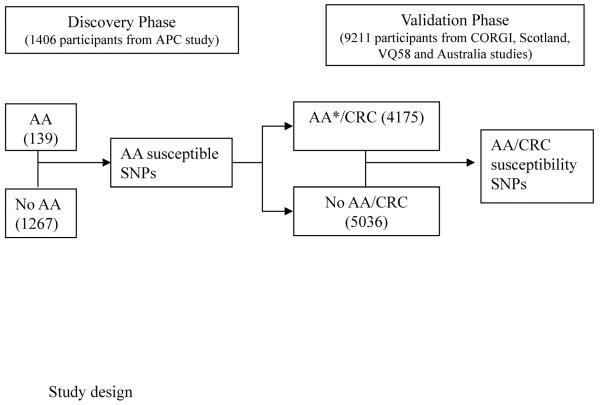
Study design and populations (Figure 1)
Figure 1.
AA: advanced adenoma *Including subjects with familial adenoma from CORGI
Discovery Phase
1,406 evaluable Caucasian patients were identified from the APC trial, a randomized, placebo-controlled study to test whether celecoxib reduced the occurrence of endoscopically detected colorectal adenomas. The endpoint advanced adenoma was defined as any adenoma with size ≥1cm, villous/tubulovillous histology, or high grade dysplasia. During the prospective follow-up period, 139 participants developed advanced adenomas identified during a scheduled colonoscopy screening exam. Detailed information regarding the trial design and primary outcomes was reported elsewhere (4).
Validation Phase
The advanced adenoma susceptibility SNPs identified from APC trial were further evaluated using GWAS data from the following four non-overlapping colorectal cancer case-control series of European ancestry (5).
-
(1)
CORGI: 931 familial colorectal adenoma or colorectal cancer cases and 929 cancer-free controls of white British origin ascertained through the COloRectal Gene Identification (CORGI) consortium. All cases had at least one first-degree relative with colorectal tumors and no mutations in the known highly-penetrant CRC genes. Controls were spouses or partners of the cases and had no personal history of CRC(6).
-
(2)
Scotland: 1003 early-onset Scottish CRC cases (<55 years) and 979 cancer-free Scottish population controls. Known Mendelian syndromes were excluded. Controls were matched by age (± 5 years), gender and area of residence(6).
-
(3)
VQ58: 1,800 British Stage II/III CRC patients from the VICTOR (N=923) and QUASAR2 (http://www.octo-oxford.org.uk/alltrials/trials/q2.html, N=877) clinical trials, together with publicly available data from 2,690 population controls from the Wellcome Trust Case Control Consortium (WTCCC) 1958 Birth Cohort (7).
-
(4)
Australia: 441 CRC cases treated in the Royal Melbourne, Western and St Francis Xavier Cabrini Hospitals in Melbourne and 438 population controls from Brisbane Twin Nevus and Genes in Myopia studies, matched to the cases using principal component analysis(6).
Thus, 4,175 familial colorectal adenoma or CRC cases and 5,036 controls were included in the validation analysis. Human Subjects Committee approval to collect and genotype whole blood samples was obtained by Brigham and Women's Hospital and the RIKEN Center for Genomic Medicine.
Genotyping and quality control
DNA was isolated from blood samples using standard methods and quantified with picogreen. For the APC cohort, genotyping was performed by the RIKEN Center for Genomic Medicine using the Illumina Human610-Quad BeadChip platform (Illumina, San Diego, CA). A white parent-child CEPH trio from the HapMap was used to check for Mendelian transmission of alleles. Chi-square test based on genotype frequencies at each SNP was used to test for deviations from Hardy-Weinberg equilibrium (HWE). Any SNP with HWE p<0.001 was excluded. Two cases and two controls were randomly chosen as duplicates for quality control (QC) of genotype concordance. A total of 28 subjects and 1,792 markers were excluded for quality control reasons, including duplicates, those that showed identity-by-descent >12.5% or were gender mismatched, samples with <98% and markers with <99% call rate or heretozygous haploids. The final Manhattan plot and QQ plot indicated the satisfactory QC process (supplement Figure 1 and 2).
For the additional susceptibility evaluation cohorts, samples were genotyped on Illumina Infinium SNP arrays, ranging from the Hap300 (for VQ58) to the Hap1M (for Australia). Details concerning genotyping and quality control for these studies have been provided previously (5). Ethics Committees approved these five studies and samples were collected in accordance with the tenets of the Declaration of Helsinki.
Among the top 19 SNPs identified from the APC trial, 12 SNPs had genotype data available from the CORGI, Scotland, VQ58 or Australia GWA studies. Nine of these SNPs were typed in all four studies (rs1381392, rs17651822, rs17781398, rs16909065, rs9582985, rs2837156, rs2837241, rs741864) and three were typed in three (rs13085889, rs1424593 and rs2837237, Supplemental Table 1).
Statistical methods
To assess the strength of association between genotype and advanced adenoma risk, a per allele unconditional logistic regression model was used to estimate odd ratios (ORs) and their corresponding 95% confidence intervals (CIs). For the APC trial, genotype-phenotype interactions were evaluated for sex, age at trial entry (≤ age 60 years vs. > age 60 years), and family history (first-degree relatives with colorectal cancer). Genotype-environment interactions were evaluated for aspirin use at baseline and treatment with celecoxib. The Breslow-Day test was used to test the homogeneity of odds ratios. PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/), STATA and SAS were used to conduct all the analysis. Genome-wide significance was defined as false discovery rate (FDR) < 0.05, which corresponds to an unadjusted p=7.4×10−7 in this analysis(8, 9).
RESULTS
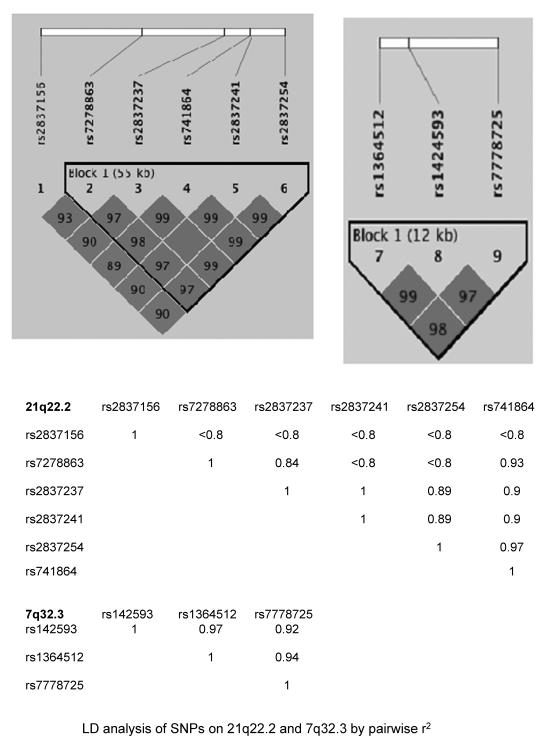
1,406 evaluable Caucasian patients were genotyped from the APC trial (4). Eight SNPs were identified by association with on-study development of advanced adenomas at a genome-wide level of significance: rs2837156, rs7278863, rs2837237, rs2837241, rs2837254, rs741864, all at 21q22.2, and rs1381392 and rs17651822, both at 3p24.1 (Table 1). The associations between the 6 SNPs in the 21q22.2 region and advanced adenoma development were all highly significant (unadjusted p=10−8–10−9) with ORs per allele ranging from 2.22 to 2.55. All 6 SNPs in the 21q22.2 region were located near the coding region for the adherens junction protein, IGSF5, and five of these SNPs (rs7278863, rs2837237, rs741864, rs741864 and rs2837241) were in strong linkage disequilibrium (r2=0.8–1, Figure 1). For the 3p24.1 signal, the OR for genotype rs1381392 was 2.01 (95% CI 1.52–2.65, unadjusted p=7.4×10−07), and that for rs17651822 was 2.16 (95% CI 1.61–2.91, unadjusted p=2.1×10−07).
Table 1.
APC trial advanced adenoma susceptibility loci*
| Chromosome Region | SNP | Position (BP) | Alleles | MAF | p value | OR | Gene | |
|---|---|---|---|---|---|---|---|---|
| 2p24.2 | rs11886781 | 18154780 | A | C | 0.08 | 9.7E-06 | 2.25(1.56,3.25) | KCNS3 EPHB1, |
| 3q22.2 | rs13085889 | 135843760 | A | C | 0.29 | 8.8E-06 | 1.77(1.37,2.29) | KY |
| 3p24.1 | rs1381392 | 28724318 | A | G | 0.18 | 7.4E-07 | 2.01(1.52,2.65) | |
| 3p24.1 | rs17651822 | 28695130 | A | G | 0.14 | 2.1E-07 | 2.16(1.61,2.91) | |
| 7p14.3 | rs17781398 | 30807966 | A | G | 0.10 | 9.0E-06 | 0.19(0.08,0.43) | FAM188b |
| 7q32.3 | rs1424593 | 131605541 | C | A | 0.50 | 9.1E-06 | 0.56 (0.44, 0.73) | PLXNA4 |
| 7q32.3 | rs1364512 | 131602384 | C | A | 0.49 | 8.6E-06 | 0.56(0.42,0.71) | PLXNA4 |
| 7q32.3 | rs7778725 | 131614936 | G | A | 0.49 | 4.0E-06 | 0.55(0.42,0.71) | PLXNA4 |
| 9q33.2 | rs16909065 | 121597606 | A | G | 0.05 | 3.6E-06 | 2.59(1.71,3.93) | |
| 9q33.2 | rs16909036 | 121587049 | G | A | 0.05 | 3.7E-06 | 2.59(1.71,3.93) | |
| 13q33.2 | rs1535989 | 104820723 | G | A | 0.11 | 8.9E-06 | 2.09(1.50,2.91) | |
| 13q33.2 | rs17654765 | 104828038 | A | G | 0.10 | 4.7E-06 | 2.14(1.53,2.98) | |
| 13q33.2 | rs9582985 | 104829133 | C | A | 0.11 | 9.3E-06 | 2.05(1.48,2.83) | |
| 21q22.2 | rs2837156 | 40048557 | G | A | 0.12 | 3.2E-07 | 2.22(1.62,3.03) | IGSF5 |
| 21q22.2 | rs7278863 | 40087578 | A | G | 0.10 | 1.4E-08 | 2.48(1.80,3.42) | IGSF5 |
| 21q22.2 | rs2837237 | 40119727 | G | A | 0.12 | 3.6E-09 | 2.48(1.82,3.38) | |
| 21q22.2 | rs2837241 | 40130476 | A | C | 0.12 | 3.7E-09 | 2.48(1.82,3.38) | |
| 21q22.2 | rs2837254 | 40143171 | A | G | 0.11 | 2.9E-09 | 2.55(1.86,3.51) | |
| 21q22.2 | rs741864 | 40129665 | A | G | 0.11 | 1.1E-08 | 2.48(1.80,3.41) | |
Total number of subjects is 1,406, of which 139 developed advanced adenomas
Eleven SNPs (rs11886781 at 2p24.2, rs13085889 at 3q22.2, rs1424593, rs1364512 and rs7778725 at 7q32.3, rs16909065 and rs16909036 at 9q33.2, rs17654765, rs1535989 and rs9582985 at 13q33.2) were associated with moderate (~2-fold) ORs for advanced adenoma detection, but the associations did not reach genome-wide significance (p ≤ 10−6). Of these 11 SNPs, 6 mapped to gene coding regions: rs11886781 to KCNS3, rs17781398 to FAM188b, rs13085889 to EPHB1 and KY, and rs1424593, rs1364512 and rs7778725 all to PLXNA4 (Table 1 and Figure 1).
There are no comparable adenoma chemoprevention cohorts currently available for validation of the APC GWAS results. We therefore further examined APC trial results using GWAS data from four non-overlapping CRC case-control series of European ancestry, one of which (CORGI) also included advanced adenoma cases (5). Among the 19 advanced adenoma risk SNPs with a nominal significance level of p≤10−6, 12 were genotyped in at least 3 of the available four CRC GWA studies (Supplemental Table 1). Allelic frequencies of each variant and the corresponding associations with CRC phenotype were accessed in each of the 4 case-control samples. The results of the meta-analysis for overall associations with CRC risk are reported in Table 2 and supplement figure 3.
Table 2.
Meta-analysis using adenoma or CRC as a composite outcome
| SNP | N | P | P(R) | OR | OR(R) | Q | I |
|---|---|---|---|---|---|---|---|
| rs13085889 | 4 | 0.1048 | 0.1048 | 1.0575 | 1.0575 | 0.5438 | 0 |
| rs1381392 | 4 | 0.4636 | 0.4636 | 1.0295 | 1.0295 | 0.4186 | 0 |
| rs1424593 | 3 | 0.405 | 0.405 | 1.027 | 1.027 | 0.4233 | 0 |
| rs1535989 | 4 | 0.012 | 0.012 | 1.1304 | 1.1304 | 0.7925 | 0 |
| rs 16909065 | 4 | 0.1273 | 0.1273 | 0.9054 | 0.9054 | 0.498 | 0 |
| rs17651822 | 4 | 0.689 | 0.689 | 1.0176 | 1.0176 | 0.8499 | 0 |
| rs17781398 | 4 | 0.7972 | 0.7972 | 1.016 | 1.016 | 0.9487 | 0 |
| rs2837156 | 4 | 0.8017 | 0.8017 | 0.9881 | 0.9881 | 0.4326 | 0 |
| rs2837210 | 4 | 0.8706 | 0.9766 | 1.0083 | 0.9983 | 0.3114 | 16.05 |
| rs2837237 | 3 | 0.3464 | 0.3464 | 0.9379 | 0.9379 | 0.413 | 0 |
| rs2837241 | 4 | 0.938 | 0.8585 | 0.9963 | 0.9907 | 0.3395 | 10.69 |
| rs741864 | 4 | 0.6248 | 0.6248 | 0.971 | 0.971 | 0.6068 | 0 |
| rs9582985 | 4 | 0.05468 | 0.05468 | 1.1188 | 1.1188 | 0.9416 | 0 |
One of the 19 SNPs identified in the APC trial, rs1535989, was replicated in the independent CRC cohorts, with an OR for CRC development of 1.12 (95% CI 1.019–1.23, p=0.019). There was no evidence of inter-study heterogeneity (Phet =0.71; I2=0.0%). An additional exploratory meta-analysis was performed, combining all five studies and using either advanced adenoma or CRC as the outcome (Table 2). SNP rs9582985 originally identified in the APC cohort showed marginally significant association with outcome (OR=1.11, p=0.055).
Clinical data from the APC trial was used to further characterize rs1535989 by examining the association of this variant with other susceptibility factors for advanced colorectal neoplasia, including age, sex, aspirin use at baseline, family history of CRC and on-study treatment with celecoxib. SNP and environmental factors interaction terms were included in the model. SNP rs1535989 showed statistically significant interactions with subjects' age (p=0.0016), sex (p=0.0057) and aspirin use at baseline (p=0.02). The associations with advanced neoplasia were stronger in older individuals (>60, OR 3.20; 95% CI 2.10–4.87), males (OR 2.74; 95% CI 1.89–3.97), and those using aspirin at baseline (OR=3.63; 95% CI 2.06–6.40) (Table 3). There were no statistically significant interactions with CRC family history or on-study treatment with celecoxib.
Table 3.
Genotype-phenotype/environment interactions for SNP rs1535989
| Phenotype | OR | Interaction (p value) | |
|---|---|---|---|
| Age | ≤60 | 0.91(0.42, 1.76) | |
| >60 | 3.20(2.10,4.87) | 0.0016 | |
| Sex | Female | 0.65(0.25, 1.68) | |
| Male | 2.74(1.89,3.97) | 0.0057 | |
| Family History | No | 2.23(1.51,3.28) | |
| Yes | 1.74(0.88,3.43) | 0.53 | |
| Prior Aspirin | No | 1.58(1.03,2.42) | |
| Use | Yes | 3.63(2.06, 6.40) | 0.02 |
| Placebo | 1.63(1.03,2.58) | ||
| Treatment | 200mg | 2.51(1.27,4.95) | |
| 400mg | 2.43(1.16,5.07) | NS | |
DISCUSSION
Among the approximately 145,000 CRC cases diagnosed per year in the United States, only 5% represent autosomal dominant predisposition syndromes, with the majority of these involving either hereditary nonpolyposis colon cancer (HNPCC) or familial adenomatous polyposis (FAP). An additional 20–25% of CRC cases show a familial association without precise genetic characterization, and the majority of CRCs occur in individuals without a family history of the disease. Self-reported family history does not accurately assess the inherited risk of advanced adenomas because patients' knowledge of their family history of colorectal adenomas is often unknown or incomplete (10). Recent GWAS from members of this collaboration have identified 18 CRC susceptibility variants with minor allele frequencies ranging from 0.07 to 0.48 that each convey a small degree of risk modification (OR per allele: 0.87–1.35) (11–15). The results presented here expand these data to address inherited susceptibility for developing advanced adenomas that represent targets for CRC prevention. In addition to the studies whose data were used here, there have been a number of other GWAS with CRC or colorectal adenomas as the primary phenotype (16–20). These have yielded a substantial number of possible susceptibility variants, most conveying modestly altered risk. A recent case-control meta-analysis from 14 studies identified SNPs on 2q32.3 (rs11903757), 1q25.3 (rs10911251), 12p13.32 (rs3217810), and 12q24.21 (rs59336) that represented odds ratios ranging from 0.84–1.15 (16).
The APC trial was designed to determine whether the selective cyclooxygenase-2 inhibitor, celecoxib, prevented adenomas in patients at high risk for CRC. Eligibility criteria required that participants have at least one prior adenoma > 6mm in size, or multiple adenomas. During the 5 years of endoscopic surveillance, 21.3% of APC trial participants randomized to placebo developed recurrent advanced adenomas (21). This rate was decreased to 12.5% in patients receiving celecoxib 200 mg twice daily (p<0.0001), however concerns over cardiovascular toxicity currently prohibit the use of celecoxib for routine CRC chemoprevention (22). Results presented here showed that advanced adenomas were twice as likely to occur in APC trial participants with variant rs1535989, and that this increased risk was not affected by celecoxib treatment. For males or older individuals, the risk was more than 3-fold higher than that for females or participants <age 60. The observed interaction between baseline aspirin use and advanced adenoma risk is particularly interesting. Aspirin use reduces the incidence of colorectal adenomas and CRC, and subjects enrolled in the APC trial who used aspirin at baseline were those who developed adenomas despite aspirin use. These individuals may therefore have constituted a higher risk subset because they were relatively resistant to aspirin chemoprevention. The analyses conducted here showed that APC trial participants who both developed adenomas while taking aspirin and had variant rs1535989 demonstrated a 3.63-fold increase in advanced adenoma risk during surveillance. If this association can be confirmed in other studies, and other variants of similar effect found, then genotyping will represent a useful method to target high risk patients for preventive treatments including more frequent colonoscopic screening.
Additional results from this GWAS suggest areas for further research concerning the molecular basis of colorectal neoplasia. Variants at rs2837156, rs7278863, rs2837237, rs2837241, rs2837254 and rs741864 are in close association at 21q22.2. Two of these SNPs, rs2837156 and rs7278863, are within the coding region of IGSF5, a gene encoding a transmembrane protein whose murine homologue, JAM4, binds to the tumor suppressor, MAGI-1 at intestinal epithelial tight junctions (23). To explore the potential effect of IGSF5 on the prognosis of CRC patients, microarray expression data from the GEO data set (GSE14333) were retrieved for 229 Dukes A, B, and C patients (24). The gene expression profile was performed with Affymetrix u133p2 platform. Our preliminary analysis of these data indicated that the overexpression of IGSF5 is associated with significantly worse relapse-free survival (unadjusted p=0.000004, bonferroni adjusted p=0.00085, supplement figure 4). In addition, rs1424593, rs1364512 and rs7778725 all involve PLXNA4, a member of the plexin family located on chromosome 7. Plexins are transmembrane, secreted, and GPI-anchored semaphorins that modulate the adhesive and migratory properties of malignant cells. The protein product of PLXNA4 forms stable complexes with FGFR1 and VEGFR-2 tyrosine kinase receptors and enhances both VEGF-induced VEGFR-2 phosphorylation and βFGF-induced cell proliferation (25). Finally, EPHB1 encodes a ligand that binds to an Eph receptor tyrosine kinase to mediate bidirectional signaling required for intestinal epithelial homeostasis (26). EphB-ephrin B interactions regulate cell adhesion, migration and positioning, and play an important role in colorectal tumor progression (27).
The limitation of our current study resides in the following two aspects. The limited number of advanced adenoma cases in the APC trial restricted the power to identify more advanced adenoma susceptibility SNPs. In addition, the CRC/adenoma cases with a strong family history of CRC were not excluded from replication datasets. This might under/overestimate the association between identified SNPs and sporadic CRC risks.
In summary, this study identified 19 SNPs associated with advanced adenoma risk at a level of p≤10−6. Of these, 12 SNPs were tested in a meta-analysis using independent datasets to evaluate their association with CRC development, and rs1535989 was also associated with increased risk of both advanced adenomas and CRC. In addition, eight of the variants identified in the APC trial mapped to coding regions of genes previously implicated in CRC progression, and warrant further study to confirm their role in modifying tissue-specific biological function.
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVANCE.
Identification of patients at highest risk of colorectal cancer is essential for providing optimal disease screening and prevention. This study uncovers germline susceptibility loci that indicate risk of disease and potential for improved understanding of disease biology.
Figure 2.
The 6 IGSF-5 related SNPs are within very tight LD region
ACKNOWLEDGEMENTS
We are grateful to all of the individuals who participated in the various studies. The APC study was funded by the US National Cancer Institute (CA-N01-95015 and HHSN261201000082C) and by Pfizer. Cancer Research UK funded the CORGI, Scotland and NSCCG studies. LGC-C is supported by the EU FP7 CHIBCHA project. IT received support from the Oxford NIHR Comprehensive Biomedical Research Centre. Core infrastructure support to the Wellcome Trust Centre for Human Genetics, Oxford was provided by grant 090532/Z/09/Z. The UK National Cancer Research Network supported the NSCCG. MD received from from the Medical Research Council (G0000657-53203), CORE and Scottish Executive Chief Scientist's Office (K/OPR/2/2/D333, CZB/4/449). This study made use of genotyping data on the 1958 Birth Cohort and NBS samples, kindly made available by the Investigators of those studies and the Wellcome Trust Case-Control Consortium 2; a full list of the investigators who contributed to the generation of the data is available from http://www.wtccc.org.uk/. The Colon Cancer Family Registry was supported by the National Cancer Institute, National Institutes of Health under Request for Application #CA-95-011, and through cooperative agreements with the Australian Colorectal Cancer Family Registry (UO1 CA097735), the USC Familial Colorectal Neoplasia Collaborative Group (UO1 CA074799), Mayo Clinic Cooperative Family Registry for Colon Cancer Studies (UO1 CA074800), Ontario Registry for Studies of Familial Colorectal Cancer (UO1 CA074783), Seattle Colorectal Cancer Family Registry (UO1 CA074794), and The University of Hawaii Colorectal Cancer Family Registry (UO1 CA074806).
The CORGI acknowledgments and investigators: The CORGI study comprised: Huw Thomas. Family Cancer Clinic, St Mark's Hospital and Imperial College, London; Eamonn Maher, Dept. of Clinical Genetics, University of Birmingham, UK; Gareth Evans, Dept. of Clinical Genetics, University of Manchester, UK; Lisa Walker and Dorothy Halliday, Oxford Regional Genetics Service, Churchill Hospital, Oxford, UK; Anneke Lucassen, Wessex Regional Genetics Service, Princess Anne Hospital, Southampton, UK; Joan Paterson, Anglia Regional Genetics Service, Addenbrooke's Hospital, Cambridge, UK; Shirley Hodgson and Tessa Homfray, South-West Thames Regional Genetics Service, St George's Hospital, Tooting, London, UK; Lucy Side, North-East Thames Regional Genetics Service, Great Ormond Street Hospital, London, UK; Louise Izatt, South-East Thames Regional Genetics Service, Guy's Hospital, London, UK; Alan Donaldson and Susan Tomkins, South-West Regional Genetics Service, Bristol, UK; Patrick Morrison, Northern Ireland Regional Genetics Service, City Hospital, Belfast, UK; Carole Brewer, South-West Regional Genetics Service, Royal Devon and Exeter Hospital, Exeter, UK; Alex Henderson, Northern Regional Genetics Service, International Centre for Life, Newcastle, UK; Rosemarie Davidson and Victoria Murday, West of Scotland Regional Genetics Service, Yorkhill Hospital, Glasgow, UK; Jaqueline Cook, Sheffield Regional Genetics Service, Children's Hospital, Sheffield, UK; Neva Haites, North of Scotland Regional Genetics Service, Foresterhill Hospital, Aberdeen, UK; Timothy Bishop and Eamonn Sheridan, Yorkshire Regional Genetics Service, St James's Hospital, Leeds, UK; Andrew Green, Republic of Ireland Genetics Service, Our Lady's Hospital for Sick Children, Dublin, Republic of Ireland; Christopher Marks, Sue Carpenter and Mary Broughton, The Royal Surrey County Hospital, Egerton Road, Guildford, Surrey; Lynn Greenhalge, Department of Clinical Genetics, Royal Liverpool Children's Hospital, Eaton Road, Alder Hay, Liverpool; and Mohnish Suri,Department of Clinical Genetics, City Hospital, Hucknall Road, Nottingham.
APC acknowledgments and investigators: The following persons participated in the APC Study:Steering Committee:M.M. Bertagnolli, E.T., Hawk, C.J. Eagle; Statistical Team: A.G. Zauber, K.M. Kim, D. Corle, R. Rosenstein, J. Tang, T. Hess, A. Wilton; Medical Monitors: W. Anderson, L. Doody; Central Pathology Review: M. Redston; Project Directors: G.M. Woloj, D. Bagheri, A. Crawford, M. Schietrum, V. Ladouceur; Data and Safety Monitoring Board: S. Rosen (chair), L. Friedman, R. Makuch, R. Phillips, P. Taylor; Principal Investigators, United States: S. Auerbach (California Professional Research, Newport Beach), C.F. Barish (Wake Research Associates, Raleigh, NC), T. Barringer (Carolinas Medical Center, Charlotte, NC), R.W. Bennetts (Northwest Gastroenterology Clinic, Portland, OR), M. Blitstein (Associates in Gastroenterology and Liver Disease, Lake Forest, IL), J. Bruggen (Wake Forest University Baptist Medical Center, Winston Salem, NC), P Carricaburu (Veterans Affairs Hospital, Sheridan, WY), D. Chung (Massachusetts General Hospital, Boston, MA), F. Colizzo (Pentucket Medical Associates, Haverhill, MA), R. Curtis (Newton-Wellesley Hospital, Newton, MA), T. Dewar (Harris Methodist Hospital Fort Worth, Ft. Worth, TX), R. DuBois (Vanderbilt University Medical Center, Nashville, TN),T. Feinstat (Gastroenterology Consultants of Sacramento, Roseville, CA), T.R. Foley (Regional Gastroenterology Associates of Lancaster, Lancaster, PA, D. Gabbaizadeh (Huntington Research Group, Huntington Station, NY), J. Geenen (Wisconsin Center for Advanced Research, Milwaukee, WI), F. Giardiello (Johns Hopkins Hospital, Baltimore, MD), A. Goetsch (nTouch Research, Huntsville, AL), M. Goldberg (Regional Gastroenterology Associates of Lancaster, Evanston, IL), J.L. Goldstein (University of Illinois at Chicago, Chicago, IL), W. Harlan, III (Asheville Gastroenterology Associates, Asheville, NC), R. Hogan (Gastrointestinal Associates, Jackson, MS), M. Kamionkowski (Gastroenterology Associates of Cleveland, Mayfield Heights, OH), M. Kelfer (Fallon Clinic, West Boylston, MA), B. Kerzner (Health Trends Research, Baltimore, MD), K. Kim (University of Chicago Medical Center, Chicago, IL), I. Klimberg (Gastroenterology Associates of Ocala, Ocala, FL), G. Koval (West Hills Gastroenterology Associates, Portland, OR), C. Krone (Advanced Clinical Therapeutics, Tucson, AZ), S.Krumholz (Waterside Clinical Research, West Palm Beach, FL), M.W. Layton (South Puget Sound Clinical Research Center, Olympia, WA), C. Lightdale (Columbia-Presbyterian Medical Center, New York, NY), P.J. Limburg (May Clinic, Rochester, MN), C. Lind (Vanderbilt University Medical Center, Nashville, TN), D. Lipkis (Institute for Health Care Assessment, San Diego, CA), M. Lloyd (Idaho Gastroenterology, Meridian, ID), D. Maccini (Spokane Digestive Disease Center, Spokane, WA), F. MacMillan, Sr. (Pentucket Medical Associates, Haverhill, MA), R. Madoff (University of Minnesota, Minneapolis, MN), A. Malik (Advanced Clinical Research, North Providence, RI), A. Markowitz (Memorial Sloane-Kettering Cancer Center, New York, NY), R. Marks (Alabama Digestive Research Center, Alabaster, AL), C. J. McDougall (Manhattan Associates, New York, NY), P. Miner (Oklahoma Foundation for Digestive Research, Oklahoma City, OK), M. Murphy (Southern Digestive and Liver Disease Institute, Savannah, GA), A. Namais (Gastrointestinal Physicians, Salem, MA), N. Nickl (University of Kentucky Medical Center, Lexington, KY), M. Pochapin (Jay Monahan Center for Gastrointestinal Health, New York, NY), R.E. Pruitt (Nashville Medical Research Institute, Nashville, TN), J Puolos (Cumberland Research Associates, Fayetteville, NC), D.S. Riff (AGMG Clinical Research, Anaheim, CA), R. Roman (South Denver Gastroenterology, Englewood, CO), L. Rubin (New Jersey Physicians, Passaic, NJ), D. Ruff (Healthcare Discoveries, San Antonio, TX), M. Safdi (Consultants for Clinical Research, Cincinnati, OH), J. Saltzman (Brigham and Women's Hospital, Boston, MA), B. Salzberg (Atlanta Gastroenterology Associates, Atlanta, GA), J.A. Sattler (Western Clinical Research, Torrence, CA), P. Schleinitz (Americas Doctors Research, Medford, OR), J. Schwartz (Northwest Gastroenterologists, Arlington Heights, IL), M. Schwartz (Jupiter Research Association, Jupiter, FL), M. Silpa (Gastroenterology Associates of the East Bay Medical Group, Berkeley, CA), D. Silvers (Drug Research Services, Metairie, LA), D. Smoot (Howard University Cancer Center, Washington, DC), S. Sontag (Veterans Affairs Medical Center, Hines, IL), R.J. Sorrell (Gastroenterology Specialties, Lincoln, NE), D. Stanton (Community Clinical Trials, Orange, CA), J. Sturgeon (Americas Doctors Research, Shawnee Mission, KS), J.P. Tracey (Hawthorne Medical Associates, North Dartmouth, MA), T. Werth (CharlotteGastroenterology and Hepatology, Charlotte, NC), C.M. Wilcox (University of Alabama at Birmingham, Birmingham, AL), R. Wohlman (Northwest Gastroenterology Associates, Bellevue, WA), S. Woods (Gastroenterology Associates of Fairfield County, Bridgeport, CT); United Kingdom: J. Burn (South Cleveland Hospital, Middlesbrough); Australia: H. Ee (Sir Charles Gairdner Hospital, Nedlands, W.A.), M. Korman (Monash Medical Centre, Clayton, Victoria), A. Lee (Concord Repatriation and General Hospital, Concord, NSW), B. Leggett (Royal Brisbane Hospital, Herston, Queensland), F. Macrae (Royal Melbourne Hospital, Melbourne, Victoria), L. Mollison (Freemantle Hospital, Freemantle, WA), N. Yeomans (Western Hospital, Footscray, Victoria), G. Young (Flinders Medical Center, Bedford, SA); Canada: G. Aumais (Hospital Maisonneuve-Rosemont, Montreal), R. Bailey (Hys Medical Center, Edmonton, Alberta), C. Bernstein (Winnipeg Health Sciences Centre, Winnipeg, Manitoba), L. Cohen (Sunnybrook and Women's Hospital, Toronto), C. Dallaire, R. Dube (Centre Hospitalier Universitaire de Quebec, Quebec), D. Morgan (McMaster University, Hamilton, Ontario), T. Sylwestrowicz (St. Paul's Hospital, Saskatoon, Saskatoon), G. Van Rosendaal (University of Calgary, Alberta), S.J. Van Zantan (Queen Elizabeth II Health Sciences Centre, Halifax, NS).
Colon Cancer Family Registry acknowledgments and investigators: David Conti and Graham Casey, University of Southern California, Los Angeles, USA; Polly Newcomb, Fred Hutchinson Cancer Research Center, Seattle USA; John Hopper and Mark Jenkins, University of Melbourne, Melbourne, Australia; Steven Gallinger, Cancer Care Ontario, Toronto, Ontario, Canada; David J. Duggan, Translational Genomics Research Institute, Phoenix, Arizona.
Grant support: We are grateful for funding from NCI N01-95015 and HHSN261201000082C (MMB, AGZ, JW), U01GM61393 (MJR), U01GM61390, and the Biobank Japan Project funded by the Japanese Ministry of Education, Culture, Sports, Science and Technology. This work is part of the NIH Pharmacogenomics Research Network-RIKEN Center for Genomic Medicine Global Alliance. Pfizer, Inc. partially funded the APC Trial. LGC-C and IT received support from Cancer Research UK and the Oxford Comprehensive Biomedical Research Centre, and the European Union (FP7 CHIBCHA Consortium). The Wellcome Trust Centre for Human Genetics is supported by a Wellcome Trust Core Grant 090532/Z/09/Z. None of the funders played a role in the design and performance of the present study.
Abbreviations
- SNP
Single nucleotide polymorphism
- CRC
Colorectal cancer
- APC
Adenoma Prevention with Celecoxib study
- GWAS
Genome-wide Association Study
- Corgi
Colorectal gene identification study
- FAP
Familial adenomatous polyposis
- NBS
National blood service
- NPS
National Polyp Study
- OR
odds ratio
- BC58
1958 Birth Cohort.
Footnotes
Disclosures: None of the authors declare conflict of interests
Authors' contributions: JW, LGC-C, AGZ, STW, IT, and MMB planned and designed the study. MK carried out sample genotyping. JW, LGC-C, AGZ, J-WC, SW and IT performed the statistical analyses. MD, OS, LL, PG, NGM, GWM, JY, PNB, AGZ, JW and MMB coordinated the sample collection and characterization of the CCFR and APC studies. YN, MJR and KM obtained resources for APC trial genotyping. IT, RSH and MD obtained funding for the CORGI, NSCCG and Scotland studies. MMB and JW wrote the manuscript.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Zauber AG, Winawer SJ, O'Brien MJ, Lansdorp-Vogelaar I, van Ballegooijen M, Hankey BF, et al. Colonoscopic polypectomy and long-term prevention of colorectal cancer deaths. New Engl J Med. 2012;366:687–96. doi: 10.1056/NEJMoa1100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saini SD, Kim HM, Schoenfeld P. Incidence of advanced adenomas at surveillance colonoscopy in patients with a personal history of colon adenomas: a meta-analysis and systematic review. Gastrointest Endosc. 2006;64:614–26. doi: 10.1016/j.gie.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 4.Bertagnolli MM, Eagle CJ, Zauber AG, Redston M, Solomon SD, Kim K, et al. Celecoxib for the prevention of sporadic colorectal adenomas. N Engl J Med. 2006;355:873–84. doi: 10.1056/NEJMoa061355. [DOI] [PubMed] [Google Scholar]
- 5.Carvajal-Carmona LG, Cazier J-B, Jones AM, Howarth K, Broderick P, Pittman A, et al. Fine-mapping of colorectal cancer susceptibility loci at 8q23.3, 16q22.1 and 19q13.11: refinement of association signals and use of in silico analysis to suggest functional variation and unexpected candidate target genes. Human Molecular Genetics. 2011;20:2879–88. doi: 10.1093/hmg/ddr190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomlinson IP, Carvajal-Carmona LG, Dobbins SE, A. T, Jones AM, Howarth K, et al. Multiple common susceptibility variants near BMP pathway loci GREM1, BMP4, and BMP2 explain part of the missing heritability of colorectal cancer. PLoS Genet. 2011;7:e1002105. doi: 10.1371/journal.pgen.1002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wellcome TCCC Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benjamini YH, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B. 1995;57:12. [Google Scholar]
- 9.Benjamini YaH, Y. The adaptive control of the false discovery rate in multiple hypotheses testing. J Behav Educ Statist. 2000;25:14. [Google Scholar]
- 10.Elias PS, Romagnuolo J, Hoffman B. Poor patient knowledge regarding family history of colon polyps: implications for the feasibility of stratified screening recommendations. Gastrointest Endosc. 2012;75:598–603. doi: 10.1016/j.gie.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Houlston RS, Cheadle J, Dobbins SE, Tenesa A, Jones AM, Howarth K, et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat Genet. 2010;42:973–7. doi: 10.1038/ng.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Houlston RS, Webb E, Broderick P, Pittman AM, Di Bernardo MC, Lubbe S, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40:1426–35. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–8. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 14.Tomlinson IP, Webb E, Carvajal-Carmona L, Broderick P, Howarth K, Pittman AM, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40:623–30. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 15.Broderick P, Carvajal-Carmona L, Pittman AM, Webb E, Howarth K, Rowan A, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007;39:1315–7. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 16.Peters U, Jiao S, Schumacher FR, Hutter CM, Aragaki AK, Baron JA, et al. Identification of Genetic Susceptibility Loci for Colorectal Tumors in a Genome-Wide Meta-analysis. Gastroenterology. 2013;144:799–807. e24. doi: 10.1053/j.gastro.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zanke BW, Greenwood CM, Rangrej J, Kustra R, Tenesa A, Farrington SM, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–94. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 18.Tenesa A, Farrington SM, Prendergast JG, Porteous ME, Walker M, Haq N, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40:631–7. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaeger E, Webb E, Howarth K, Carvajal-Carmona L, Rowan A, Broderick P, et al. Common genetic variants at the CRAC1 (HMPS) locus on chromosome 15q13.3 influence colorectal cancer risk. Nat Genet. 2008;40:26–8. doi: 10.1038/ng.2007.41. [DOI] [PubMed] [Google Scholar]
- 20.Dunlop MG, Dobbins SE, Farrington SM, Jones AM, Palles C, Whiffin N, et al. Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk. Nat Genet. 2012;44:770–6. doi: 10.1038/ng.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertagnolli MM, Eagle CJ, Zauber AG, Redston M, Breazna A, Kim K, et al. Five-year efficacy and safety analysis of the Adenoma Prevention with Celecoxib Trial. Cancer Prev Res (Phila) 2009;2:310–21. doi: 10.1158/1940-6207.CAPR-08-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Solomon SD, McMurray JJ, Pfeffer MA, Wittes J, Fowler R, Finn P, et al. Cardiovascular risk associated with celecoxib in a clinical trial for colorectal adenoma prevention. N Engl J Med. 2005;352:1071–80. doi: 10.1056/NEJMoa050405. [DOI] [PubMed] [Google Scholar]
- 23.Hirabayashi S, Tajima M, Yao I, Nishimura W, Mori H, Hata Y. JAM4, a junctional cell adhesion molecule interacting with a tight junction protein, MAGI-1. Mol Cell Biol. 2003;23:4267–82. doi: 10.1128/MCB.23.12.4267-4282.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jorissen RN, Gibbs P, Christie M, Prakash S, Lipton L, Desai J, et al. Metastasis-Associated Gene Expression Changes Predict Poor Outcomes in Patients with Dukes Stage B and C Colorectal Cancer. Clin Cancer Res. 2009;15:7642–51. doi: 10.1158/1078-0432.CCR-09-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kigel B, Rabinowicz N, Varshavsky A, Kessler O, Neufeld G. Plexin-A4 promotes tumor progression and tumor angiogenesis by enhancement of VEGF and bFGF signaling. Blood. 2011;118:4285–96. doi: 10.1182/blood-2011-03-341388. [DOI] [PubMed] [Google Scholar]
- 26.Batlle E, Bacani J, Begthel H, Jonkheer S, Gregorieff A, van de Born M, et al. EphB receptor activity suppresses colorectal cancer progression. Nature. 2005;435:1126–30. doi: 10.1038/nature03626. [DOI] [PubMed] [Google Scholar]
- 27.Cortina C, Palomo-Ponce S, Iglesias M, Fernandez-Masip JL, Vivancos A, Whissell G, et al. EphB-ephrin-B interactions suppress colorectal cancer progression by compartmentalizing tumor cells. Nat Genet. 2007;39:1376–83. doi: 10.1038/ng.2007.11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.