Abstract
Human cytochrome P450 2D6 (CYP2D6) is involved in metabolism of approximately 25% of pharmaceutical drugs. Inactivation of CYP2D6 can lead to adverse drug interactions. Four inactivators of CYP2D6 have previously been identified: 5-Fluoro-2-[4-[(2-phenyl-1H-imidazol-5-yl)methyl]-1-piperazinyl]pyrimidine (SCH66712), (1-[(2-ethyl-4-methyl-1H(-EMTPP-imidazol-5-yl)-methyl]-4-[4-(trifluoromethyl)-2-pyridinyl]piperazine (EMTPP), paroxetine, and 3,4-methylenedioxymethamphetamine (MDMA). All four contain planar, aromatic groups as well as basic nitrogens common to CYP2D6 substrates. SCH66712 and EMTPP also contain piperazine groups and substituted imidazole rings that are common in pharmaceutical agents, though neither of these compounds is clinically relevant. Paroxetine and MDMA contain methylenedioxyphenyls. SCH66712 and EMTPP are both known protein adductors while paroxetine and MDMA are probable heme modifiers. The current study shows that each inactivator displays Type I binding with Ks values that vary by 2-orders of magnitude with lower Ks values associated with greater inactivation. Comparison of KI, kinact, and partition ratio values shows SCH66712 is the most potent inactivator. Molecular modeling experiments using AutoDock identify Phe120 as a key interaction for all four inactivators with face-to-face and edge-to-face pi interactions apparent. Distance between the ligand and heme iron correlates with potency of inhibition. Ligand conformations were scored according to their binding energies as calculated by AutoDock and correlation was observed between molecular models and Ks values.
Keywords: CYP2D6, mechanism-based inactivation, paroxetine, MDMA, EMTPP, SCH66712
Introduction
Cytochrome P450 enzymes (CYPs) are a family of heme-containing monooxygenases responsible for metabolism of a variety of small organic compounds. In humans, CYPs are known to metabolize sterols, fatty acids, eicosanoids, vitamins, and xenobiotics such as pharmaceuticals (1). Over 80% of pharmaceuticals are metabolized by CYPs (1). Loss of specific drug-metabolizing CYP activity is a major concern and can lead to adverse drug interactions. In vivo, the enzyme activity that is lost by inactivation by a mechanism-based inactivator can only be recovered by synthesis of new enzyme.
Mechanism-based enzyme inactivators are those inhibitors that require metabolic activation to form a reactive electrophile that then becomes covalently attached to the enzyme that formed it. For CYPs, inactivation can occur by protein modification, heme modification, or heme destruction (2). Criteria for mechanism-based inhibition of P450s include concentration-, time-, and NADPH-dependent inhibition, among others (2).
While only a minor constituent of total liver CYP protein, CYP2D6 is a major drug-metabolizing enzyme in humans, metabolizing nearly 25% of current pharmaceutical drugs (1). CYP2D6 is also involved in metabolism of a variety of other xenobiotics including some illicit drugs such as amphetamine analogs. Common structural features of CYP2D6 substrates include compounds with aromatic rings and frequently basic nitrogens (3-7). Further, many of the CYP2D6 substrates are members of pharmaceutical classes with narrow therapeutic indices such as antiarrhythmics, antihypertensives, and antidepressants. Thus, inactivation of CYP2D6 may result in serious clinical side effects as exemplified by debrisoquine metabolism by individuals with a CYP2D6 poor-metabolizer phenotype (8). Mechanism-based inactivation of CYPs can lead to the same phenotype as a poor-metabolizer phenotype.
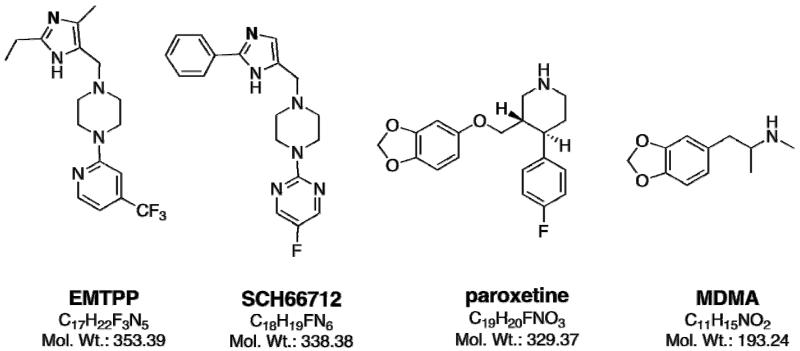
To date, only a handful of mechanism-based inhibitors of CYP2D6 have been identified including two that contain substituted imidazole rings (SCH66712 and EMTPP) and two that are methylenedioxyphenyls (paroxetine and MDMA) (Figure 1) (9-13). Additionally, substituted methylenedioxyphenyls containing arylamides isolated from Piper nigrum (black pepper) have been reported as mechanism-based inactivators of CYP2D6 (14). Another compound, metoclopramide, has also been identified as a potential mechanism-based inhibitor of CYP2D6 (15) though our studies have not confirmed that finding1 (unpublished observations).
Figure 1.
Structure of EMTPP, SCH 66712, paroxetine, and MDMA with molecular formulas and weights indicated for each.
In the current study, we examined existing kinetic data and expanded those studies to allow for more comprehensive, comparative examination of the kinetic features and the structural basis for varying potencies in inhibition of four of the known mechanism-based inhibitors of CYP2D6 - SCH66712, EMTPP, paroxetine, and MDMA. By studying a combination of known CYP2D6 mechanism-based inactivators by molecular modeling coupled to kinetic data, our goal was to better understand the relationship between kinetic parameters and structural elements important for CYP2D6 catalytic inactivation.
METHODS
Reagents
Human CYP2D6 with P450 reductase (Supersomes™) were purchased from BD-Gentest (Woburn, MA). Purified, recombinant human CYP2D6 and recombinant P450 NADPH-reductase were a generous gift from Dr. F. P. Guengerich (Vanderbilt University, Nashville, TN) and were used for spectral binding titrations described below; all other assays used Supersomes. All solvents were HPLC grade and purchased from Sigma-Aldrich (St. Louis, MO). MDMA was obtained from the National Institute on Drug Abuse (Rockville, MD). EMTPP was purchased from Interchim, Inc. (San Pedro, CA). Paroxetine and all other reagents were purchased from Sigma-Aldrich (St. Louis, MO).
Spectral Binding Titrations
Spectral binding titration studies were performed with recombinant, purified CYP2D6 (1 μM) in 100 mM potassium phosphate buffer, pH 7.4, to a final volume of 2 mL. The solution was evenly divided between two cuvettes and the experiments were performed at room temperature by titrating in aliquots of paroxetine (0.25 - 50 μM) or EMTPP (0.1 – 200 μM) to the sample cuvette with the equivalent volume of solvent control added to the reference cuvette. A baseline of the reference cuvette was recorded (250-500 nm) on a Cary 300 dual-beam spectrophotometer (Varian, Inc., Walnut Creek, CA). The ligand was then added to the sample cuvette and solvent to the reference cuvette and the spectra were recorded (350-500 nm) after each addition. The difference in absorbance between the wavelength maximum and minimum was plotted against the concentration of ligand, and the data were analyzed by nonlinear regression methods with KaleidaGraph (Synergy Software, Reading, PA). The dissociation constant, Ks, was determined using the following quadratic velocity equation or tight-binding equation:
where S is the substrate concentration, E is the total enzyme concentration, and Ks is the spectral dissociation constant for the reaction .
Partition Ratios
Primary reaction mixtures containing 0-240 μM paroxetine or MDMA and 20 pmol of CYP2D6 Supersomes in 100 mM potassium phosphate buffer, pH 7.4 (final volume of 100 μL), were preincubated in a 37 °C shaking water bath. After 5 minutes, the primary reactions were initiated with the addition of NADPH-generating system (5 mM glucose 6-phosphate, 0.5 mM NADP+, and 0.5 units/mL glucose 6-phosphate dehydrogenase) and incubated at 37 °C. To allow the inactivation to go to completion, inactivation assays were incubated for 60 minutes. Aliquots of 10 μL were then removed from the primary reactions and added to the secondary reaction mixtures containing 100 μM bufuralol and NADPH-generating system in 100 mM potassium phosphate buffer, pH 7.4, with a final volume of 200 μL. The secondary mixtures (in triplicate) were incubated for 10 minutes at 37 °C and quenched with 15 μL of 70% perchloric acid. Reaction mixtures were centrifuged (2000 × g, 5 minutes) to remove the precipitated enzyme and aliquots of the recovered supernatants were directly injected onto a Waters Alliance e2695 HPLC for analysis. The formation if the 1′-hydroxybufuralol product was quantified by HPLC as previously described (13).
Molecular Modeling and Docking Simulations
The structures of the four inactivator ligands used in this study are shown in Figure 1. The three-dimensional structures of the ligands for docking studies were built in Spartan 4.0 (Wavefunction, Inc., Irvine, CA) with all hydrogen atoms added and energy minimized. AutoDock 4.0 was used to perform molecular modeling and docking (http://autodock.scripps.edu; (16, 17). The protein structure used in these studies was CYP2D6 (PDB: 2F9Q). Solvent molecules were removed and the heme was retained. The Fe atom of the heme was assigned a charge of +3. Residues within 5 Å of the heme iron were identified and set as flexible residues for simulations. For the protein structure, charges were calculated by the Gasteiger-Marsili method. The grid maps were calculated using AutoGrid. The dimensions of the grid box were set to 40 × 40 × 40 Å, and the grid spacing was set to 0.375 Å. Docking simulations were performed using the Lamarckian genetic algorithm. Each docking experiment was performed 100 times, resulting in 100 docked conformations. The consensus docking conformations of the models were obtained by visual inspection and docking scores.
RESULTS AND DISCUSSION
Due to its role in metabolism of a large number of pharmaceuticals, CYP2D6 inactivation has important implications for potential drug-drug interactions and adverse drug events. There are only a handful of known mechanism-based inactivators of CYP2D6, but commonalities in their inactivation of CYP2D6 may provide insight into not only catalysis by CYP2D6 but also drug moieties involved in adverse drug events. The mechanism-based inactivators of CYP2D6 studied here include EMTPP, SCH66712, paroxetine, and MDMA (Figure 1). These four inactivators represent two different classes of chemical compounds – those with substituted imidazoles and piperazine rings (EMTPP and SCH66712) and those with methylenedioxyphenyls (paroxetine and MDMA). We note some individuals consume antidepressants and illicit drugs such as paroxetine and MDMA simultaneously that can lead to further adverse drug events (18, 19).
Potency of mechanism-based inactivation is dependent on kinetic parameters kinact and KI as well as on partition ratio. For all four inactivators, kinetic constants KI and kinact have been determined previously by other investigators, though the model systems used varied (Table 1). The value of kinact/KI is a measure of inhibition potency. Since kinact is a measure of the rate of inactivation and KI the concentration of inhibitor that gives one-half the rate of inactivation, the larger the value for the ratio, the greater the inhibition. Comparison of kinact/KI ratios indicates that SCH66712 is the most potent inactivator with a ratio of kinact/KI of 582 ml/min-μmol. The other three inactivators have ratios an order of magnitude lower between 16-35 ml/min-μmol. In the literature there are two reports of kinact and KI values for paroxetine. Values determined by Bertelsen et al. (10) are shown on Table 1. Obach et al. (20) report a similar kinact value for paroxetine, 0.17 min−1, but a value for KI (0.81 μM) that is an order of magnitude less than Bertelsen et al. Both groups used similar approaches in HLM with dextromethorphan as reporter and Kitz-Wilson analysis for determination of KI, but incubation times and reporter concentrations varied. We note that given this discrepancy, it is possible that paroxetine is a more potent inactivator than represented in the current study; it would be worthwhile to repeat the inactivation assays with paroxetine in a recombinant system to understand the differences in reported values. Obach et al. also report kinact and KI for MDMA as 0.38 min−1 and 6.3 μM, respectively; these values are similar to the values shown in Table 1 as reported by Heydari et al. (11).
Table 1.
KI and kinact values for inactivation of CYP2D6.
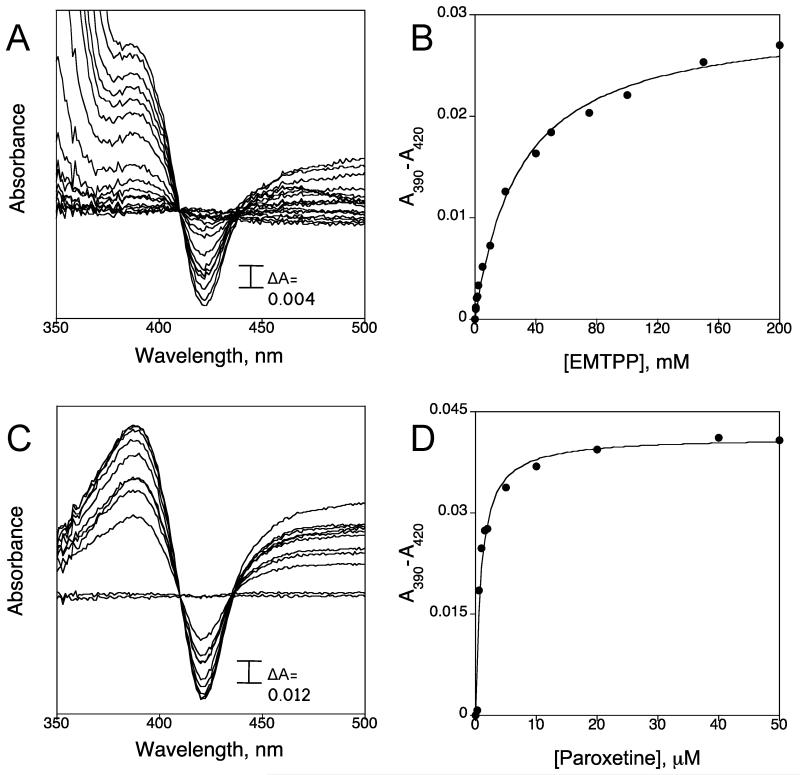
Spectral binding constants, Ks, for SCH66712 and MDMA have been determined previously and both compounds showed Type I binding as predicted for a mechanism-based inactivator that binds as a substrate prior to inactivation (13, 21). In the present study we measured the binding constants for EMTPP and paroxetine by titration of CYP2D6 with increasing concentrations of EMTPP or paroxetine. We found that both EMTPP and paroxetine showed Type I binding and Ks values were 29.76 ± 3.04 μM and 0.88 ± 0.18 μM, respectively (Figure 2 and Table 2). The range of values for Ks among the four inactivators does not show a pattern based on common structural features. However, there is a trend for lower spectral binding constant and greater inactivation since SCH66712 and paroxetine show the lowest spectral binding constants and largest values for the kinact/KI ratio (Tables 1 and 2). The spectral binding constant for MDMA was previously fitted with a curved hyperbolic equation rather than tight-binding equation and the corrected Ks could be lower than the value reported (22).
Figure 2.
Spectral binding titration of CYP2D6 and determination of spectral binding parameter Ks. (A) Titration of CYP2D6 with EMTPP and (B) plot of ΔA430-395 (from panel A) vs. concentration of EMTPP. (C) Titration of CYP2D6 with paroxetine and (D) plot of ΔA430-395 (from panel C) vs. concentration of paroxetine.
Table 2.
Spectral Dissociation Constants for Inactivators of CYP2D6. 1
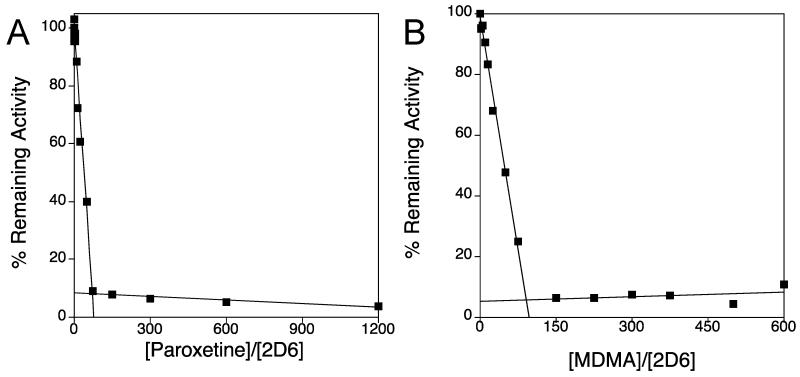
Partition ratio is a measure of the number of molecules metabolized per molecule of CYP2D6 inactivated. Partition ratios for SCH66712 and EMTPP are 3 and 99, respectively (12, 13) (Table 3). For comparison, partition ratios for paroxetine and MDMA inactivation of CYP2D6 were determined in the present study. CYP2D6 was incubated with various concentrations of either paroxetine or MDMA over 60 minutes to allow the inactivation to progress until essentially complete. The percentage of the activity remaining was plotted as a function of the molar ratio of paroxetine or MDMA to CYP2D6. The turnover number (partition ratio +1) was estimated from the intercept of the linear regression line obtained from the lower ratios of inactivator to CYP2D6 with the straight line derived from the higher ratios of inactivator to CYP2D6 as described previously (23). With this method, the partition ratio values for paroxetine and MDMA were 70 and 91, respectively (Figure 3; Table 3). These values are similar to that of EMTPP and again an order of magnitude different from the partition ratio of SCH66712. Lim et al. (24) reported apparent partition ratios for mechanism-based inactivators of various drug-metabolizing CYPs, including CYP2D6, by use of an automated screening strategy in human liver microsomes (HLM). From their analysis, they found the partition ratio for EMTPP and paroxetine with CYP2D6 to be 213.5 and 5.6, respectively. These values are different from those measured in recombinant systems, particularly for paroxetine. In our study with recombinant CYP2D6 the observed partition ratio for paroxetine was an order of magnitude greater at 70. Lim et al. clarify that their automated screening strategy is only meant to identify potentially potent inactivators that can then be further studied in time-and concentration-dependent assays. For paroxetine, the higher partition ratio values reported here versus Lim et al. also contributes to understanding the safety record for paroxetine as a clinical drug even though it is a mechanism-based inactivator.
Table 3.
Partition Ratio Values for Inactivators of CYP2D6.1
Figure 3.
Loss of CYP2D6 activity as a function of the ratio of inhibitor to CYP2D6. CYP2D6 was incubated with varying concentrations of (A) paroxetine or (B) MDMA for 60 minutes to allow for complete inactivation. The partition ratio was estimated to be 70 for paroxetine and 91 for MDMA based on the intercept of the linear regression line from the lower ratios and the straight line obtained from higher ratios.
We also note that based on previously reported values for kinact and KI in HLM (14), the two mechanism-based arylamine inactivators of CYP2D6 from Piper nigrum have kinact/KI ratios of 120 and 90 ml/min-μmol suggesting that they are the most potent clinically relevant inactivators of CYP2D6 since SCH66712 is not a clinically relevant drug or natural product. In most contexts the daily dose or consumption of these arylamines would be considerably less than that of a pharmaceutical agent thus lowering the potential for adverse drug effects from their consumption. However, in some traditional medicines, black pepper is used for medicinal purposes (such as in ayurvedic medicine, i.e. traditional Indian medicine, and in traditional Indonesian medicine). In fact, “Trikatu,” an ayurvedic medicine made of equal parts of P. nigrum, P. lungum and dried rhizomes of Z. officinale, is used to increase bioavailability of some pharmaceutical drugs (25, 26). While the mechanism of increased bioavailability has not been established, it seems likely that inactivation of P450s, such as CYP2D6, could be the cause for the observed increases in bioavailability reported in the literature (25-28).
A series of molecular modeling studies with each inactivator and CYP2D6 was performed to better understand the relationship between binding and inactivation. Previous modeling studies in our laboratory with SCH66712 and EMTPP have shown that molecular docking experiments are consistent with observed metabolite formation and therefore may provide insight into the binding events that lead to inactivation (13).
The lowest energy binding conformations were chosen for analysis (Figure 5). These binding conformations were also the most frequently observed orientations with respect to the moiety pointing toward the heme group. That is, while other binding orientations were observed, only the lowest energy orientation is shown. We note that observation of multiple binding orientations is consistent with known metabolite profiles for each compound and the orientations shown are consistent with the major metabolites of each compound (Figure 4 and Supplemental Figure 1).
Figure 5.
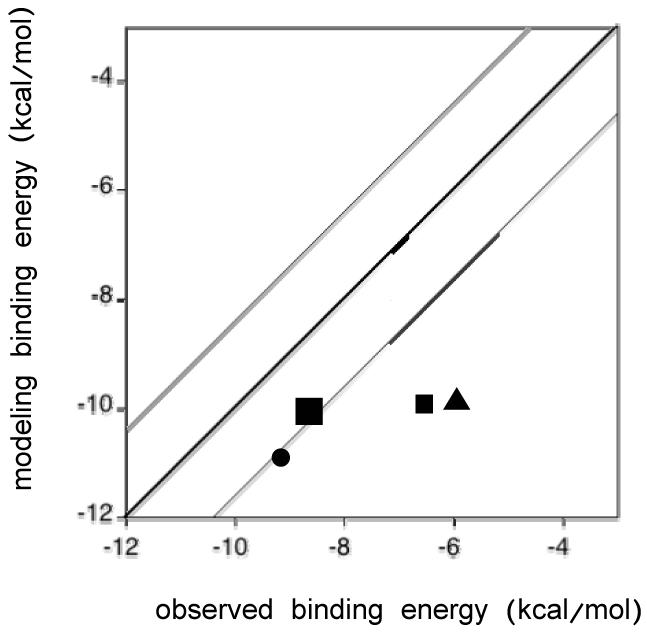
Molecular modeling vs. observed binding energies for each docked structure. The bold line represents a perfect correlation between molecular modeling and observed binding energies (Table 4). The other lines represent one standard deviation from the bold line.
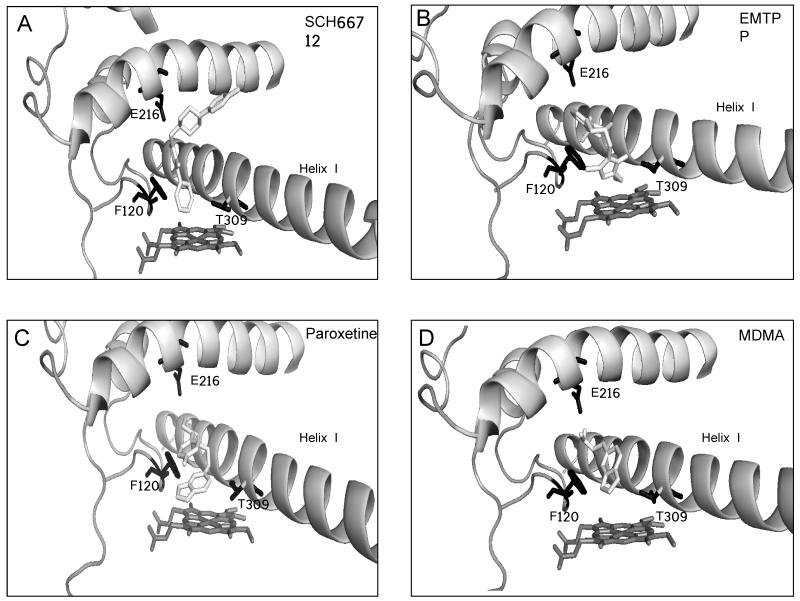
Figure 4.
Molecular modeling of inactivators bound to CYP2D6. AutoDock was used to model binding in the active site of each inactivator in the conformation believed to lead to inactivation of CYP2D6 as described in the Materials and Methods. Docking simulation with (A) SCH66712, (B) EMTPP, (C) paroxetine, and (D) MDMA. Active site amino acids Phe120, Glu216, and Thr309 are shown in black.
CYP2D6 has two phenylalanine residues in the binding pocket – Phe120 and Phe483. These aromatic groups have been postulated to be involved in substrate orientation and substrate recognition, respectively. Modeling experiments show pistacking interactions between all inactivators and Phe120 but no apparent interactions with Phe483 in the binding site (Figure 4). This is consistent with the role of Phe120 in substrate orientation at the active site (6). The distances between the docked ligands and the heme iron were calculated (Table 4). The phenyl ring of SCH66712 - the postulated site for inactivation (13), is only 2.1 Å from the heme iron, as previously shown (13), and the closest orientation of all four inactivators (Table 4). The distances for EMTPP, paroxetine, and MDMA were 2.9, 5.2, and 5.2 Å, respectively. This is consistent with kinetic data that show SCH66712 is the most potent inhibitor of the four and consistent with the trend observed by Sridhar et al. for the most potent arylacetylenes mechanism-based inactivators of other CYPs being bound closest to the heme iron (29). The arylacetylene inactivators are heme-adductors while two of the inactivators of CYP2D6 (SCH66712 and EMTPP) are protein-adductors and two are postulated heme-adductors via carbene intermediates (paroxetine and MDMA) (10-13). Since our data support the observation that closer binding to heme leads to more potent inactivation, it may be that results of modeling interactions with the heme iron as a predictor of inactivation potency is independent of the type of inactivation (protein vs. heme).
Table 4.
Summary of Docking Results Related to Binding Energy.
| Inhibitor | ΔGexp1 (kcal/mol) |
ΔGcalc (kcal/mol) model |
Site of reaction2 |
Phe120 π interactions with inhibitor |
Distance to heme iron (Å) |
|---|---|---|---|---|---|
| EMTPP | −6.42 | −9.98 | ethyl | face to face | 2.9 |
| SCH66712 | −9.10 | −10.93 | phenyl | edge to face | 2.1 |
| Paroxetine | −8.59 | −10.12 | methylene | edge to face | 5.2 |
| MDMA | −6.04 | −9.93 | methylene | edge to face | 5.2 |
ΔGexp was calculated using spectral binding constants, Ks, and the relationship: ΔGexp = RT ln Ks.
Site of known oxidation and also site of either known or putative electrophile formation that leads to enzyme inactivation.
Though distance to the heme iron and inactivation potency differs, the binding orientations of the two piperazine-containing compounds, SCH66712 and EMTPP, are similar. For SCH66712, pi-stacking occurs between Phe120 and its imidazole ring. The same is true for EMTPP with additional pi-stacking between Phe120 and the heteroaromatic ring of EMTPP that is not seen with SCH66712 due to its more elongated structure. SCH66712 is the only inactivator that has interactions that extend to residues above helix I; perhaps the additional interactions allow SCH66712 more opportunity to react and inactivate CYP2D6. Residues in this region between helix I and helix F include Leu213, Val308, Leu484, and Ala482 (data not shown). Modeling with both piperazine-containing compounds indicates that Thr309 is the closest nucleophile to the active site and a highly plausible target for inactivation. Others have shown by mutation studies with a Thr309Val mutant that Thr309 is involved in CYP2D6 oxidation reactions but has little effect on substrate affinity (22).
For paroxetine and MDMA, the methylene group of the methylenedioxyphenyl moiety is closest to the heme, consistent with known metabolism at the methylene carbon. In both cases Phe120 forms an edge-to-face interaction with the methylenedioxyphenyl group and the distance to the heme is 5.2 Å. The edge-to-face orientation is believed to be a more favorable orientation than edge-to-edge (30) though, as stated above, Sridhar et al. found that the distance to the heme iron correlated with potency of inhibition better than specific type of piinteractions (29). Overall, pi-pi interactions are contributors to inactivation that should be considered complimentary to distance between inactivator and heme iron for CYP2D6.
To further confirm validity of the molecular models, the binding energies from the models were compared to the experimentally determined values calculated using spectral binding constants, Ks, for each inactivator and the relationship: ΔGexp = RT ln Ks, where R is the gas constant and T is absolute temperature (298 K) (Table 4). The calculated energies from the molecular models were then plotted against the observed binding energies measured from spectral binding (Figure 5). The binding energies from the molecular models for SCH66712 and paroxetine were closer to the calculated energies than were the energies for EMTPP and MDMA. Overall, comparison of binding energies shows that the molecular model tends to show lower binding energy than the experimentally measured binding energy. Given that the structure of CYP2D6 used for the molecular modeling was determined in the absence of ligand, it is not unexpected that the binding energies may not be accurately predicted as positions of active site amino acids would likely change in the presence of ligand (31).
CONCLUSIONS
Based on both the collection of previous findings and the findings presented here for the established inactivators of CYP2D6, the more potent inactivators not only have the largest values for kinact/KI ratios and lowest values for partition ratio, as would be expected, but also have lower values for Ks and closer interaction with heme iron in molecular models. When considering potency of inactivation, measurement of all these parameters should be considered. Further, since this group of inactivators represents both protein and heme adductors, it also appears that these trends may apply to both types of inactivators, though the data set for inactivators of CYP2D6 is limited. Thus, confirmation of these findings with new inactivators of CYP2D6 as they are identified should be of interest.
Supplementary Material
Acknowledgement
We also thank Drs. F. P. Guengerich, P. Hollenberg, H. Zhang, L. Kuhn, and S. Ferguson-Miller for helpful comments. We thank Logan Kinch for help with PyMol. This work was supported by the National Institutes of Health (L.L.F.) [Grants 1R15GM086767-01, 3R15BM086767-01S1]; a grant to Kalamazoo College from the Howard Hughes Medical Institute [52006304] through the Precollege and Undergraduate Science Education Program; and the Hutchcroft Fund of Kalamazoo College.
Abbreviations used
- SCH66712
5-Fluoro-2-[4-[(2-phenyl-1H-imidazol-5-yl)methyl]-1-piperazinyl]pyrimidine
- CYP
cytochrome P450 enzyme
- HLM
human liver microsomes
- HPLC
high performance liquid chromatography
- EMTPP
(1-[(2-ethyl-4-methyl-1H-imidazol-5-yl)-methyl]-4-[4-(trifluoromethyl)-2-pyridinyl]piperazine
- MDMA
3,4-methylenedioxymethamphetamine
Footnotes
Bolles, A., Livezey, M., Nagy, L., Briggs, E., and Furge, L.L. Unpublished results.
Supporting Information Available. This material is available online at xxxx.
Authorship Contributions
Participated in research design: Furge, Livezey, Nagy, Diffenderfer, Arthur, Hsi
Conducted experiments: Furge, Livezey, Nagy, Diffenderfer, Arthur, Hsi
Performed data analysis: Furge, Livezey, Nagy, Diffenderfer, Arthur, Hsi, Holton
Wrote or contributed to the writing of the manuscript: Furge, Livezey, Nagy, Diffenderfer
Other: Furge acquired funding for the research
References
- (1).Guengerich FP. Cytochromes P450, drugs, and diseases. Mol Interv. 2003;3:194–204. doi: 10.1124/mi.3.4.194. [DOI] [PubMed] [Google Scholar]
- (2).Hollenberg PF, Kent UM, Bumpus NN. Mechanism-based inactivation of human cytochromes p450s: experimental characterization, reactive intermediates, and clinical implications. Chem Res Toxicol. 2008;21:189–205. doi: 10.1021/tx7002504. [DOI] [PubMed] [Google Scholar]
- (3).Wolff T, Distlerath LM, Worthington MT, Groopman JD, Hammons GJ, Kadlubar FF, Prough RA, Martin MV, Guengerich FP. Substrate specificity of human liver cytochrome P-450 debrisoquine 4-hydroxylase probed using immunochemical inhibition and chemical modeling. Cancer Res. 1985;45:2116–22. [PubMed] [Google Scholar]
- (4).Islam SA, Wolf CR, Lennard MS, Sternberg MJ. A three-dimensional molecular template for substrates of human cytochrome P450 involved in debrisoquine 4-hydroxylation. Carcinogenesis. 1991;12:2211–9. doi: 10.1093/carcin/12.12.2211. [DOI] [PubMed] [Google Scholar]
- (5).Koymans L, Vermeulen NP, van Acker SA, te Koppele JM, Heykants JJ, Lavrijsen K, Meuldermans W, Donne-Op den Kelder GM. A predictive model for substrates of cytochrome P450-debrisoquine (2D6) Chem Res Toxicol. 1992;5:211–9. doi: 10.1021/tx00026a010. [DOI] [PubMed] [Google Scholar]
- (6).Rowland P, Blaney FE, Smyth MG, Jones JJ, Leydon VR, Oxbrow AK, Lewis CJ, Tennant MG, Modi S, Eggleston DS, Chenery RJ, Bridges AM. Crystal structure of human cytochrome P450 2D6. J Biol Chem. 2006;281:7614–22. doi: 10.1074/jbc.M511232200. [DOI] [PubMed] [Google Scholar]
- (7).Guengerich FP, Miller GP, Hanna IH, Martin MV, Leger S, Black C, Chauret N, Silva JM, Trimble LA, Yergey JA, Nicoll-Griffith DA. Diversity in the oxidation of substrates by cytochrome P450 2D6: lack of an obligatory role of aspartate 301-substrate electrostatic bonding. Biochemistry. 2002;41:11025–34. doi: 10.1021/bi020341k. [DOI] [PubMed] [Google Scholar]
- (8).Gonzalez FJ, Skoda RC, Kimura S, Umeno M, Zanger UM, Nebert DW, Gelboin HV, Hardwick JP, Meyer UA. Characterization of the common genetic defect in humans deficient in debrisoquine metabolism. Nature. 1988;331:442–6. doi: 10.1038/331442a0. [DOI] [PubMed] [Google Scholar]
- (9).Palamanda JR, Casciano CN, Norton LA, Clement RP, Favreau LV, Lin C, Nomeir AA. Mechanism-based inactivation of CYP2D6 by 5-fluoro-2-[4-[(2-phenyl-1H-imidazol-5-yl)methyl]-1-piperazinyl]pyrimidine. Drug Metab Dispos. 2001;29:863–7. [PubMed] [Google Scholar]
- (10).Bertelsen KM, Venkatakrishnan K, Von Moltke LL, Obach RS, Greenblatt DJ. Apparent mechanism-based inhibition of human CYP2D6 in vitro by paroxetine: comparison with fluoxetine and quinidine. Drug Metab Dispos. 2003;31:289–93. doi: 10.1124/dmd.31.3.289. [DOI] [PubMed] [Google Scholar]
- (11).Heydari A, Yeo KR, Lennard MS, Ellis SW, Tucker GT, Rostami-Hodjegan A. Mechanism-based inactivation of CYP2D6 by methylenedioxymethamphetamine. Drug Metab Dispos. 2004;32:1213–7. doi: 10.1124/dmd.104.001180. [DOI] [PubMed] [Google Scholar]
- (12).Hutzler JM, Steenwyk RC, Smith EB, Walker GS, Wienkers LC. Mechanism-based inactivation of cytochrome P450 2D6 by 1-[(2-ethyl-4-methyl-1H-imidazol-5-yl)methyl]-4-[4-(trifluoromethyl)-2-pyridinyl]piperazine: kinetic characterization and evidence for apoprotein adduction. Chem Res Toxicol. 2004;17:174–84. doi: 10.1021/tx034199f. [DOI] [PubMed] [Google Scholar]
- (13).Nagy LD, Mocny CS, Diffenderfer LE, Hsi DJ, Butler BF, Arthur EJ, Fletke KJ, Palamanda JR, Nomeir AA, Furge LL. Substituted imidazole of 5-fluoro-2-[4-[(2-phenyl-1H-imidazol-5-yl)methyl]-1-piperazinyl]pyrimidine Inactivates cytochrome P450 2D6 by protein adduction. Drug Metab Dispos. 2011;39:974–83. doi: 10.1124/dmd.110.037630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Subehan, Usia T, Kadota S, Tezuka Y. Mechanism-based inhibition of human liver microsomal cytochrome P450 2D6 (CYP2D6) by alkamides of Piper nigrum. Planta Med. 2006;72:527–32. doi: 10.1055/s-2006-931558. [DOI] [PubMed] [Google Scholar]
- (15).Desta Z, Wu GM, Morocho AM, Flockhart DA. The gastroprokinetic and antiemetic drug metoclopramide is a substrate and inhibitor of cytochrome P450 2D6. Drug Metab Dispos. 2002;30:336–43. doi: 10.1124/dmd.30.3.336. [DOI] [PubMed] [Google Scholar]
- (16).Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and empirical binding free energy function. J. Computational Chemistry. 1998;19:1639–1662. [Google Scholar]
- (17).Huey R, Morris GM, Olson AJ, Goodsell DS. A semiempirical free energy force field with charge-based desolvation. J. Computational Chemistry. 2007;28:1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- (18).Farre M, Abanades S, Roset PN, Peiro AM, Torrens M, O’Mathuna B, Segura M, de la Torre R. Pharmacological interaction between 3,4-methylenedioxymethamphetamine (ecstasy) and paroxetine: pharmacological effects and pharmacokinetics. J Pharmacol Exp Ther. 2007;323:954–62. doi: 10.1124/jpet.107.129056. [DOI] [PubMed] [Google Scholar]
- (19).Ramamoorthy Y, Yu AM, Suh N, Haining RL, Tyndale RF, Sellers EM. Reduced (+/−)-3,4-methylenedioxymethamphetamine (“Ecstasy”) metabolism with cytochrome P450 2D6 inhibitors and pharmacogenetic variants in vitro. Biochem Pharmacol. 2002;63:2111–9. doi: 10.1016/s0006-2952(02)01028-6. [DOI] [PubMed] [Google Scholar]
- (20).Obach RS, Walsky RL, Venkatakrishnan K. Mechanism-based inactivation of human cytochrome p450 enzymes and the prediction of drug-drug interactions. Drug Metab Dispos. 2007;35:246–55. doi: 10.1124/dmd.106.012633. [DOI] [PubMed] [Google Scholar]
- (21).Keizers PH, de Graaf C, de Kanter FJ, Oostenbrink C, Feenstra KA, Commandeur JN, Vermeulen NP. Metabolic regio- and stereoselectivity of cytochrome P450 2D6 towards 3,4-methylenedioxy-Nalkylamphetamines: in silico predictions and experimental validation. J Med Chem. 2005;48:6117–27. doi: 10.1021/jm050338+. [DOI] [PubMed] [Google Scholar]
- (22).Keizers PH, Schraven LH, de Graaf C, Hidestrand M, Ingelman-Sundberg M, van Dijk BR, Vermeulen NP, Commandeur JN. Role of the conserved threonine 309 in mechanism of oxidation by cytochrome P450 2D6. Biochem Biophys Res Commun. 2005;338:1065–74. doi: 10.1016/j.bbrc.2005.10.066. [DOI] [PubMed] [Google Scholar]
- (23).Silverman RB. Mechanism-based Enzyme Inactivation: Chemistry and Enzymology. Vol. 1. CRC Press; Boca Raton, FL: 1988. [Google Scholar]
- (24).Lim HK, Duczak N, Jr., Brougham L, Elliot M, Patel K, Chan K. Automated screening with confirmation of mechanism-based inactivation of CYP3A4, CYP2C9, CYP2C19, CYP2D6, and CYP1A2 in pooled human liver microsomes. Drug Metab Dispos. 2005;33:1211–9. doi: 10.1124/dmd.104.003475. [DOI] [PubMed] [Google Scholar]
- (25).Atal CK, Zutshi U, Rao PG. Scientific evidence on the role of Ayurvedic herbals on bioavailability of drugs. J Ethnopharmacol. 1981;4:229–32. doi: 10.1016/0378-8741(81)90037-4. [DOI] [PubMed] [Google Scholar]
- (26).Lala LG, D’Mello PM, Naik SR. Pharmacokinetic and pharmacodynamic studies on interaction of “Trikatu” with diclofenac sodium. J Ethnopharmacol. 2004;91:277–80. doi: 10.1016/j.jep.2003.12.027. [DOI] [PubMed] [Google Scholar]
- (27).Subehan, Usia T, Iwata H, Kadota S, Tezuka Y. Mechanism-based inhibition of CYP3A4 and CYP2D6 by Indonesian medicinal plants. J Ethnopharmacol. 2006;105:449–55. doi: 10.1016/j.jep.2005.12.001. [DOI] [PubMed] [Google Scholar]
- (28).Subehan, Usia T, Kadota S, Tezuka Y. Constituents of Zingiber aromaticum and their CYP3A4 and CYP2D6 inhibitory activity. Chem Pharm Bull (Tokyo) 2005;53:333–5. doi: 10.1248/cpb.53.333. [DOI] [PubMed] [Google Scholar]
- (29).Sridhar J, Jin P, Liu J, Foroozesh M, Stevens CL. In silico studies of polyaromatic hydrocarbon inhibitors of cytochrome P450 enzymes 1A1, 1A2, 2A6, and 2B1. Chem Res Toxicol. 2010;23:600–7. doi: 10.1021/tx900348v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Hunter CA, Singh J, Thornton JM. Pi-pi interactions: the geometry and energetics of phenylalanine-phenylalanine interactions in proteins. J Mol Biol. 1991;218:837–46. doi: 10.1016/0022-2836(91)90271-7. [DOI] [PubMed] [Google Scholar]
- (31).Ito Y, Kondo H, Goldfarb PS, Lewis DF. Analysis of CYP2D6 substrate interactions by computational methods. J Mol Graph Model. 2008;26:947–56. doi: 10.1016/j.jmgm.2007.07.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.