Abstract
Human induced pluripotent stem cells (hiPSCs) have been shown to differentiate along the retinal lineage in a manner that mimics normal mammalian development. Under certain culture conditions hiPSCs form optic vesicle-like structures (OVs), which contain proliferating progenitors capable of yielding all neural retina (NR) cell types over time. Such observations imply conserved roles for regulators of retinogenesis in hiPSC-derived cultures and the developing embryo. However, whether and to what extent this assumption holds true has remained largely uninvestigated. We examined the role of a key NR transcription factor, Visual System Homeobox 2 (VSX2), using hiPSCs derived from a patient with microphthalmia caused by an R200Q mutation in the VSX2 homeodomain region. No differences were noted between (R200Q)VSX2 and sibling control hiPSCs prior to OV generation. Thereafter, (R200Q)VSX2 hiPSC-OVs displayed a significant growth deficit compared to control hiPSC-OVs, as well as increased production of retinal pigmented epithelium (RPE) at the expense of NR cell derivatives. Furthermore, (R200Q)VSX2 hiPSC-OVs failed to produce bipolar cells, a distinctive feature previously observed in Vsx2 mutant mice. (R200Q)VSX2 hiPSC-OVs also demonstrated delayed photoreceptor maturation, which could be overcome via exogenous expression of wildtype VSX2 at early stages of retinal differentiation. Finally, RNAseq analysis on isolated hiPSC-OVs implicated key transcription factors and extracellular signaling pathways as potential downstream effectors of VSX2-mediated gene regulation. Our results establish hiPSC-OVs as versatile model systems to study retinal development at stages not previously accessible in humans, and support the bona fide nature of hiPSC-OV-derived retinal progeny.
Keywords: retina, transcription factors, homeobox, genes, neurogenesis, VSX2 protein, human, human induced pluripotent stem cells
Introduction
Studies have highlighted the utility of human pluripotent stem cells (hPSCs) in disease pathophysiology research, drug and gene therapy discovery, and cell-based transplantation. While the focus for hPSC technology remains on the study and treatment of disease, hPSCs also have the potential to recapitulate many, if not all, stages of human cellular development [1–4]. However, the depth to which hPSCs can model cell and tissue development on a molecular level is currently unclear.
To date, most reports on targeted differentiation of specific cell types from hPSCs have focused on the effects of exogenous signaling factors [5–11], leaving the role(s) of key transcriptional control proteins mostly unexplored. Both human embryonic and induced pluripotent stem cells (hESCs and hiPSCs) are well-suited for such studies. However, hiPSCs possess the added benefit of clinical correlation; that is, knowledge of the phenotypic impact of developmental gene defects in affected human patients. Therefore, hiPSCs seem particularly ideal for modeling inherited developmental disorders of known genetic origin.
As a candidate for assessing the validity of in vitro hiPSC-based developmental studies, the retinal lineage offers significant advantages. The retina is composed of a limited number of major cell types, many of which can be readily distinguished in culture. Furthermore, mammalian retinal development is highly conserved, and numerous cellular and molecular events underlying retinogenesis have been studied in detail in multiple organisms. Similarly, hiPSCs have been shown to produce retinal progeny in a predictable, stepwise fashion [1, 2, 4]. The added ability to isolate optic vesicle-like (OV) populations from hiPSCs enhances the potential to investigate developmental processes, beginning at a very early stage of retinal differentiation [2, 4, 12]. Recently published observations suggest that mechanisms governing retinal differentiation in hiPSCs may be similar to those present in vivo [2, 4, 12]; however, this assumption has not been directly studied. Results from such studies would increase our knowledge of human retinogenesis, establish a developmental profile for hiPSC-derived retinal progeny, and perhaps reveal avenues to improve retinal cell production for therapeutic applications.
To examine intrinsic mechanisms of retinal differentiation in hiPSCs, we reprogrammed T lymphocytes from a patient with a mutation in the transcription factor Visual Systems Homeobox 2 (VSX2, also known as CHX10). VSX2 is the earliest known and most highly selective marker of multipotent neural retina progenitor cells (NRPCs) in the OV [13, 14]. Isolated cultures of hiPSC-derived OVs initially express VSX2 in most cells, with the remainder expressing markers of retinal ganglion cells, the earliest born cell type in the retina [2, 4, 12]. As such, retinal identity can be assigned to all OV cell derivatives with high reliability.
In vertebrates, VSX2 is expressed initially in the distal OV, where it is believed to pattern the naïve OV into the neural retina (NR), at least in part by transcriptional repression of the OV- and RPE-associated gene Microphthalmia-associated Transcription Factor (MITF) [15]. In addition, VSX2 is postulated to indirectly modulate NRPC proliferation [16, 17], perhaps also through repression of MITF. Disruption of Vsx2 function in animal models causes severe defects of the eye and retina, including microphthalmia, reduced NR thickness, and ectopic RPE differentiation in the NR [15, 17–23]. The importance of VSX2 in human retinogenesis is evident in individuals born blind as a result of a mutation in the DNA-binding homeodomain region of the gene [24–26]. These exceedingly rare patients have a pure ocular phenotype that includes microphthalmia and a thin retina, although the precise consequences of VSX2 mutations on human retinal development remain unknown. The robust, retina-specific phenotype observed in both transgenic animals and genotyped patients make VSX2 mutant hiPSC-OVs a prime resource to test the potential of hiPSCs to model intrinsic developmental mechanisms.
To probe their capacity to model retinogenesis, hiPSCs were generated from a patient with an R200Q mutation in the VSX2 homeodomain [24], which in mice results in a fully expressed protein that lacks the ability to bind DNA [23]. As a wildtype (WT) control, we also derived hiPSCs from an unaffected brother. Developmental repercussions of the mutation were examined by comparing isolated OVs from the (R200Q)VSX2 and WT hiPSC lines. Prior to and immediately after the initiation of VSX2 expression in differentiating cultures, (R200Q)VSX2 and WT hiPSCs were indistinguishable, with both generating early VSX2+ hiPSC-OVs as previously described [2, 4, 12]. However, with time the (R200Q)VSX2 hiPSC-OVs exhibited profoundly decreased growth and enhanced RPE production at the expense of NR, although all NR cell types were produced with the exception of bipolar cells. (R200Q)VSX2 hiPSC-OVs also demonstrated delayed photoreceptor maturation, a finding that was rescued by early overexpression of wildtype VSX2. Comparative RNAseq analysis performed at day 20 (d20) and d30 of differentiation, which corresponds to the optic vesicle and cup stages of development, respectively, revealed putative mechanisms for the VSX2-mediated effects on human retinogenesis. Together, our findings establish hiPSCs as a dynamic tool to study intrinsic factors involved in the regulation of retinal development from its earliest stages.
Methods
hiPSC Generation and Characterization
Activated T-cell derived-hiPSCs were generated and characterized as previously described from a male patient with an R200Q mutation in VSX2 and an unaffected brother [4, 24, 27]. Samples were obtained according to the Helsinki Declaration, with written informed consent and approval from the Institutional Review Board (IRB) at the University of Wisconsin-Madison. Methods for hiPSC reprogramming and characterization are summarized in Supporting Information Materials and Methods and Supporting Information Fig. S1. Up to three clonal hiPSC lines from each subject were used for experimental procedures to assess reproducibility (WT-1,-2, and -3; Mut-1,-2, and -3).
Targeted Retinal Differentiation of hiPSCs
Retinal cell differentiation was performed according to our previously established protocol [2, 4] (summarized in Supporting Information Fig. S2). Briefly, pluripotent hiPSC colonies were enzymatically lifted and grown for 4 days in embryoid body (EB) medium (media formulations are included in Supplemental Information), whereupon EB medium was substituted with neural induction medium (NIM). On d6, suspended cell aggregates were reattached to laminin-coated plates and grown in NIM for 10 more days. At d16, neural clusters were mechanically lifted and grown in retinal differentiation medium (RDM). At d20, OVs were isolated as previously reported [2, 4].
Gene and protein expression analyses
Immunocytochemistry (ICC), RT-PCR, qRT-PCR, flow cytometry and Western blot analyses were performed as previously reported ([2, 4, 28, 29], Supporting Information Materials and Methods). Antibodies are listed in Supporting Information Table 1, and primer sequences are provided in Supporting Information Table 2.
Cell Counts
Stereology was performed with a Zeiss Axioplan 2 microscope (Zeiss, Gottingen, Germany, http://microscopy.zeiss.com) and Stereo Investigator 10-MBF Bioscience software (MBF Biosciences, Williston, VT, www.mbfbiosciences.com). At minimum, 20 counting frames were randomly selected for analysis, which ranged in area between 10 μm2 and 30 μm2 depending upon cell density. To evaluate count precision, coefficient of error (CE) was calculated using Stereo Investigator software (CE < 0.3 for inclusion). Multiple hiPSC-OVs were counted in three biological replicates for each time point examined. For photoreceptor cell counts, only nonpigmented (i.e., containing NR and not RPE) hiPSC-OVs were used.
VSX2 overexpression
The human VSX2 coding sequence was PCR-amplified from adult NR-derived cDNA using the following primers: 5′-atgacggggaaagcag-3′ and 5′-ctaagccatgtcctccagct-3′. The resulting PCR product was cloned into the pSIN-WP-mpgk lentiviral shuttle backbone [30] and lentivirus was produced via transfection of 293T cells [31]. Virus was 40× concentrated by ultracentrifugation and working titers were determined by infecting HEK293T cells and performing ICC for VSX2 on fixed cells 48 hr post-infection. These working titers were compared to those obtained with a control mpgk-GFP lentivirus, and equal amounts of virus were used for hiPSC infection. Control infections were monitored for GFP fluorescence and infection efficiency was >70%.
Comparative RNAseq
RNAseq was performed as reported previously [32]. Samples were prepared for sequencing with the Illumina TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA; http://www.illumina.com) and quantified with a Qubit fluorometer (Life Technologies, Carlsbad, CA; www.lifetechnologies.com). Four samples were pooled per lane (8pM final loading concentration) and sequenced on an Illumina HiSeq using the HiSeq 2000 SR multiplex recipe. RNAseq data was analyzed with GeneSifter software (Perkin Elmer, Waltham, MA; http://www.geospiza.com/Products/AnalysisEdition.shtml) and data was normalized to mapped reads, with a quality score criterion of 50 (at minimum, one group pass).
Results
(R200Q)VSX2 and WT control hiPSC lines are indistinguishable prior to the induction of VSX2 expression
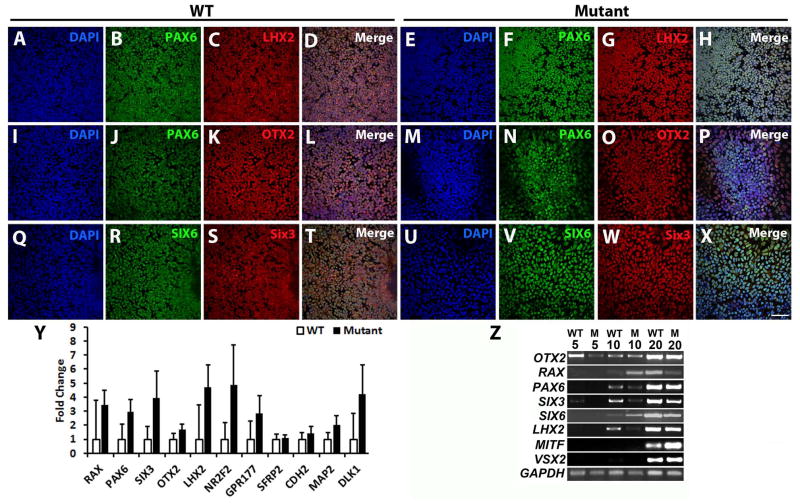
To evaluate the role of VSX2, (R200Q)VSX2 (mutant) and unaffected sibling control (WT) hiPSC lines were differentiated towards a retinal lineage using established protocols [1, 2, 4]. At d10 of differentiation, large regions within mutant and WT hiPSC cultures expressed the anterior neuroectoderm (AN) and eye field (EF) transcription factors PAX6, LHX2, OTX2, SIX3, and SIX6 (Fig. 1A-X). qRT-PCR analysis at d10 revealed no statistically significant differences in expression of AN/EF transcription factors, including RAX, PAX6, SIX3, OTX2, LHX2, and NR2F2 (Fig. 1Y), or additional AN genes such as GPR177, SFRP2, CDH2, MAP2, and DLK1. RT-PCR analysis demonstrated similar upregulation of AN/EF transcription factors in the mutant and WT hiPSC lines over the first 20 days of differentiation (Fig. 1Z). OTX2 was expressed at d0 (data not shown) and throughout differentiation, while PAX6, SIX3, LHX2, RAX, and SIX6 were expressed by d10. VSX2 and MITF were also expressed at d20 in mutant and WT cultures, indicating appropriate developmental progression from the EF fate seen at d10 to an early neural retina progenitor cell (NRPC) fate.
Figure 1. The (R200Q)VSX2 mutation does not affect anterior neuroectoderm/eye field specification in differentiating hiPSCs.
Representative immunocytochemical analysis of WT (A–D, I–L, Q–T) and (R200Q)VSX2 mutant (E–H, M–P, U–X) hiPSC cultures at day 10 of differentiation. Cell nuclei were identified with DAPI (A, E, I, M, Q, U). In both WT and mutant cultures, tightly packed neural colonies uniformly expressed the eye field transcription factors PAX6 (WT: B, J and mutant: F, N); LHX2 (WT: C and mutant: G); OTX2 (WT: K and mutant: O); SIX6 (WT: R and mutant: V); and SIX3 (WT: S and mutant: W). Merged images are shown in panels D, L, and T (WT) and H, P, and X (mutant). Scale bar = 50 μm. (Y) qRT-PCR analysis revealed similar expression levels of key eye field genes in WT and mutant cultures at day 10 (n=3 separate hiPSC lines for both groups with 3 biological replicates per line). (Z) Representative RT-PCR analysis demonstrated upregulation of critical eye field and optic vesicle transcription factors in WT and mutant (M) cultures over time.
(R200Q)VSX2 and WT control hiPSCs generate VSX2+ optic vesicle-like structures
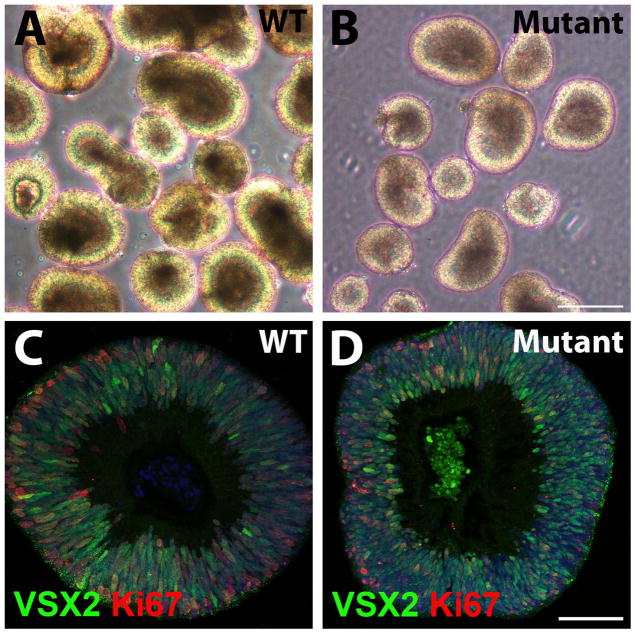
Optic vesicle-like structures (OVs) readily formed from WT and mutant hiPSCs by d20, permitting their isolation using established methods [2, 4]. WT and mutant hiPSC-OVs were morphologically indistinguishable (Fig. 2A,B), with hiPSC-OVs from all lines displaying robust nuclear expression of VSX2, a definitive marker for NRPCs (Fig. 2C,D). The VSX2+ NRPCs were highly proliferative as determined by co-expression of Ki-67. Of note, the expression pattern of VSX2 and Ki-67 in hiPSC-OVs was reminiscent of that observed in the neuroblastic layer of the developing human retina (Supporting Information Fig. S3A).
Figure 2. WT and (R200Q)VSX2 mutant hiPSC lines produce optic vesicle-like structures (hiPSC-OVs), allowing purification of VSX2+ neural retina progenitor cells (NRPCs).
(A, B) Representative micrographs of WT (A) and mutant (B) hiPSC-OVs isolated at day 20 of differentiation. At this stage, hiPSC-OVs are morphologically indistinguishable between WT and mutant cultures. Scale bar in panel B = 250 μm (also applies to panel A). (C, D) Both WT (C) and mutant (D) hiPSC-OVs were comprised of VSX2+ NRPCs that were proliferative, as shown by Ki-67 expression. Scale bar in panel D = 50 μm (also applies to panel C).
(R200Q)VSX2 hiPSC-OVs demonstrate growth retardation and preferential differentiation toward an RPE fate
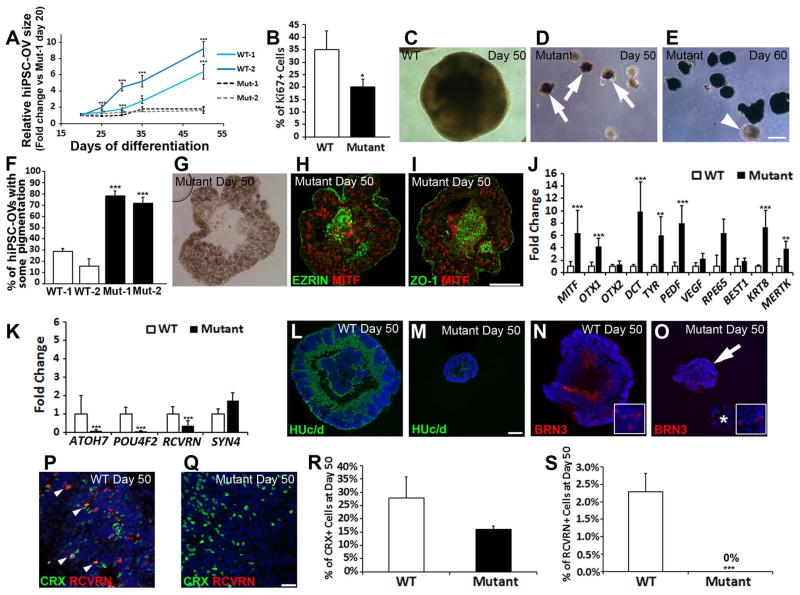
Although (R200Q)VSX2 and WT hiPSC-OVs were similar in size when first isolated (Fig. 2A,B), growth of mutant hiPSC-OVs lagged behind WT hiPSC-OVs, resulting in drastically smaller mutant hiPSC-OVs by d50 in all lines tested (Fig. 3A). To compare levels of proliferation, Ki-67 was quantified via stereology at d30, which revealed fewer Ki-67+ cells in mutant hiPSC-OVs (Fig. 3B; mutant: 20.09±3.18% vs. WT: 35.12±7.34%; P=0.042; n=7 hiPSC-OVs for both groups), implicating reduced proliferation as a contributing factor in the growth retardation of mutant hiPSC-OV cultures. In addition to demonstrating drastic size differences between (R200Q)VSX2 and WT hiPSC-OVs at d50 (Fig. 3C,D), light microscopy revealed that the majority of mutant hiPSC-OVs acquired pigmentation by d50 (Fig. 3D). By d60, dense pigmentation throughout most (R200Q)VSX2 hiPSC-OVs was evident (Fig. 3E), although some never pigmented (Fig. 3E, arrowhead). By comparison, WT hiPSC-OVs infrequently became pigmented (Fig. 3F; Mut-1: 78.18±4.49%, Mut-2: 71.88±4.89% vs. WT-1: 28.66±2.96%, WT-2: 15.59±6.63%; p<0.001). Furthermore, when hiPSCs were maintained as adherent cultures in RPE promoting conditions, increased pigmentation was observed in (R200Q)VSX2 vs. WT hiPSC cultures, further suggesting a propensity for mutant hiPSCs to produce RPE (Supporting Information Fig. S4).
Figure 3. The (R200Q)VSX2 mutation leads to a microphthalmia-like phenotype in hiPSC-OVs and an increase in RPE differentiation at the expense of neural retina progeny.
(A) Starting with equivalently sized hiPSC-OVs at day 20, WT hiPSC-OVs, but not their mutant counterparts, experienced significant growth over time (two hiPSC lines tested per group: WT-1, -2 and Mut-1, -2; data was normalized to the Mut-1 line at day 20). (B) At day 30, proliferation was reduced in mutant hiPSC-OVs, as determined by stereological counts of Ki-67+ nuclei. (C, D) Light microscopy demonstrated the typical size and appearance of WT (C) and mutant (D) hiPSC-OVs at day 50. WT hiPSC-OVs grew considerably larger than mutant hiPSC-OVs, and many mutant hiPSC-OVs developed pigmentation (arrows) by day 50. (E) Pigmentation of mutant hiPSC-OVs became more pronounced over time (day 60 shown), although some mutant hiPSC-OVs remained nonpigmented indefinitely (arrowhead). Scale bar in panel E = 250 μm (also applies to panels C and D). (F) Quantification of the percentage of WT and mutant hiPSC-OVs containing pigmentation at day 50. (G) Brightfield image of a sectioned pigmented mutant hiPSC-OV at day 50. (H, I) Immunostaining of day 50 mutant hiPSC-OV sections showed that the majority of cells expressed the RPE transcription factor MITF, as well as the RPE markers EZRIN and the tight junction protein ZO-1. Scale bar in panel I = 100 μm (also applies to panels G and H). (J, K) qRT-PCR analysis comparing the expression levels of RPE (J) and selected neural retina (K) genes in WT vs. mutant hiPSC-OVs at day 55 (n=2 separate hiPSC lines for both groups with 3 biological replicates per line). (L–O) Immunostaining revealed a higher prevalence of HUc/d+ postmitotic neurons (L, M) and BRN3+ ganglion cells (N, O) in WT vs. mutant hiPSC-OV sections at day 50. The asterisk in panel O demarcates a pigmented hiPSC-OV lacking BRN3 expression. Insets in panels N and O demonstrate the nuclear expression of BRN3. Scale bar in panel M = 100 μm (applies to panels L–O). (P,Q) CRX was expressed in both the WT (P) and mutant (Q) hiPSC-OVs at day 50. However, the photoreceptor marker RCVRN was only expressed in WT hiPSC-OVs at this time point. Scale bar in panel Q = 50 μm. (also applies to panel P). (R, S) Stereological analysis of CRX+ (R) and RCVRN+ (S) nuclei from nonpigmented spheres at day 50. *p < 0.05, ** p < 0.01, ***p < 0.001.
To analyze differences in gene and protein expression between (R200Q)VSX2 and WT hiPSC-OVs, qRT-PCR and ICC were performed. Nuclei within pigmented mutant hiPSC-OVs expressed MITF (Fig. 3G–I) and other characteristic RPE proteins including EZRIN and ZO-1 (Fig. 3H,I). In addition, numerous RPE signature genes were significantly upregulated in mutant hiPSC-OVs compared to WT hiPSC-OVs (Fig. 3J). Notably, expression of MITF, a direct target of VSX2-mediated gene repression [15, 20, 33] was upregulated in mutant vs. WT hiPSC-OVs at d55, as were the MITF targets DCT and TYR, and the RPE specification gene OTX1 [34–36]. Conversely, qRT-PCR analysis showed decreased NR gene expression in (R200Q)VSX2 hiPSC-OVs compared to WT hiPSC-OVs. This included reduced expression of the ganglion cell genes ATOH7 and POU4F2 and the photoreceptor gene RCVRN, although the horizontal cell gene SYN4 was unchanged (Fig. 3K). These findings were consistent with the preferential production of RPE over NR progeny in (R200Q)VSX2 hiPSC-OVs relative to WT hiPSC-OVs.
Photoreceptor maturation is delayed in (R200Q)VSX2 hiPSC-OVs
While differentiation toward an RPE fate predominated in (R200Q)VSX2 hiPSC-OVs, some mutant OVs maintained a NR appearance. These nonpigmented mutant hiPSC-OVs could be manually separated from pigmented hiPSC-OVs, facilitating comparison of NR differentiation in mutant and WT hiPSC lines. Immunocytochemistry analysis performed on d50 hiPSC-OVs (Fig. 3L–Q) showed that HUc/d and BRN3, markers for postmitotic neurons and ganglion cells, respectively, were expressed in many cells within WT hiPSC-OVs (Fig. 3L,N) and in some centrally located cells within the smaller, nonpigmented mutant hiPSC-OVs (Fig. 3M,O). However, pigmented mutant hiPSC-OVs were devoid of such cells (Fig. 3O, asterisk). Also by d50, CRX+ cells were present in both mutant and WT hiPSC-OV cultures (Fig. 3P,Q), but there was a striking difference in the expression of RCVRN. A small number of CRX+ cells co-expressed RCVRN in WT hiPSC-OVs at d50 (Fig. 3P, arrowheads), whereas no RCVRN staining was found in mutant hiPSC-OVs at this time point (Fig 3Q,S; WT: 2.29±0.52% vs. mutant: 0%, p<0.001, n=8 for both groups). CRX+ cells trended higher in WT vs. mutant hiPSC-OV cultures at d50, but did not reach statistical significance (Fig. 3R; WT: 27.90±7.95%, mutant: 15.93±1.32%, p=0.091, n=9 for both groups).
To assess the role of VSX2 later during NR differentiation, nonpigmented (R200Q)VSX2 and WT hiPSC-OVs were differentiated for an additional 30 days (d80). As with the earlier time points, CRX+ cells were present at d80 in mutant and WT hiPSC-OVs (Fig. 4A–D; Supporting Information Fig. 5A,B). However, unlike mutant hiPSC-OVs, WT hiPSC-OVs also demonstrated a large increase in RCVRN expression at d80 (Fig. 4A–D; Supporting Information Fig. S5A,B). Stereological analysis revealed approximately half the number of CRX+ cells in mutant compared to WT hiPSC-OVs (Fig. 4E; WT: 26.09±4.07% vs. mutant: 12.83±1.73%, p=0.005, n=10 for both groups), while the number of RCVRN+ photoreceptors was ~100-fold reduced in mutant hiPSC-OV cultures (Fig. 4F; WT: 12.08±1.99% vs. mutant = 0.12±0.04%, p<0.001, n=9 for both groups). qRT-PCR analysis confirmed a significant reduction in CRX and RCVRN gene expression in (R200Q)VSX2 vs. WT hiPSC-OVs (Fig. 4G) . In addition, expression of rod-specific NRL, s-cone opsin (OPN1SW), and the phototransduction gene PDE6B were greatly diminished in mutant hiPSC-OVs.
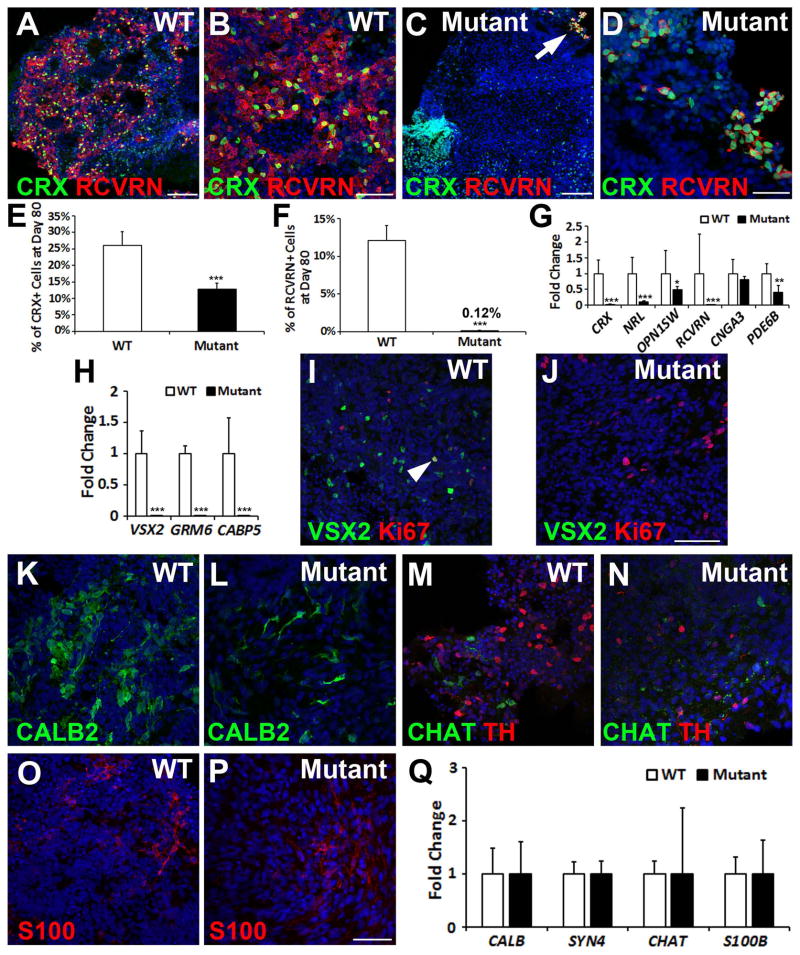
Figure 4. The (R200Q)VSX2 mutation delays photoreceptor maturation and prevents bipolar cell differentiation.
(A–D) At day 80, numerous CRX+ cells co-expressed RCVRN in WT hiPSC-OVs (A, B), whereas CRX+ nuclei in mutant hiPSC-OVs rarely co-expressed RCVRN (C, D) (arrow in panel C is shown at a higher magnification in panel D). Scale bars = 100 μm (A, C); 50 μm (B, D). (E) Stereological counts confirmed a reduction in the percentage of CRX+ nuclei in mutant hiPSC-OVs at day 80. (F) Also at day 80, mutant hiPSC-OVs showed a ~100-fold reduction in the percentage of RCVRN+ nuclei relative to WT hiPSC-OVs. Of note, all pigmented hiPSC-OVs were excluded from these studies. (G) qRT-PCR analysis revealed reduced levels of expression for most photoreceptor genes in mutant hiPSC-OVs compared to WT hiPSC-OVs at day 80. (H) The bipolar cell genes VSX2, GRM6, and CABP5 were not expressed in mutant hiPSC-OVs, as shown by qRT-PCR. (I) Immunocytochemical analysis demonstrated persistence of VSX2+ cells in WT hiPSC-OVs at day 80, most of which were Ki-67 negative, indicative of bipolar cell differentiation. However, occasional VSX2+/Ki-67+ NRPCs were also present (arrowhead). (J) In contrast, VSX2 was no longer expressed in mutant hiPSC-OVs at day 80, confirming a lack of bipolar cell generation. Scale bar in panel J = 100 μm (also applies to panel I). (K–P) Both WT and mutant hiPSC-OVs produced neural retina cell types other than bipolar cells, including CALB2+ neurons (K,L), CHAT+ and TH+ amacrine cells (M,N), and S100+ glia (O,P). Scale bar in panel P = 50 μm (also applies to K–O). (Q) qRT-PCR analysis comparing neural retina gene expression in WT vs. mutant hiPSC-OVs at day 80. *p < 0.05, ** p < 0.01, ***p < 0.001.
Bipolar cell genesis is absent in (R200Q)VSX2 hiPSC-OVs
In distinct contrast to WT hiPSC-OVs, VSX2 expression was no longer detectable by qRT-PCR in (R200Q)VSX2 hiPSC-OVs by d80 (Fig. 4H); similarly, the bipolar cell-specific genes GRM6 and CABP5 were not expressed in mutant hiPSC-OVs. Immunocytochemical analysis performed on WT hiPSC-OVs at d80 confirmed VSX2 expression (Fig. 4I, Supporting Information Fig. S5C). The majority of these cells were negative for Ki-67, consistent with bipolar cell identity in the adult human retina (Supporting Information Fig. S3). Mutant hiPSC-OVs, on the other hand, completely lacked VSX2 expression at d80 of differentiation (Fig. 4J, Supporting Information Fig. S5D).
(R200Q)VSX2 and WT hiPSC-OV cultures were also examined for the production of other major NR cell types at d80. CALB2 (also known as Calretinin), a gene expressed in amacrine and ganglion cells, was expressed in all hiPSC-OV cultures (Fig. 4K, L; Supporting Information S5E,F), as were specific subtypes of amacrine cells, including cholinergic (CHAT+) and dopaminergic (TH+) amacrine cells (Fig. 4M,N, Supporting Information Fig. S5G,H). S100+ glia were also present in both mutant and WT hiPSC-OVs (Fig. 4O,P, Supporting Information Fig. S5I,J). qRT-PCR analysis confirmed gene expression of numerous NR cell types in both mutant and WT hiPSC-OV cultures (Fig. 4Q). Together, these results demonstrate that all major classes of NR cells are produced in (R200Q)VSX2 hiPSC-OVs, with the exception of bipolar cells. Remaining Ki-67+ cells in mutant and WT hiPSC-OVs co-expressed PAX6, indicating the continued presence of NRPCs and/or proliferative glia at d80 (Supporting Information Fig. S5K,L).
Exogenous expression of wildtype VSX2 early during retinal differentiation partially rescues the (R200Q)VSX2 hiPSC-OV phenotype
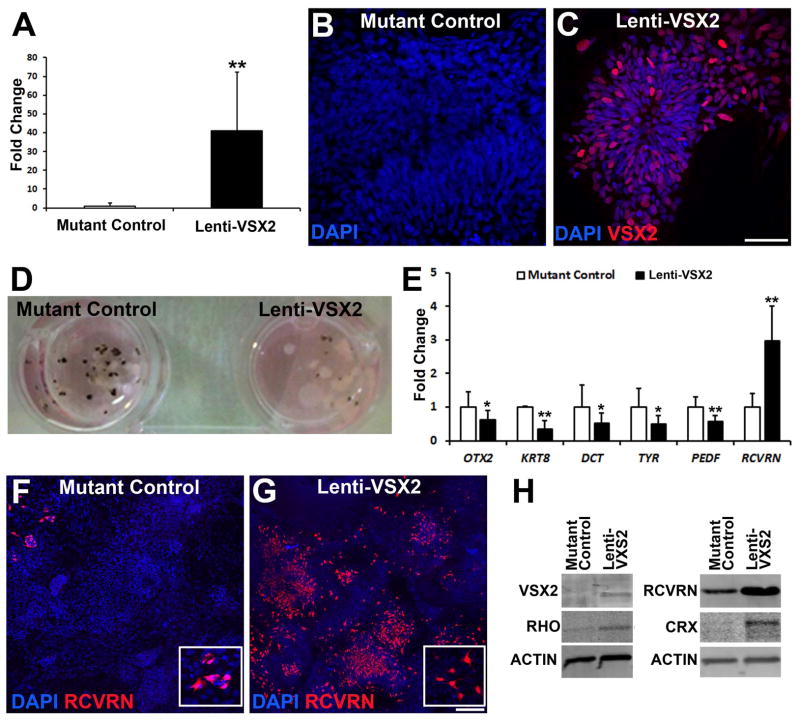
In an effort to rescue the effects of the (R200Q)VSX2 mutation on later stages of retinal development, we exogenously expressed WT VSX2 in differentiating mutant hiPSCs beginning at d14, a time point corresponding to the initial expression of VSX2 in WT hiPSC-OVs (data not shown). (R200Q)VSX2 hiPSC cultures were infected with a lentiviral VSX2 overexpression construct and left adherent to the plates, since infected hiPSC-OVs did not survive well after lifting. As a control, an equal number of culture wells within the same six well plate were infected with a lenti-GFP overexpression construct. Lenti-VSX2 overexpression led to a large and sustained upregulation of VSX2 when compared to control (Fig. 5A). By d80, VSX2 was absent in lenti-GFP control mutant hiPSC cultures (Fig. 5B); however, VSX2 remained abundant at this time in mutant hiPSCs overexpressing WT VSX2 (Fig. 5C), indicating continued expression of the transgene. The phenotypic consequences of VSX2 overexpression in (R200Q)VSX2 cultures were apparent by d70, when a drastic reduction in pigmentation could be seen compared to lenti-GFP control cultures (Fig. 5D). Decreased pigmentation corresponded with reduced expression of RPE signature genes and increased RCVRN expression (Fig. 5E). Indeed, mutant cultures infected with WT lenti-VSX2 generated large numbers of RCVRN+ photoreceptors as opposed to the sparse RCVRN expression seen in lenti-GFP infected mutant hiPSCs (Fig. 5F,G; Supporting Information Fig. S6A–D). Western blot analysis at d100confirmed VSX2 overexpression, as well as increased expression of CRX, RCVRN, and Rhodopsin (RHO) in WT lenti-VSX2 infected (R200Q)VSX2 hiPSC cultures (Fig. 5H). However, expression of mature bipolar cell markers (GRM6 and CABP5) was not restored in lenti-VSX2 transduced mutant hiPSC-OVs. These data suggest that exogenous expression of WT VSX2 can partially reverse the mutant phenotype of (R200Q)VSX2 hiPSCs-OVs, and further support a role for VSX2 in photoreceptor development.
Figure 5. Exogenous expression of WT VSX2 reduces RPE production and enhances photoreceptor development in (R200Q)VSX2 hiPSC-OVs.
(A) Overall VSX2 expression was significantly increased in day 70 mutant hiPSC-OVs after transduction at day 14 with a WT VSX2-expressing lentiviral construct (Lenti-VSX2). Mutant control hiPSC-OVs were transduced at day 14 with a lenti-GFP construct. (B,C) At day 80, VSX2 was absent in lenti-GFP infected mutant hiPSC-OV controls (B), but remained expressed in mutant hiPSC-OV cells infected with lenti-WT VSX2 (C). Scale bar = 50 μm in panel C (also applies to panel B). (D) By day 70, exogenous expression of WT VSX2 in mutant hiPSC-OVs resulted in reduced pigmentation compared to lenti-GFP infected mutant hiPSC-OV controls. (E) qRT-PCR analysis at day 70 demonstrated reduced expression of characteristic RPE genes and increased expression of the photoreceptor gene RCVRN in lenti-WT infected vs. lenti-GFP infected mutant hiPSC-OVs. (F) At day 80, few RCVRN+ photoreceptors were present in lenti-GFP infected mutant hiPSC-OV control cultures. (G) Exogenous expression of WT VSX2 rescued RCVRN expression in mutant hiPSC-OVs. Insets in panels F and G demonstrate the cytoplasmic nature of RCVRN expression. Scale bar in panel G = 100 μm (also applies to panel F). (H) Western blot analysis at day 100 revealed increased expression of the photoreceptor genes RHO, CRX, and RCVRN in lenti-WT VSX2 infected mutant hiPSC-OVs when compared to lenti-GFP infected mutant hiPSC-OV control cultures. *p < 0.05, ** p < 0.01.
Transcriptome analysis reveals altered expression of developmental signaling molecules in (R200Q)VSX2 hiPSC-OVs
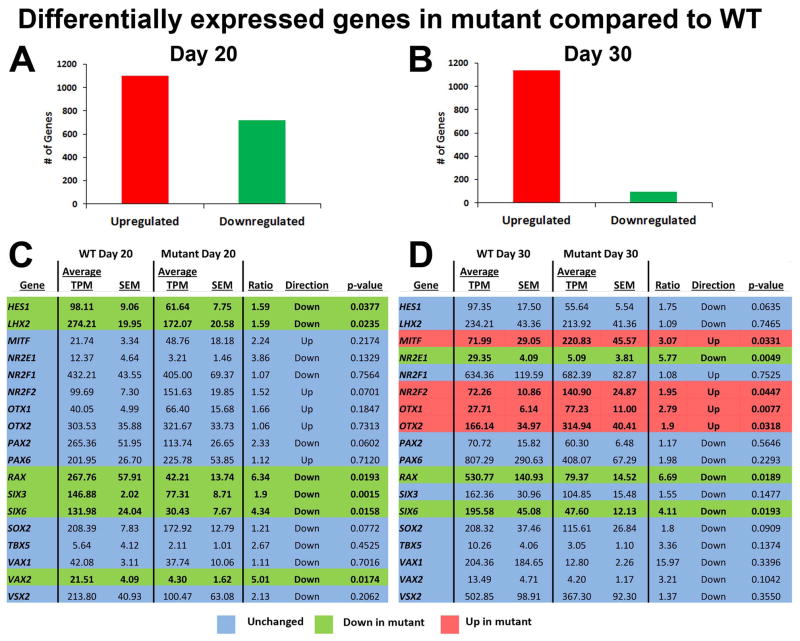
Results presented thus far support a conserved role for VSX2 in differentiating hiPSCs that involves early maintenance and proliferation of NRPCs and later production and/or development of bipolar cells and photoreceptors. To further probe the impact of (R200Q)VSX2 on gene expression during early retinal differentiation in hiPSCs, comparative RNAseq analysis was performed on d20 and d30 mutant and WT hiPSC-OVs (n=3 for both at d20, n=4 at d30). These time points in vitro correspond to the optic vesicle and early optic cup stages of differentiation in the developing human embryo, respectively [37, 38]. Expression analysis (Supporting Information Table 3) on d20 hiPSC-OVs revealed 1818 differentially expressed genes (1098 upregulated and 720 downregulated genes in mutant vs. WT hiPSC-OVs, Fig. 6A). In d30 hiPSC-OVs, 1236 genes were differentially expressed, with the vast majority (92%) being increased in mutant cultures (1140 upregulated and 96 downregulated genes in mutant vs. WT hiPSC-OVs, Fig. 6B).
Figure 6. Comparative RNAseq analysis of WT and (R200Q)VSX2 hiPSC-OVs: differences in expression of genes involved in early retinal differentiation.
(R200Q)VSX2 mutant hiPSC-OVs were subjected to high throughput transcriptome analysis. Differential expression analysis performed with GeneSifter software revealed the total number of genes significantly changed (p < 0.05) in mutant hiPSC-OVs compared to WT at day 20 (A) and day 30 (B). At day 30, the majority of such genes were upregulated in mutant hiPSCs-OVs, consistent with a transcriptional repressor function for VSX2. Expression levels of selected genes involved in retinal differentiation were also compared between the two groups at day 20 (C) and day 30 (D), with genes up- or downregulated in mutant hiPSC-OVs highlighted in red or green, respectively. Unchanged genes are highlighted in blue. MITF, NR2F2, OTX1, and OTX2, transcription factors involved in RPE development, were upregulated in mutant hiPSC-OVs at day 30. TPM=transcripts per million, SEM=standard error of the mean.
To compare expression of early retinal developmental genes in mutant and WT VSX2 hiPSC-OVs, transcription factors critical for OV and early optic cup differentiation [39–42] were analyzed using RNAseq data (Fig. 6C,D). To verify RNAseq data, qRT-PCR analysis was performed on a panel of genes expressed in the early retina (Supporting Information Fig. S7). Consistent with PCR and immunocytochemistry results, there were no significant differences in expression of the OV transcription factors VSX2 and PAX6, or the early OV/RPE marker MITF, in d20 mutant vs. WT hiPSC-OVs (Fig. 6C). However, other markers of eye field and early NR differentiation were significantly reduced in mutant OVs, including RAX, SIX3, SIX6, and LHX2 (Fig. 6C), with mutant hiPSC-OVs maintaining significant downregulation of RAX and SIX6 at d30 relative to WT hiPSC-OVs (Fig. 6D). Also at d30, genes involved in early RPE fate determination, including MITF, NR2F2, OTX1, and OTX2, were upregulated in mutant hiPSC-OVs (Fig. 6D). Conversely, expression of the ganglion cell genes ATOH7 and POU4F2 were reduced in mutant hiPSC-OVs (Supporting Information Fig. 7). Combined, these findings provide further evidence for a relative NR-to-RPE fate switch resulting from the (R200Q)VSX2 mutation.
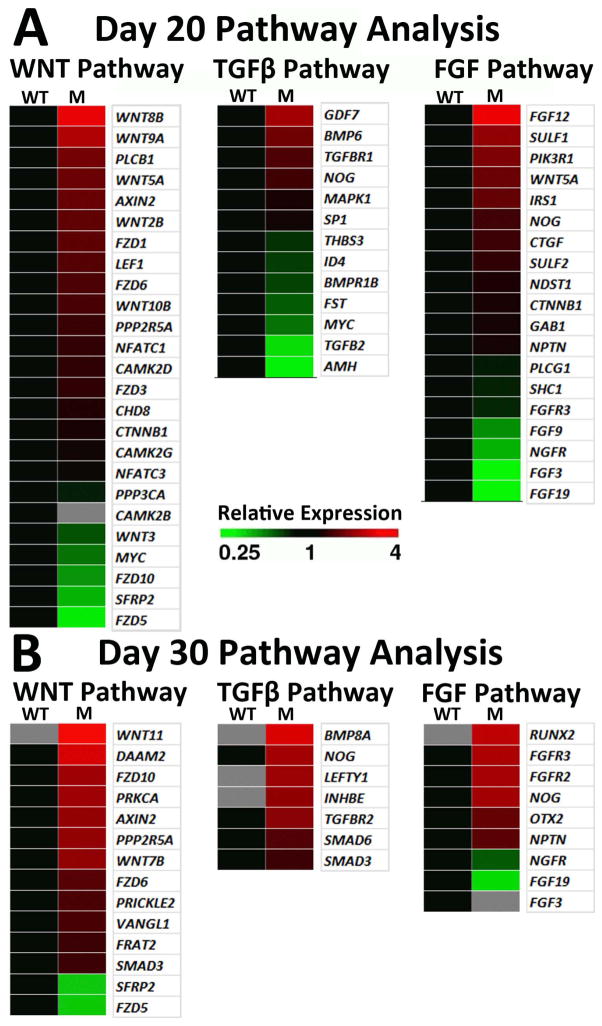
Comparative RNAseq analysis also demonstrated that key signaling pathways were altered in (R200Q)VSX2 hiPSC-OVs. KEGG and Gene Ontology search functions revealed that the majority of WNT pathway genes, which are known to be important for RPE specification and differentiation [43–45], were upregulated at d20 and d30 of differentiation in mutant vs. WT hiPSC-OVs (Fig. 7A, B). Numerous WNTs, FZD receptors, and downstream effectors were differentially regulated in the VSX2 mutant, indicating robust engagement of WNT signaling (Fig. 7A,B). In addition, significant increases in TGFβ family gene expression was seen in mutant vs. WT hiPSC-OV cultures at d30 (Fig. 7B), consistent with the pro-RPE role of this signaling pathway [41, 46]. Unlike WNT and TGFβ signaling pathway genes, the pro-NR FGFs 3, 9, and 19 [47–50] were downregulated in mutant vs. WT hiPSC-OVs at d20 (Fig. 7A), with FGF3 and 19 also decreased or absent at d30 (Fig. 7B). Of note, two FGF receptors that bind FGF3, 9, and/or 19, FGFR2 and FGFR3, were upregulated at d30 in the (R200Q)VSX2 hiPSC-OVs, suggesting a compensatory response for decreased FGF expression in the mutant. The reciprocal downregulation of pro-NR FGFs and upregulation of pro-RPE WNT and TGFβ family genes suggest putative mechanisms for the RPE cell fate bias seen in (R200Q) VSX2 hiPSC-OVs.
Figure 7. Comparative RNAseq signaling pathway analysis suggests potential mechanisms for the RPE cell fate bias in (R200Q)VSX hiPSC-OVs.
WNT, TGFβ, and FGF pathway analyses were performed at day 20 (A) and day 30 (B) of differentiation. Grey squares denote genes that were not expressed. Up- (red) and down- (green) regulation of mutant hiPSC-OV gene expression is shown relative to WT hiPSCs at the same time point. Pathway analyses were performed with GeneSifter software, using KEGG search terms for WNT and TGFβ, while FGF pathway analysis was performed with Gene Ontology (GO) search terms.
Discussion
In this report, we sought to determine the utility of patient-specific hiPSCs as platforms to study intrinsic regulators of retinogenesis, beginning at developmental time points inaccessible to investigation in humans. The homeodomain transcription factor VSX2 is an appealing focus for such a study, as it is the earliest specific marker of the neuroretinal lineage and is expressed in both multipotent NRPCs and late-born bipolar cells [13, 14]. Thus, its potential influence spans the breadth of human retinal development. In addition, VSX2+ NRPCs comprise the vast majority of the cells initially present in hiPSC-OVs; therefore, all experiments were assured of a consistent, highly enriched starting population of cells expressing the protein of interest. Lastly, VSX2 has demonstrated certain conserved functions across multiple vertebrate species, providing a reference to compare findings from patient-specific hiPSCs. The use of information from animal models to predict results in hiPSCs is bolstered by the fact that VSX2 mutations cause severe microphthalmia in both mice and humans [15, 17, 18, 20, 23–25, 51]. However, other observed effects of VSX2 mutations on mammalian retinogenesis, such as perturbed maintenance of the NR and RPE domains and abnormal development of photoreceptors and bipolar cells, have not been confirmed in humans.
The microphthalmic patient recruited for this study harbored a missense mutation in VSX2 that altered the Arg200 residue in its homeodomain region [24]. Amino acid substitutions at this position were shown in animal studies and/or heterologous overexpression systems to abolish DNA binding and transcriptional repression without affecting VSX2 expression [23, 24]. Consistent with this last finding, OVs from both the patient and WT sibling hiPSC lines initially contained equivalent amounts of VSX2+ NRPCs. Also as expected, no differences were observed in mutant and WT hiPSC cultures prior to the onset of VSX2 expression. However, with further differentiation, mutant hiPSC-OVs displayed developmental abnormalities similar to those found in the orJ (VSX2−/−) and (R200Q)Vsx2 mouse models [15, 17, 18, 20, 23]. Of note, (R200Q)Vsx2 mice phenocopy orJ null mutant mice in most respects [23], further demonstrating the critical importance of the Arg200 residue for Vsx2 functions.
The first observable difference between (R200Q)VSX2 and WT hiPSC-OVs was the profoundly slower growth of the mutant OVs. This microphthalmia-like phenotype arose at least in part from decreased NRPC proliferation, a finding also seen in orJ and (R200Q)Vsx2 mice [17, 23]. The mechanism underlying the reduced proliferation likely involves the absence of VSX2-mediated transcriptional repression of MITF, a basic helix-loop-helix transcription factor that, among other important roles, can act to limit cell division [15, 33]. In (R200Q)VSX2 hiPSC-OVs, elevation in MITF transcript levels occurred following the onset of VSX2 expression, which was also when differences in growth rates between mutant and WT hiPSC-OVs first became apparent. Thus, it is plausible that VSX2 influences NRPC proliferation in hiPSC-OVs in much the same manner as it does in the developing embryo.
In addition to growth and proliferation disparities, (R200Q)VSX2 and WT hiPSC-OVs differed in their fate potential. When cultured in isolation, excised OVs from developing vertebrate embryos possess a default tendency to generate NR cell types as opposed to RPE [52]. A similar trend has been noted for hESC- and hiPSC-OVs that are separated from mixed stem cell cultures at early stages of differentiation and maintained in minimal medium without RPE-promoting factors [2, 4, 12]. However, unlike all WT hiPSC-OVs that we have studied to date, (R200Q)VSX2 hiPSC-OVs preferentially differentiated into RPE. This finding is similar to that published with Vsx2 mutant animal models, including the (R200Q)Vsx2 mouse, where ectopic pigmented RPE cells were found in the NR, albeit with variable frequency depending upon the background mouse strain [15, 20, 23]. In these mouse models, lack of Vsx2-mediated repression of Mitf was thought to contribute to the encroachment of RPE into the NR domain, since Mitf also regulates differentiation and development of RPE cells and is responsible for transcription of melanogenesis genes [15, 33, 35, 45]. Consistent with this hypothesis, upregulation of MITF expression in (R200Q)VSX2 hiPSC-OVs correlated with higher RPE gene expression levels and increased pigmentation. However, not all findings in (R200Q)VSX2 hiPSC-OVs mirrored those of the (R200Q)Vsx2 mouse model. For example, unlike the (R200Q)Vsx2 mouse [23], OVs from (R200Q)VSX2 hiPSCs demonstrated an upregulation of the pro-RPE transcription factor gene OTX1. Even so, it is remarkable how closely the mutant hiPSC-OV phenotype mimicked that of the developing (R200Q)Vsx2 mouse OV given that the latter is subject to influences from surrounding tissues that are unaccounted for in culture.
Although RPE production was favored in (R200Q)VSX2 hiPSC-OV cultures, differentiation toward NR still occurred. All major NR cell types were produced in mutant hiPSC-OV cultures except bipolar cells, a hallmark of Vsx2 mutant mouse models [15, 17–19, 23, 53]. This finding reflects a specific role for VSX2 in the production of human bipolar cells. Of note, the total absence of bipolar cell genesis was unique to the (R200Q)VSX2 hiPSC lines used in this study, as all other hPSC lines we have examined thus far generate this NR cell type ([2, 4], data not shown). Developmental defects were also seen in other NR cell types. In particular, photoreceptor maturation was profoundly delayed in (R200Q)VSX2 hiPSC-OVs, as evidenced by the near absence of RCVRN expression in mutant hiPSC-OVs at d80 of differentiation, a time when RCVRN+ photoreceptors abound in WT cultures. This finding was consistent with the overall lag in neurogenesis found in the (R200Q)Vsx2 mouse, including an absence of Otx2+ cone photoreceptor precursors at E12.5, suggestive of delayed photoreceptor differentiation [23]. Similar findings have been observed in the orJ mouse [54].
Additional evidence in support of multiple roles for VSX2 in hiPSC-OV differentiation was provided by overexpressing WT VSX2 in (R200Q)VSX2 hiPSC cultures. In addition to reversing the RPE differentiation bias of mutant hiPSC cultures, lentiviral overexpression of WT VSX2 restored the timing of CRX, RCVRN, and RHODOPSIN expression to that of WT hiPSC-OVs. However, exogenous expression of WT VSX2 was not sufficient to re-establish bipolar cell genesis in mutant hiPSC-OVs. The reason for this shortcoming is unclear, but may be due to nonphysiological regulation and/or premature silencing of the VSX2 transgene.
Once we validated the use of hiPSCs to study the role of VSX2 in human retinal development, we utilized the scalability of our system to generate the quantity of hiPSC-OVs needed for high throughput RNA sequencing. Among the gene transcript levels altered in mutant hiPSC-OVs, almost all were upregulated relative to WT hiPSC-OVs by d30, consistent with VSX2 functioning as a transcriptional repressor during early human retinogenesis [15, 17–23]. RNAseq analysis also implicated several key extracellular signaling pathways in the NR-to-RPE fate shift observed in (R200Q)VSX2 hiPSC-OVs. Increased expression of pro-RPE WNT and TGFβ pathway genes was clearly evident in mutant vs. WT hiPSC-OVs, as was a corresponding decrease in specific FGFs involved in early NR determination (e.g., FGF3, 9, and 19). The effects of (R200Q)VSX2 on FGF, WNT, and TGFβ family gene expression in hiPSC-OVs suggest that VSX2 acts upstream of these key signaling molecules to aid in the establishment and/or maintenance of the NR domain. The fact that exogenous administration of the TGFβ family member Activin-A can override the default NR program in isolated WT hiPSC-OVs and induce RPE differentiation supports this notion [2].
Since the hiPSC-OVs used in this study were isolated and grown in minimal medium, our results only take into account autonomous influences on retinal cell differentiation. In contrast, OVs within the developing embryo are exposed to factors secreted from periocular mesenchyme and surface ectoderm [39–42]. The relative contributions of intrinsic and extrinsic signaling in the establishment of the NR and RPE domains in vivo remains the subject of investigation, but our data provides further evidence that human OVs expressing functional VSX2 preferentially differentiate along the NR lineage, and that little or no outside instruction is required for this process to occur.
Ultimately, the potential to generate patient-specific hiPSC-OVs not only offers an opportunity to study the dynamics of early human retinal development, but also to use that knowledge to improve methods of RPE or NR cell production. For example, results herein show that antagonizing or eliminating VSX2 function in hiPSCs leads to more efficient generation of RPE, a cell type currently in clinical trials for the treatment of certain blinding disorders [55]. On a broader level, the observation that hiPSC-OVs are subject to many of the same principles and processes that govern OV development in vivo supports the authenticity of retinal cell products derived from hiPSCs.
Conclusion
hiPSC-OVs generated from a patient with an (R200Q)VSX2 mutation and an unaffected sibling provided a unique and flexible platform to model early human retinogenesis in vitro. More specifically, the high degree of fidelity shown between OVs from (R200Q)VSX2 hiPSCs and previously published Vsx2 mutant mouse models offers compelling evidence for the use of hiPSCs to study cell intrinsic events underlying early human retinal development. This validation may expand the scope of future hiPSC studies beyond the confines of human conditions that have well-established animal models.
Supplementary Material
Supporting Information Figure S1: Generation and characterization of WT and (R200Q)VSX2 hiPSC clones. (A) Three WT and three (R200Q)VSX2 mutant clonal hiPSCs lines were characterized (WT-1, -2, -3 and Mut-1, -2, -3). All hiPSC lines formed colonies that expressed the pluripotency genes NANOG, TRA1-60, OCT4, and SSEA-4. Scale bar = 200 μm. (B) The vast majority of hiPSCs expressed pluripotency markers as shown by representative FACS analyses of WT and mutant hiPSC lines. (C) All hiPSC lines were sequenced and verified for the presence or absence of the appropriate VSX2 homeodomain mutation at codon 200. The guanine to adenine switch in codon 200 leads to an arginine to glutamine missense mutation (R200Q) in the mutant hiPSC lines (representative sequencing analysis shown for one WT and one mutant hiPSC line). (D) All hiPSC lines were karyotypically normal (representative G-banding analysis shown). (E) Genomic DNA PCR analysis demonstrated genomic integration of reprogramming vectors in mutant (M) and WT hiPSCs (one reprogramming transgene from each bicistronic vector was analyzed). Reprogramming vector plasmids (+ control) served as positive PCR amplification controls, while WA09 human embryonic stem cells (hESCs) and peripheral blood mononuclear cells (PBMCs) served as negative controls. (F) Characteristic pluripotency genes were expressed in WT and mutant hiPSCs as shown by RT-PCR. hESCs and PBMCs served as positive and negative PCR amplification controls, respectively. (G) Reprogramming factor transgenes were silenced and endogenous pluripotency genes were upregulated in hiPSCs. Reprogramming plasmids and hESCs served as positive controls for transgene-mediated and endogenous pluripotency gene amplification, respectively. (H) Teratoma analysis confirmed the competency of each hiPSC line to produce all three germ layers, including gut and respiratory epithelium (endoderm), cartilage (mesoderm), and neural tissue (ectoderm) (one representative WT and mutant hiPSC line shown). Scale bar = 100 μm.
Supporting Information Figure S2: Schematic depicting the differentiation protocol and the cell types present at the time points analyzed in this study. Of note, while small, superficial patches of pigmented RPE develop in a minority of WT hiPSC-OVs, most mutant hiPSC-OVs become uniformly pigmented with time. However, some mutant hiPSC-OVs remain pigmentation-free, permitting comparisons of NR differentiation in mutant vs. WT hiPSC-OVs. RPC: retinal progenitor cell, RGC: retinal ganglion cell, PR: photoreceptor.
Supporting Information Figure S3: VSX2 expression in human prenatal and adult retina. (A) VSX2 was expressed in the neuroblastic layer (NBL) of day 85 human prenatal retina. Numerous VSX2+ cells co-expressed Ki-67, indicative of proliferating NRPCs. (B) In a 71 year-old adult retina, VSX2 expression was confined to bipolar cells within the inner nuclear layer (INL). These adult VSX2+ cells were non-proliferative (i.e., Ki-67 negative). ONL = outer nuclear layer; GCL = ganglion cell layer. Scale bar = 50 μm.
Supporting Information Figure S4: (R200Q)VSX2 hiPSCs differentiated as adherent cultures displayed greater pigmentation than WT hiPSCs. Instead of lifting neural colonies at day 16 to generate OVs, (R200Q)VSX2 and WT hiPSCs were left adherent to culture plates to promote RPE differentiation. A representative six well plate is shown at day 60 of differentiation (top three wells = WT hiPSCs; lower three wells = (R200Q)VSX2 mutant hiPSCs).
Supporting Information Figure S5: Examination of additional WT and mutant hiPSC lines confirmed the effects of the (R200Q)VSX2 mutation on photoreceptor maturation and bipolar cell differentiation. (A) Similar to the WT-1 hiPSC line shown in Fig. 4A and B, the WT-3 hiPSC line produced numerous CRX+/RCVRN+ photoreceptors by day 80. (B) The Mut-3 hiPSC line also produced CRX+ cells; however, RCVRN+ cells were notably absent. (C) VSX2+ nuclei were predominately Ki-67-negative, indicative of bipolar cell differentiation in WT-3 hiPSC-OVs. (D) VSX2 expression was absent in Mut-3 hiPSC-OVs by day 80, although some Ki-67 cells persisted. (E–J) WT-3 and Mut-3 hiPSC-OVs produced other neural retina cell types, including CALB2+ neurons (E,F), TH+ amacrine cells (G,H), and S100+ glia (I,J). (K,L) Most of the Ki-67+ cells co-labeled with PAX6, indicating the continued presence of neural retina progenitors cells and/or proliferative glia in both the WT-3 (K) and Mut-3 lines (L). Scale bar = 50 μm for panel B (also applies to panel A) and 50 μm for panel L (applies to panels C–L).
Supporting Information Figure S6: Exogenous expression of WT VSX2 in the Mut-2 hiPSC line increased production of CRX+/RCVRN+ photoreceptors. (A) Similar to the Mut-1 (R200Q)VSX2 hiPSC line shown in Fig. 5F, RCVRN expression was absent at day 100 in Mut-2 (R200Q)VSX2 hiPSCs infected at day 14 with a lenti-GFP control vector (mutant control). (B) After infection at day 14 with lenti-WT VSX2 (Lenti-VSX2), Mut-2 hiPSC-OVs expressed numerous CRX+/RCVRN+ photoreceptors at day 100, similar to the Mut-1 hiPSC line shown in Fig. 5G. Scale bar in panel B = 100 μm (also applies to panel A). (C) High magnification of CRX+/RCVRN+ cells following exogenous expression of WT VSX2 in Mut-2 hiPSC-OVs. (D) RCVRN+ cells in lenti-WT VSX2 infected mutant-2 hiPSC-OVs were VSX2 negative, showing that the WT VSX2-expressing transgene was downregulated in differentiating photoreceptors. Scale bar in panel D = 50 μm (also applies to panel C).
Supporting Information Figure S7: qRT-PCR validation of RNAseq data. qRT-PCR was performed for selected retinal differentiation and developmental signaling genes at day 20 (A) and day 30 (B) of differentiation and compared to the corresponding RNAseq data for each gene. Results were normalized to WT hiPSC-OV expression data for each gene (denoted by the green dashed line). All genes analyzed demonstrated similar trends with qRT-PCR and RNAseq analyses. *p < 0.05, ** p < 0.01, ***p < 0.001.
Acknowledgments
We thank the patients and their family for their blood sample donations, and Jennifer Bolin and Angela Elwell for technical assistance. This work was funded by NIH R01EY21218 and P30HD03352, the Foundation Fighting Blindness Wynn-Gund Translational Research Acceleration Program, the Retina Research Foundation Emmett A. Humble Distinguished Directorship, the McPherson Eye Research Institute Sandra Lemke Trout Chair in Eye Research, the Muskingum County Community Foundation, and the David and Nancy Walsh Family Fund.
Footnotes
Author contribution summary
M. Joseph Phillips: Conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript
Enio T. Perez: collection and/or assembly of data, final approval of manuscript
Jessica M. Martin: collection and/or assembly of data, final approval of manuscript
Kyle A. Wallace: collection and/or assembly of data, final approval of manuscript
Elizabeth E. Capowski: collection and/or assembly of data, data analysis and interpretation, final approval of manuscript
Ruchira Singh: collection and/or assembly of data, data analysis and interpretation, final approval of manuscript
Lynda S. Wright: collection and/or assembly of data, data analysis and interpretation, final approval of manuscript
Eric M. Clarke: collection and/or assembly of data, final approval of manuscript
Samantha T. Reshel: collection and/or assembly of data, final approval of manuscript
Patrick M. Barney: collection and/or assembly of data, final approval of manuscript
Ron Stewart: collection and/or assembly of data, data analysis and interpretation, final approval of manuscript
Sarah J. Dickerson: Provision of study material or patients, collection and/or assembly of data
Michael M. Miller: Provision of study material or patients, collection and/or assembly of data
E. Ferda Percin: Provision of study material or patients, final approval of manuscript
James A. Thomson: Data analysis and interpretation, final approval of manuscript
David M. Gamm: Conception and design, financial support, data analysis and interpretation, manuscript writing, final approval of manuscript
References
- 1.Meyer JS, Shearer RL, Capowski EE, et al. Modeling early retinal development with human embryonic and induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2009;106:16698–16703. doi: 10.1073/pnas.0905245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer JS, Howden SE, Wallace KA, et al. Optic vesicle-like structures derived from human pluripotent stem cells facilitate a customized approach to retinal disease treatment. Stem Cells. 2011;29:1206–1218. doi: 10.1002/stem.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakano T, Ando S, Takata N, et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 4.Phillips MJ, Wallace KA, Dickerson SJ, et al. Blood-derived human iPS cells generate optic vesicle-like structures with the capacity to form retinal laminae and develop synapses. Invest Ophthalmol Vis Sci. 2012;53:2007–2019. doi: 10.1167/iovs.11-9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osakada F, Jin ZB, Hirami Y, et al. In vitro differentiation of retinal cells from human pluripotent stem cells by small-molecule induction. J Cell Sci. 2009;122:3169–3179. doi: 10.1242/jcs.050393. [DOI] [PubMed] [Google Scholar]
- 6.Lamba DA, McUsic A, Hirata RK, et al. Generation, purification and transplantation of photoreceptors derived from human induced pluripotent stem cells. PLoS One. 2010;5:e8763. doi: 10.1371/journal.pone.0008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mellough CB, Sernagor E, Moreno-Gimeno I, et al. Efficient stage-specific differentiation of human pluripotent stem cells toward retinal photoreceptor cells. Stem Cells. 2012;30:673–686. doi: 10.1002/stem.1037. [DOI] [PubMed] [Google Scholar]
- 8.Boucherie C, Mukherjee S, Henckaerts E, et al. Brief report: self-organizing neuroepithelium from human pluripotent stem cells facilitates derivation of photoreceptors. Stem Cells. 2013;31:408–414. doi: 10.1002/stem.1268. [DOI] [PubMed] [Google Scholar]
- 9.Buchholz DE, Pennington BO, Croze RH, et al. Rapid and efficient directed differentiation of human pluripotent stem cells into retinal pigmented epithelium. Stem Cells Transl Med. 2013;2:384–393. doi: 10.5966/sctm.2012-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maruotti J, Wahlin K, Gorrell D, et al. A simple and scalable process for the differentiation of retinal pigment epithelium from human pluripotent stem cells. Stem Cells Transl Med. 2013;2:341–354. doi: 10.5966/sctm.2012-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rowland TJ, Blaschke AJ, Buchholz DE, et al. Differentiation of human pluripotent stem cells to retinal pigmented epithelium in defined conditions using purified extracellular matrix proteins. J Tissue Eng Regen Med. 2013;7:642–653. doi: 10.1002/term.1458. [DOI] [PubMed] [Google Scholar]
- 12.Sridhar A, Steward MM, Meyer JS. Nonxenogeneic growth and retinal differentiation of human induced pluripotent stem cells. Stem Cells Transl Med. 2013;2:255–264. doi: 10.5966/sctm.2012-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu IS, Chen JD, Ploder L, et al. Developmental expression of a novel murine homeobox gene (Chx10): evidence for roles in determination of the neuroretina and inner nuclear layer. Neuron. 1994;13:377–393. doi: 10.1016/0896-6273(94)90354-9. [DOI] [PubMed] [Google Scholar]
- 14.Liang L, Sandell JH. Focus on molecules: homeobox protein Chx10. Exp Eye Res. 2008;86:541–542. doi: 10.1016/j.exer.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 15.Horsford DJ, Nguyen MT, Sellar GC, et al. Chx10 repression of Mitf is required for the maintenance of mammalian neuroretinal identity. Development. 2005;132:177–187. doi: 10.1242/dev.01571. [DOI] [PubMed] [Google Scholar]
- 16.Dyer MA. Regulation of proliferation, cell fate specification and differentiation by the homeodomain proteins Prox1, Six3, and Chx10 in the developing retina. Cell Cycle. 2003;2:350–357. [PubMed] [Google Scholar]
- 17.Green ES, Stubbs JL, Levine EM. Genetic rescue of cell number in a mouse model of microphthalmia: interactions between Chx10 and G1-phase cell cycle regulators. Development. 2003;130:539–552. doi: 10.1242/dev.00275. [DOI] [PubMed] [Google Scholar]
- 18.Burmeister M, Novak J, Liang MY, et al. Ocular retardation mouse caused by Chx10 homeobox null allele: impaired retinal progenitor proliferation and bipolar cell differentiation. Nat Genet. 1996;12:376–384. doi: 10.1038/ng0496-376. [DOI] [PubMed] [Google Scholar]
- 19.Barabino SM, Spada F, Cotelli F, et al. Inactivation of the zebrafish homologue of Chx10 by antisense oligonucleotides causes eye malformations similar to the ocular retardation phenotype. Mech Dev. 1997;63:133–143. doi: 10.1016/s0925-4773(97)00036-1. [DOI] [PubMed] [Google Scholar]
- 20.Rowan S, Chen CM, Young TL, et al. Transdifferentiation of the retina into pigmented cells in ocular retardation mice defines a new function of the homeodomain gene Chx10. Development. 2004;131:5139–5152. doi: 10.1242/dev.01300. [DOI] [PubMed] [Google Scholar]
- 21.Wong G, Conger SB, Burmeister M. Mapping of genetic modifiers affecting the eye phenotype of ocular retardation (Chx10or-J) mice. Mamm Genome. 2006;17:518–525. doi: 10.1007/s00335-005-0159-z. [DOI] [PubMed] [Google Scholar]
- 22.Vitorino M, Jusuf PR, Maurus D, et al. Vsx2 in the zebrafish retina: restricted lineages through derepression. Neural Dev. 2009;4:14. doi: 10.1186/1749-8104-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou C, Levine EM. Vsx2 controls eye organogenesis and retinal progenitor identity via homeodomain and non-homeodomain residues required for high affinity DNA binding. PLoS Genet. 2012;8:e1002924. doi: 10.1371/journal.pgen.1002924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferda Percin E, Ploder LA, Yu JJ, et al. Human microphthalmia associated with mutations in the retinal homeobox gene CHX10. Nat Genet. 2000;25:397–401. doi: 10.1038/78071. [DOI] [PubMed] [Google Scholar]
- 25.Faiyaz-Ul-Haque M, Zaidi SH, Al-Mureikhi MS, et al. Mutations in the CHX10 gene in non-syndromic microphthalmia/anophthalmia patients from Qatar. Clin Genet. 2007;72:164–166. doi: 10.1111/j.1399-0004.2007.00846.x. [DOI] [PubMed] [Google Scholar]
- 26.Reis LM, Khan A, Kariminejad A, et al. VSX2 mutations in autosomal recessive microphthalmia. Mol Vis. 2011;17:2527–2532. [PMC free article] [PubMed] [Google Scholar]
- 27.Brown ME, Rondon E, Rajesh D, et al. Derivation of induced pluripotent stem cells from human peripheral blood T lymphocytes. PLoS One. 2010;5:e11373. doi: 10.1371/journal.pone.0011373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh R, Phillips MJ, Kuai D, et al. Functional Analysis of Serially Expanded Human iPS Cell-Derived RPE Cultures. Invest Ophthalmol Vis Sci. 2013;54:6767–6778. doi: 10.1167/iovs.13-11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh R, Shen W, Kuai D, et al. iPS cell modeling of Best disease: insights into the pathophysiology of an inherited macular degeneration. Hum Mol Genet. 2013;22:593–607. doi: 10.1093/hmg/dds469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Capowski EE, Schneider BL, Ebert AD, et al. Lentiviral vector-mediated genetic modification of human neural progenitor cells for ex vivo gene therapy. J Neurosci Methods. 2007;163:338–349. doi: 10.1016/j.jneumeth.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 31.Zufferey R, Nagy D, Mandel RJ, et al. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 32.Stewart R, Rascon CA, Tian S, et al. Comparative RNA-seq analysis in the unsequenced axolotl: the oncogene burst highlights early gene expression in the blastema. PLoS Comput Biol. 2013;9:e1002936. doi: 10.1371/journal.pcbi.1002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bharti K, Liu W, Csermely T, et al. Alternative promoter use in eye development: the complex role and regulation of the transcription factor MITF. Development. 2008;135:1169–1178. doi: 10.1242/dev.014142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez-Morales JR, Signore M, Acampora D, et al. Otx genes are required for tissue specification in the developing eye. Development. 2001;128:2019–2030. doi: 10.1242/dev.128.11.2019. [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Morales JR, Rodrigo I, Bovolenta P. Eye development: a view from the retina pigmented epithelium. Bioessays. 2004;26:766–777. doi: 10.1002/bies.20064. [DOI] [PubMed] [Google Scholar]
- 36.Lane BM, Lister JA. Otx but not Mitf transcription factors are required for zebrafish retinal pigment epithelium development. PLoS One. 2012;7:e49357. doi: 10.1371/journal.pone.0049357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barishak YR. Embryology of the eye and its adnexae. Dev Ophthalmol. 1992;24:1–142. [PubMed] [Google Scholar]
- 38.Finlay BL. The developing and evolving retina: using time to organize form. Brain Res. 2008;1192:5–16. doi: 10.1016/j.brainres.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 39.Adler R, Canto-Soler MV. Molecular mechanisms of optic vesicle development: complexities, ambiguities and controversies. Dev Biol. 2007;305:1–13. doi: 10.1016/j.ydbio.2007.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bassett EA, Wallace VA. Cell fate determination in the vertebrate retina. Trends Neurosci. 2012;35:565–573. doi: 10.1016/j.tins.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 41.Fuhrmann S. Eye morphogenesis and patterning of the optic vesicle. Curr Top Dev Biol. 2010;93:61–84. doi: 10.1016/B978-0-12-385044-7.00003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graw J. Eye development. Curr Top Dev Biol. 2010;90:343–386. doi: 10.1016/S0070-2153(10)90010-0. [DOI] [PubMed] [Google Scholar]
- 43.Fujimura N, Taketo MM, Mori M, et al. Spatial and temporal regulation of Wnt/beta-catenin signaling is essential for development of the retinal pigment epithelium. Dev Biol. 2009;334:31–45. doi: 10.1016/j.ydbio.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 44.Westenskow P, Piccolo S, Fuhrmann S. Beta-catenin controls differentiation of the retinal pigment epithelium in the mouse optic cup by regulating Mitf and Otx2 expression. Development. 2009;136:2505–2510. doi: 10.1242/dev.032136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bharti K, Gasper M, Ou J, et al. A regulatory loop involving PAX6, MITF, and WNT signaling controls retinal pigment epithelium development. PLoS Genet. 2012;8:e1002757. doi: 10.1371/journal.pgen.1002757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fuhrmann S, Zou C, Levine EM. Retinal pigment epithelium development, plasticity, and tissue homeostasis. Exp Eye Res. 2013 doi: 10.1016/j.exer.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pittack C, Grunwald GB, Reh TA. Fibroblast growth factors are necessary for neural retina but not pigmented epithelium differentiation in chick embryos. Development. 1997;124:805–816. doi: 10.1242/dev.124.4.805. [DOI] [PubMed] [Google Scholar]
- 48.Zhao S, Hung FC, Colvin JS, et al. Patterning the optic neuroepithelium by FGF signaling and Ras activation. Development. 2001;128:5051–5060. doi: 10.1242/dev.128.24.5051. [DOI] [PubMed] [Google Scholar]
- 49.Esteve P, Bovolenta P. Secreted inducers in vertebrate eye development: more functions for old morphogens. Curr Opin Neurobiol. 2006;16:13–19. doi: 10.1016/j.conb.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 50.Nakayama Y, Miyake A, Nakagawa Y, et al. Fgf19 is required for zebrafish lens and retina development. Dev Biol. 2008;313:752–766. doi: 10.1016/j.ydbio.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 51.Zhou J, Kherani F, Bardakjian TM, et al. Identification of novel mutations and sequence variants in the SOX2 and CHX10 genes in patients with anophthalmia/microphthalmia. Mol Vis. 2008;14:583–592. [PMC free article] [PubMed] [Google Scholar]
- 52.Fuhrmann S, Levine EM, Reh TA. Extraocular mesenchyme patterns the optic vesicle during early eye development in the embryonic chick. Development. 2000;127:4599–4609. doi: 10.1242/dev.127.21.4599. [DOI] [PubMed] [Google Scholar]
- 53.Bone-Larson C, Basu S, Radel JD, et al. Partial rescue of the ocular retardation phenotype by genetic modifiers. J Neurobiol. 2000;42:232–247. doi: 10.1002/(sici)1097-4695(20000205)42:2<232::aid-neu7>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 54.Rutherford AD, Dhomen N, Smith HK, et al. Delayed expression of the Crx gene and photoreceptor development in the Chx10-deficient retina. Invest Ophthalmol Vis Sci. 2004;45:375–384. doi: 10.1167/iovs.03-0332. [DOI] [PubMed] [Google Scholar]
- 55.Schwartz SD, Hubschman JP, Heilwell G, et al. Embryonic stem cell trials for macular degeneration: a preliminary report. Lancet. 2012;379:713–720. doi: 10.1016/S0140-6736(12)60028-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure S1: Generation and characterization of WT and (R200Q)VSX2 hiPSC clones. (A) Three WT and three (R200Q)VSX2 mutant clonal hiPSCs lines were characterized (WT-1, -2, -3 and Mut-1, -2, -3). All hiPSC lines formed colonies that expressed the pluripotency genes NANOG, TRA1-60, OCT4, and SSEA-4. Scale bar = 200 μm. (B) The vast majority of hiPSCs expressed pluripotency markers as shown by representative FACS analyses of WT and mutant hiPSC lines. (C) All hiPSC lines were sequenced and verified for the presence or absence of the appropriate VSX2 homeodomain mutation at codon 200. The guanine to adenine switch in codon 200 leads to an arginine to glutamine missense mutation (R200Q) in the mutant hiPSC lines (representative sequencing analysis shown for one WT and one mutant hiPSC line). (D) All hiPSC lines were karyotypically normal (representative G-banding analysis shown). (E) Genomic DNA PCR analysis demonstrated genomic integration of reprogramming vectors in mutant (M) and WT hiPSCs (one reprogramming transgene from each bicistronic vector was analyzed). Reprogramming vector plasmids (+ control) served as positive PCR amplification controls, while WA09 human embryonic stem cells (hESCs) and peripheral blood mononuclear cells (PBMCs) served as negative controls. (F) Characteristic pluripotency genes were expressed in WT and mutant hiPSCs as shown by RT-PCR. hESCs and PBMCs served as positive and negative PCR amplification controls, respectively. (G) Reprogramming factor transgenes were silenced and endogenous pluripotency genes were upregulated in hiPSCs. Reprogramming plasmids and hESCs served as positive controls for transgene-mediated and endogenous pluripotency gene amplification, respectively. (H) Teratoma analysis confirmed the competency of each hiPSC line to produce all three germ layers, including gut and respiratory epithelium (endoderm), cartilage (mesoderm), and neural tissue (ectoderm) (one representative WT and mutant hiPSC line shown). Scale bar = 100 μm.
Supporting Information Figure S2: Schematic depicting the differentiation protocol and the cell types present at the time points analyzed in this study. Of note, while small, superficial patches of pigmented RPE develop in a minority of WT hiPSC-OVs, most mutant hiPSC-OVs become uniformly pigmented with time. However, some mutant hiPSC-OVs remain pigmentation-free, permitting comparisons of NR differentiation in mutant vs. WT hiPSC-OVs. RPC: retinal progenitor cell, RGC: retinal ganglion cell, PR: photoreceptor.
Supporting Information Figure S3: VSX2 expression in human prenatal and adult retina. (A) VSX2 was expressed in the neuroblastic layer (NBL) of day 85 human prenatal retina. Numerous VSX2+ cells co-expressed Ki-67, indicative of proliferating NRPCs. (B) In a 71 year-old adult retina, VSX2 expression was confined to bipolar cells within the inner nuclear layer (INL). These adult VSX2+ cells were non-proliferative (i.e., Ki-67 negative). ONL = outer nuclear layer; GCL = ganglion cell layer. Scale bar = 50 μm.
Supporting Information Figure S4: (R200Q)VSX2 hiPSCs differentiated as adherent cultures displayed greater pigmentation than WT hiPSCs. Instead of lifting neural colonies at day 16 to generate OVs, (R200Q)VSX2 and WT hiPSCs were left adherent to culture plates to promote RPE differentiation. A representative six well plate is shown at day 60 of differentiation (top three wells = WT hiPSCs; lower three wells = (R200Q)VSX2 mutant hiPSCs).
Supporting Information Figure S5: Examination of additional WT and mutant hiPSC lines confirmed the effects of the (R200Q)VSX2 mutation on photoreceptor maturation and bipolar cell differentiation. (A) Similar to the WT-1 hiPSC line shown in Fig. 4A and B, the WT-3 hiPSC line produced numerous CRX+/RCVRN+ photoreceptors by day 80. (B) The Mut-3 hiPSC line also produced CRX+ cells; however, RCVRN+ cells were notably absent. (C) VSX2+ nuclei were predominately Ki-67-negative, indicative of bipolar cell differentiation in WT-3 hiPSC-OVs. (D) VSX2 expression was absent in Mut-3 hiPSC-OVs by day 80, although some Ki-67 cells persisted. (E–J) WT-3 and Mut-3 hiPSC-OVs produced other neural retina cell types, including CALB2+ neurons (E,F), TH+ amacrine cells (G,H), and S100+ glia (I,J). (K,L) Most of the Ki-67+ cells co-labeled with PAX6, indicating the continued presence of neural retina progenitors cells and/or proliferative glia in both the WT-3 (K) and Mut-3 lines (L). Scale bar = 50 μm for panel B (also applies to panel A) and 50 μm for panel L (applies to panels C–L).
Supporting Information Figure S6: Exogenous expression of WT VSX2 in the Mut-2 hiPSC line increased production of CRX+/RCVRN+ photoreceptors. (A) Similar to the Mut-1 (R200Q)VSX2 hiPSC line shown in Fig. 5F, RCVRN expression was absent at day 100 in Mut-2 (R200Q)VSX2 hiPSCs infected at day 14 with a lenti-GFP control vector (mutant control). (B) After infection at day 14 with lenti-WT VSX2 (Lenti-VSX2), Mut-2 hiPSC-OVs expressed numerous CRX+/RCVRN+ photoreceptors at day 100, similar to the Mut-1 hiPSC line shown in Fig. 5G. Scale bar in panel B = 100 μm (also applies to panel A). (C) High magnification of CRX+/RCVRN+ cells following exogenous expression of WT VSX2 in Mut-2 hiPSC-OVs. (D) RCVRN+ cells in lenti-WT VSX2 infected mutant-2 hiPSC-OVs were VSX2 negative, showing that the WT VSX2-expressing transgene was downregulated in differentiating photoreceptors. Scale bar in panel D = 50 μm (also applies to panel C).
Supporting Information Figure S7: qRT-PCR validation of RNAseq data. qRT-PCR was performed for selected retinal differentiation and developmental signaling genes at day 20 (A) and day 30 (B) of differentiation and compared to the corresponding RNAseq data for each gene. Results were normalized to WT hiPSC-OV expression data for each gene (denoted by the green dashed line). All genes analyzed demonstrated similar trends with qRT-PCR and RNAseq analyses. *p < 0.05, ** p < 0.01, ***p < 0.001.