Abstract
Although much is known about genetic variation in human and African great ape (chimpanzee, bonobo, and gorilla) genomes, substantially less is known about variation in gene-expression profiles within and among these species. This information is necessary for defining transcriptional regulatory networks that contribute to complex phenotypes unique to humans or the African great apes. We took a systematic approach to this problem by investigating gene-expression profiles in well-defined cell populations from humans, bonobos, and gorillas. By comparing these profiles from 18 human and 21 African great ape primary fibroblast cell lines, we found that gene-expression patterns could predict the species, but not the age, of the fibroblast donor. Several differentially expressed genes among human and African great ape fibroblasts involved the extracellular matrix, metabolic pathways, signal transduction, stress responses, as well as inherited overgrowth and neurological disorders. These gene-expression patterns could represent molecular adaptations that influenced the development of species-specific traits in humans and the African great apes.
A major goal of human evolutionary biology is to elucidate the key environmental and genetic factors that have influenced the development of complex phenotypes that differentiate humans from other primates. On the physiological level, these phenotypes include the development of large brains and a unique musculoskeletal system. In turn, these physiological changes have contributed to the emergence of higher cognitive functions and other distinguishing characteristics such as bipedal locomotion.
By comparing the genomes of humans and their closest living relatives, the African great apes (chimpanzees, bonobos, and gorillas), it will be possible to generate a list of candidate genetic changes that could have contributed to the appearance of modern humans 200,000 years ago (Stringer 2002). Although the human and chimpanzee (as well as bonobo) genomes are >98.5% identical on the nucleotide level (Chen and Li 2001) (95% if the lengths of insertions and deletions are taken into account [Britten 2002]), the total number of nucleotide differences (>40 million) is enormous. The major challenge in the rapidly approaching human and chimpanzee genome sequence era is to determine the small subset of sequence differences that have phenotypic significance related to species-specific traits. This critical subset of sequence differences directly or indirectly alters the temporal and spatial distribution of distinct protein activities during development and into adulthood.
Thus far, differential gene expression, gene loss, gene duplication, and natural selection have received the most attention as the molecular mechanisms responsible for the differences in protein activity in these species. The differential gene-expression theory is based on the fact that the sequences of human and African great ape proteins are highly similar, and thus, it is less likely that these changes lead to specific human phenotypes (King and Wilson 1975). The gene-loss theory states that loss-of-function mutations play a key role in human evolution (Olson 1999; Olson and Varki 2003). An example is the inactivation of the CMP-Neu5AC hydroxylase (CMAH) gene in the human lineage caused by the insertion of a human-specific Alu element (Chou et al. 1998, 2002; Irie et al. 1998). Gene duplication presents a powerful means to change the activities and gene-expression patterns of family members (Gu et al. 2002; Samonte and Eichler 2002). Segmental duplications (1–200-kb DNA segments with 90%–100% sequence identity) comprise ∼5% of the human genome and contain high-copy number repeats as well as gene sequences with intron-exon structures (Samonte and Eichler 2002). Lastly, natural selection could play a major role in altering protein activity by rapidly fixing genomic segments containing advantageous alleles (selective sweeps) or by accelerating the rate of amino acid substitutions (positive selection; Kreitman 2000). For example, the region encompassing the FOXP2 gene, responsible for a rare inherited human speech and language disorder, underwent a selective sweep in recent human history (Enard et al. 2002b; Zhang et al. 2002), whereas the morpheus gene family shows positive selection in human and African great ape lineages (Eichler et al. 2001; Johnson et al. 2001).
Recently, the differential gene-expression hypothesis was tested by Enard and colleagues when they used microarrays to compare expression patterns of human and chimpanzee brain (gray matter from the left prefrontal lobe), blood leukocytes, and liver (Enard et al. 2002a). The authors reported accelerated rate of gene-expression change in human brain relative to the chimpanzee but not in leukocytes or liver. Additional analysis of this data set using orangutan as an out-group, indicates that the brain-specific rate acceleration is mainly caused by the overexpression of a group of genes in the human lineage (Gu and Gu 2003).
Although these studies yield important insights into human and African great ape evolution, examining tissue samples from a limited number of individuals can be problematic. Variability in gene expression due to gender, age, and cause of death of the donor as well as the methodology for collecting tissues must be accounted for in order to most effectively design and interpret such experiments. Furthermore, it is difficult to analyze tissues known to have a different abundance of specific cell types among these species. For example, the human brain contains a significantly higher number of spindle cells in the anterior cingulated cortex relative to those of the African great apes (Nimchinsky et al. 1999). Thus, it is difficult to ascertain the relative contributions of the cellular composition of tissues from each species and gene-expression changes within distinct cell populations for each differentially expressed gene.
Here, we begin to address the issue of identifying transcriptional differences within well-defined cell populations from these species. We compared gene-expression profiles from 18 human, 10 bonobo, and 11 gorilla-cultured primary fibroblast cell lines. We identified groups of genes that show robust differences in gene-expression levels among these species and correlated the location of these changes with known chromosomal changes in hominoid genomes. On the basis of their known functions, several candidate genes that could influence the development of traits unique to humans or the African great apes were uncovered.
RESULTS
Experimental Design
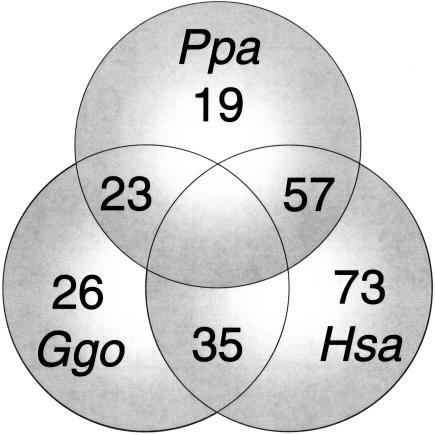
We extracted total RNA from early-passage primary fibroblast cell lines from 18 human, 10 bonobo, and 11 gorilla donors of known age and gender. All of the cell lines have similar doubling times, ranging from 1.6–2.8 days (Table 1). We performed gene-expression analysis of these samples using Affymetrix U95Av2 microarrays designed to evaluate the abundance of >10,000 different human transcripts. We averaged expression data from these individual cell cultures to create composite human, bonobo, and gorilla expression profiles. We then compared the levels of each transcript in each species. In our discussions of differentially expressed genes in humans, bonobos, and gorillas, we report ratios based on the lower bound of the 95% confidence interval (Fig. 1). This provides a rigorous means to identify genes that are predicted by microarray analysis to be differentially expressed among these species.
Table 1.
Identity of Fibroblast Cell Lines
| Namea | Donorb | DT | Namec | Donorb | DT |
|---|---|---|---|---|---|
| AG13153 | Hsa 30 M | 2.3 | KB6884 | Ppa 02 F | 2.0 |
| AG07999 | Hsa 32 F | 2.5 | KB5275 | Ppa 02 M | 2.4 |
| AG05838 | Hsa 36 F | 2.5 | KB10025 | Ppa 04 M | 2.4 |
| AG06235 | Hsa 42 M | 1.7 | KB7033 | Ppa 04 F | 2.1 |
| AG11745d | Hsa 43 F | 1.8 | KB4512 | Ppa 09 F | 2.6 |
| AG13927 | Hsa 45 F | 2.8 | KB5828 | Ppa 12 M | 2.7 |
| AG04446 | Hsa 48 M | 2.2 | KB7998 | Ppa 17 M | 2.0 |
| AG10942 | Hsa 50 M | 2.6 | KB8025 | Ppa 19 M | 2.0 |
| AG13968 | Hsa 55 F | 2.3 | KB8024 | Ppa 20 F | 2.3 |
| AG12988 | Hsa 56 F | 2.3 | KB7189 | Ppa 20 F | 2.8 |
| AG05844e | Hsa 57 F | 2.1 | KB8840 | Ggo 02 F | 2.5 |
| AG11362e | Hsa 63 F | 2.1 | KB9047 | Ggo 3 M | 2.4 |
| AG04659f | Hsa 65 M | 1.7 | KB8592 | Ggo 18 M | 1.9 |
| AG13144e | Hsa 69 F | 2.8 | KB6268 | Ggo 19 F | 2.0 |
| AG05283f | Hsa 69 M | 1.7 | KB5047 | Ggo 19 F | 2.0 |
| AG06952g | Hsa 73 F | 2.4 | KB7500 | Ggo 19 M | 1.8 |
| AG05414d | Hsa 73 M | 2.3 | KB7801 | Ggo 24 M | 1.9 |
| AG11020g | Hsa 79 F | 2.3 | KB8021 | Ggo 26 F | 2.4 |
| AG13151h | Hsa 54 M | 4.4 | KB7621 | Ggo 26 M | 2.3 |
| KB8594 | Ggo 30 F | 1.6 | |||
| KB6278 | Ggo 35 F | 1.9 |
Coriell Cell Repository ID
Donor species (Hsa = Homo sapiens, Ppa = Pan pansicus, Ggo = Gorilla gorilla), age, and gender. DT = doubling time of fibroblast
CRES ID
Cell lines with the same letter were established from the same donor at different ages
Was not used in the gene expression comparison due to its slow growth
Figure 1.
Interspecies gene-expression variation. A Venn diagram showing the number of genes that are differentially expressed greater than twofold (lower bound of 95% CI) among human (Hsa), bonobo (Ppa), and gorilla (Ggo) fibroblast cell lines is given.
Confirmatory Northern Blot and Sequencing Analysis
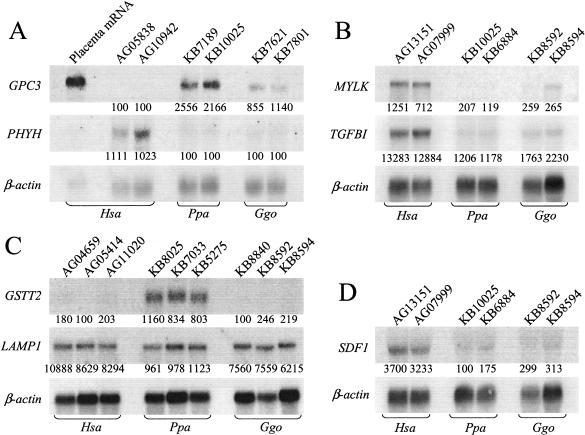
We used Northern blot analysis to confirm 14/17 genes predicted by microarray analysis to show greater than twofold differential gene expression between bonobos and humans (Fig. 2; Supplemental Fig. 1, available online at www.genome.org). This is in agreement with other reports examining chimpanzee gene-expression levels using similar assays (Enard et al. 2002a). The disagreements between microarray and Northern blot analysis (LAMP1, AKAP2, and SPTLC1) (Fig. 2C; Suppl. Fig. 1) were caused by sequence differences between African great ape transcripts and the probes in the microarray (Suppl. Table 1). This is not unexpected, as mismatches between African great ape RNA samples and oligonucleotide probes designed to interrogate human transcripts will cause these microarrays to underestimate the relative abundance of certain African great ape transcripts (Methods; Hacia et al. 1998, 1999; Enard et al. 2002a).
Figure 2.
Confirmatory Northern blot analysis of microarray results. (A–D) Fibroblast cell line names are given at the top of each lane. At the bottom of each lane is the expression level score assigned by microarray analysis. To the left of each strip is the identity of the gene interrogated. In each panel, the relative abundance of β-actin for each blot is given to normalize for loading abundance. Signal gain and contrast in each strip is adjusted for clarity.
We determined partial sequences from 21 bonobo genes predicted in Table 4 (see below) to be significantly overexpressed in humans relative to bonobos and noted the location and number of mismatches for all probes interrogating these genes. A total of 154/336 probes (45.8%) contained at least one bonobo-specific mismatch located anywhere with the target (Suppl. Table 2). Next, we excluded any probe having a mismatch with the bonobo transcript from the analysis. We found 12/21 (57%) (SLC16A3, GBE1, MMP3, PPP1R3C, STC2, XAP4, CGI-150, SEMA5A, CLOCK, ALDH1A3, HLA-E, and POLR2J) of the re-evaluated genes still display greater than or equal to twofold (lower bound of 95% confidence interval) over-expression in humans relative to bonobos (Table 4, below, and Suppl. Table 3). This agrees with estimates based on confirmatory Northern blot analysis on genes up-regulated in humans relative to bonobos (4/7, 57%) in Table 4, below. Overall, these mismatches may partially explain why 73 genes were up- and only 23 were down-regulated greater than or equal to twofold in human relative to both African great ape cell lines (Fig. 1).
Table 4.
Selected Genes Up-regulated in Human Relative to Bonobo Cells
| H/P
|
H/G
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| LL | Genea | Hsa | Ppa | Ggo | LB | UB | LB | UB | Function |
| 9123 | SLC16A3f,g | 2291 | 113 | 146 | 15.3 | 26.3 | 11.4 | 22.2 | Monocarboxylate transporter |
| 10558 | SPTLC1b,f | 1617 | 102 | 438 | 14.1 | 17.7 | 3.1 | 4.4 | Ceramide synthesis |
| 11217 | AKAP2b,f | 3119 | 162 | 168 | 14.1 | 27.5 | 14.1 | 24.6 | Signal transduction |
| 6387 | SDF1c | 3192 | 158 | 234 | 12.6 | 30.1 | 8.3 | 21.7 | Signal transduction |
| 2632 | GBE1f,g | 3722 | 271 | 1347 | 10.8 | 18.0 | 2.2 | 3.7 | Glycogen synthesis |
| 3916 | LAMP1b,f | 10435 | 1018 | 8376 | 9.0 | 11.7 | 1.0 | 1.6 | Membrane glycoprotein |
| 7045 | TGFB1c | 12159 | 1038 | 1862 | 7.7 | 20.9 | 4.7 | 9.2 | Integrin ligand |
| 4314 | MMP3g | 6451 | 451 | 2333 | 6.5 | 50.2 | 1.1 | N.D. | Proteoglycanase |
| 5507 | PPP1R3Cg | 1219 | 169 | 515 | 6.3 | 12.0 | 2.1 | 4.0 | Protein phosphotase |
| 8614 | STC2g | 1844 | 222 | 153 | 5.7 | 12.3 | 8.6 | 16.5 | Calcium homeostasis |
| 4276 | MICAe | 713 | 111 | 153 | 5.2 | 7.9 | 3.7 | 6.0 | Immune system |
| 5264 | PHYHc,f | 591 | 100 | 104 | 4.8 | 7.1 | 4.6 | 6.9 | Phytanic acid metabolism |
| 8878 | SQSTM1f,h | 2896 | 517 | 745 | 4.5 | 7.1 | 2.8 | 6.0 | Ubiquitin-binding protein |
| 10616 | XAP4f,g | 589 | 115 | 108 | 4.1 | 6.6 | 4.7 | 6.4 | Unknown |
| 1305 | COL13A1 | 809 | 142 | 107 | 4.0 | 8.7 | 6.1 | 9.1 | Structural |
| 51031 | CGI-150d,g | 448 | 100 | 406 | 4.0 | 5.1 | -1.1 | 1.3 | Unknown |
| 9037 | SEMA5Ag | 1941 | 312 | 788 | 4.0 | 11.3 | 1.4 | 6.1 | Cell adhesion |
| 10921 | RNPS1h | 2479 | 492 | 1703 | 3.9 | 6.6 | 1.1 | 2.0 | RNA processing |
| 4638 | MYLKc,d | 1416 | 182 | 343 | 3.7 | 19.8 | 2.2 | 6.7 | Myosin light-chain kinase |
| 1611 | DAPh | 3084 | 727 | 3618 | 3.7 | 4.9 | 1.1 | -1.5 | Mediates apoptosis |
| 25800 | LIV-1f,h | 563 | 129 | 405 | 3.4 | 5.7 | 1.2 | 1.7 | Zinc transport |
| 9575 | CLOCKg | 659 | 157 | 591 | 3.4 | 5.2 | -1.1 | 1.4 | Transcription factor |
| 11072 | DUSP14e,h | 2145 | 515 | 489 | 3.3 | 5.3 | 3.5 | 5.4 | Dual-specific phosphatase |
| 9551 | ATP5J2f,h | 3861 | 1005 | 1123 | 3.3 | 4.5 | 3.0 | 3.9 | Proton transport |
| 220 | ALDH1A3g | 544 | 108 | 138 | 3.1 | 7.1 | 2.4 | 5.7 | Aldehyde detoxification |
| 7001 | PRDX2e,h | 1718 | 420 | 745 | 3.1 | 5.9 | 1.7 | 3.6 | Peroxide reductase |
| 2802 | GOLGA3h | 1233 | 350 | 353 | 3.0 | 4.2 | 3.1 | 3.9 | Golgi membrane |
| 3133 | HLA-Ef,g | 876 | 191 | 293 | 3.0 | 7.8 | 2.0 | 4.9 | Immune system |
| 1832 | DSPe,h | 1011 | 201 | 100 | 3.0 | 7.6 | 6.3 | 14.1 | Intercellular junctions |
| 5439 | POLR2Jf,g | 724 | 198 | 193 | 3.0 | 4.8 | 3.0 | 4.9 | RNA polymerase II subunit |
Genes show lower bound of 95% confidence interval of greater than or equal to 3.0-fold change
Disagrees with Northern blot analysis
Confirmed by Northern blot analysis
Greater than or equal to 2.0-fold up-regulated in human brain relative to chimpanzee brain
Greater than or equal to 2.0-fold up-regulated in human liver relative to chimpanzee liver
Greater than or equal to 2.0-fold up-regulated in human liver and brain relative to chimpanzee liver and brain
Gene still overexpressed greater than or equal to 2.0-fold when bonobo sequence is considered
Gene is not overexpressed by greater than or equal to 2.0-fold when bonobo sequence is considered
Chromosomal Location of Differentially Expressed Genes
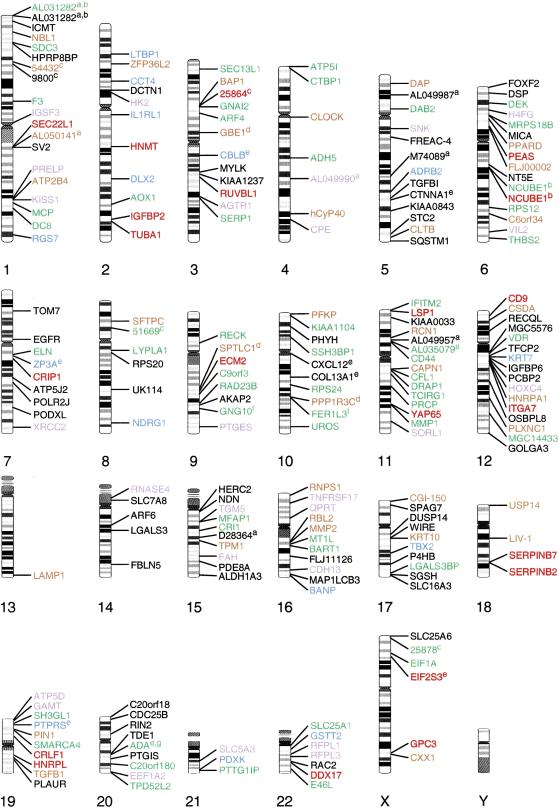
Although human and African great ape chromosomal organization is strikingly similar, a chromosomal fusion, several large-scale rearrangements, and other more subtle changes have created sequences unique to humans (Yunis and Prakash 1982; Johnson et al. 2001). These changes include the telomeric fusion of two ancestral great ape chromosomes (Fan et al. 2002), nine pericentric inversions (Yunis and Prakash 1982; Kehrer-Sawatzki et al. 2002), and paracentric inversions (Samonte and Eichler 2002). To test the hypothesis that these organizational changes could affect the regulation of nearby genes, we mapped the positions of differentially expressed genes onto the human karyotype and compared their location with large-scale rearrangements among human and African great apes genomes (Fig. 3) (Yunis and Prakash 1982; Gagneux and Varki 2001; Hacia 2001). To improve the specificity of this analysis, we only considered chromosomal rearrangements with finely mapped breakpoints. We found no evidence of gene clusters with similar expression patterns among these species located near the breakpoints of 2q13–2q14.1 (the region encompassing sequences involved in telomere-to-telomere fusion event that lead to the formation of human chromosome 2 [Fan et al. 2002]), the pericentric inversions involving human chromosomal regions 4p14–4q21 (Nickerson and Nelson 1998), 12p12–12q15 (Nickerson and Nelson 1998), and 17p13–17q21.3 (Kehrer-Sawatzki et al. 2002), and the gorilla-specific reciprocal translocation t(4;19) (Stankiewicz et al. 2001).
Figure 3.
Chromosomal locations of differentially expressed genes among humans and the African great apes. The locations of such genes (up-regulated greater than twofold [lower bound of 95% CI] in at least one species relative to the others) are indicated on the human karyotype on the basis of the human genome reference sequence. Gene names are given in gold (up in Hsa and Ggo), black (up in Hsa), green (up in Hsa and Ppa), purple (up in Ggo), red (up in Ppa and Ggo), and blue (up in Ppa). (a) GenBank Accession Number; (b) two different probe tilings interrogating the same gene give different outcomes; (c) LocusLink ID; (d) Hsa greater than twofold up-regulated relative to Ggo; (e) two different probe tilings interrogating the same gene give the same outcome; (f) Hsa greater than twofold up-regulated relative to Ppa; (g) Ppa greater than twofold up-regulated relative to Hsa.
In no case were two or more genes located nearby the cytogenetic change greater than twofold differentially expressed among species. This implies that these cytogenetic changes did not result in long-range changes in gene-expression patterns. Furthermore, there was no overabundance of such genes near centromeric and telomeric regions (Fig. 3), which are known to be enriched for segmental duplications (Yunis and Prakash 1982; Samonte and Eichler 2002).
We next searched for large genomic regions that over- or under-represent differentially expressed genes among these species. We screened for such genes located within five identified genes of one another (Fig. 3, Suppl. Table 4). Most notably, the serpinB7 and serpinB2 genes (18q21.33) are both up-regulated in bonobos and gorillas relative to humans (Irving et al. 2000). This could indicate changes in the expression and/or copy number of these family members in hominoid evolution.
Upon visual inspection, differentially expressed genes appear to be unevenly distributed across human chromosomes (Fig. 3). For example, there is only one such gene on human chromosome 13 (LAMP1). Interestingly, Northern blot analysis showed LAMP1 expression levels were even across all three species (Fig. 2C). To determine whether the apparent uneven distribution is statistically significant, we calculated the 95% confidence interval (CI) for the number of genes predicted to be differentially expressed on each chromosome on the basis of the number of probe tilings per chromosome (Methods). In each case (including chromosome 13), the observed number of genes was within the 95% CI of what is expected based upon a random distribution of these differentially expressed genes (Suppl. Table 5).
Lineage-Specific Gene Induction and Repression
Differentially expressed genes in human and chimpanzee brains tend to be the result of significant up- rather than down-regulation in humans relative to chimpanzees (Gu and Gu 2003). To determine whether this is the case in our fibroblast data set, we analyzed human and African great ape geneexpression profiles using a method used previously to examine human and great ape brain and liver gene-expression data (Gu and Gu 2003; Table 2). Using gorilla as an out-group, we determined which differentially expressed genes between humans and bonobos (shown in Fig. 1) were lineage specific. A gene was considered human lineage specific if the gorilla expression level was significantly different from human but not bonobo. Likewise, a gene was considered bonobo lineage specific if the gorilla expression level was significantly different from bonobo but not human. The human and bonobo lineage-specific genes were further divided into induced (upregulated) or repressed (down-regulated) in their respective species.
Table 2.
Induction and Repression of Human and Bonobo Lineage-Specific Genes
| Species | Lineage-Specific Genesa | Induced Genesb | Repressed Genesc | Induced/Repressedd |
|---|---|---|---|---|
| Human | 64 | 46 | 18 | 2.56 |
| Bonobo | 29 | 11 | 18 | 0.61 |
Calculated using gorilla expression values as an out-group
Number of lineage-specific genes up-regulated
Number of lineage-specific genes down-regulated
Ratio of induced to repressed lineage-specific genes
Next, we calculated the ratios of induced to repressed lineage-specific genes in humans and in bonobos (Table 2). We found the number of lineage-specific genes resulting from induction in humans (46) was greater than the number resulting from repression (18). In contrast, the number of genes induced in the bonobo lineage (11) was slightly less than the number repressed (18). The overabundance of induced genes in the human lineage is statistically significant (P < 0.05). However, one must consider that 12/28 (43%) of the genes (3/7 based on Northern blot analysis and 9/21 based on sequencing analysis) predicted by microarray analysis to be up-regulated in humans relative to bonobos were artifacts resulting from mismatches between bonobo mRNA and the microarray probes (discussed above). Therefore, the numbers of genes predicted by microarray analysis to be induced in the human lineage and repressed in the bonobo lineage are likely to be overestimates. The statistical significance of these findings will depend strongly on the magnitude of these overestimates. Overall, the lineage-specific gene-expression ratios more closely resemble those found in human and chimpanzee livers than brains (Gu and Gu 2003).
Hierarchical Clustering Analysis of Samples
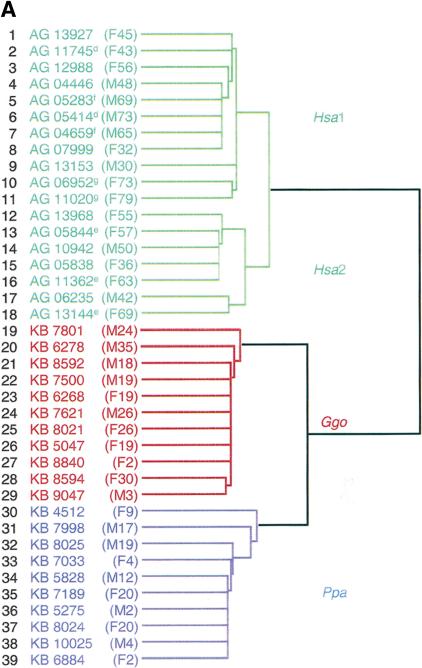
The fibroblast cell lines clustered according to the species, but not the age or gender, of the donor (Fig. 4). This clustering pattern may be influenced by sequence changes that cause the microarray data to indicate that certain transcripts (i.e., AKAP2, SPTLC1, and LAMP1) are always at low expression levels in bonobos or gorillas. However, such transcripts are not always differentially expressed among these species (Fig. 2C; Suppl. Fig. 1). This can bias the results of clustering samples across species, but should have a lesser effect on sample clustering within a given species, as these fixed sequence differences will appear in every member of the species.
Figure 4.
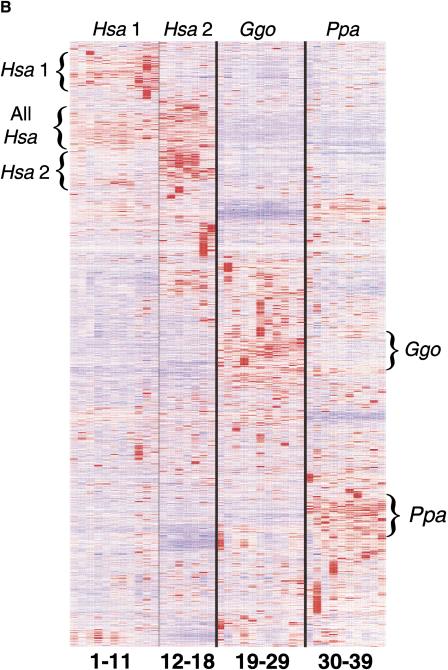
Hierarchical clustering analysis of human and African great ape fibroblast cell line expression patterns. (A) Dendrogram generated from the average linkage hierarchical clustering of geneexpression patterns from all 39 fibroblast cell lines. The cluster tree is based on expression levels of >5300 genes, whose coefficient of variation is >0.3 across all cell lines. The age and gender of the fibroblast donor is given in parenthesis. Human (Hsa), bonobo (Ppa), and gorilla (Ggo) cell lines are depicted by green, blue, and red lines, respectively. Samples with the same superscript are from the same donor at different ages. Human samples fall into Hsa1 and Hsa2 sub-branches. (B) Gene clusters derived from the analysis of all cell lines. Each horizontal strip represents the expression patterns of one gene. Relative high and low expressers within a single gene are depicted in red and blue, respectively. Human, gorilla, and bonobo samples are listed at the top of the column, whereas the numerical identity of each cell line (Fig. 4A) is listed at bottom. Gene clusters are noted on the side of the diagram by the name of the species with the highest expression levels.
Whereas bonobo and gorilla samples clustered without major subdivisions, the human fibroblast samples divided into two (Hsa1 and Hsa2) sub-branches (Fig. 4A). Thirty-two genes, similar to the mitosis-related genes reported to be associated with donor age in human fibroblast cell lines (Ly et al. 2000), were greater than or equal to 2.5-fold differentially expressed (lower bound of 95% CI, P ≤ 0.005) between these two sub-branches (Suppl. Table 6). However, there was no correlation between donor age and the samples in the two human sub-branches in this study. Furthermore, regardless of donor age, African great ape samples showed consistent expression levels of these mitosis-related genes.
To discern possible reasons for the clustering patterns among human samples, we cultured four human cell lines (AG15153, AG06235, AG04446, AG05414) again and repeated the microarray analysis. Upon hierarchical clustering with the original samples, the four cell lines clustered in human group Hsa1, even though one of them (AG06235) was originally in human group Hsa2 (data not shown). This suggests that the human subdivisions based on mitosis-related genes are most likely caused by subtle changes in culture conditions and not fibroblast subpopulations or donor age (Schneider et al. 1978; Ly et al. 2000; Chang et al. 2002).
Genes with robust differences in expression among bonobo and human cell lines are listed in Tables 3 and 4 and shown in Figure 4B. All genes listed in Tables 3 and 4 showed consistent expression differences across all human and bonobo cell lines (P ≤ 0.005). Both human groups showed at least 2.5-fold differential expression relative to the bonobo samples for all genes listed (Suppl. Table 7). Therefore, the expression levels of these genes are affected primarily by interspecies variation. A subset of these genes shows similar differential gene-expression patterns between human and chimpanzee brain and/or liver (Enard et al. 2002a; Tables 3 and 4).
Table 3.
Selected Genes Up-regulated in Bonobo Relative to Human Cells
|
H/Pd
|
H/Gd
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| LLa | Geneb | Hsac | Ppac | Ggoc | LBe | UBe | LBe | UBe | Function |
| 1396 | CRIP1 | 254 | 5129 | 1800 | -13.2 | -32.1 | -4.2 | -11.7 | Zinc carrier |
| 10521 | DDX17i | 252 | 2669 | 2825 | -7.2 | -19.1 | -7.1 | -20.8 | Putative RNA helicase |
| 2719 | GPC3f,g | 107 | 1175 | 499 | -6.1 | -16.0 | -2.6 | -6.8 | Membrane proteoglycan |
| 3191 | HNRPLh | 990 | 6218 | 3412 | -5.2 | -7.5 | -2.6 | -4.3 | RNA processing |
| 2953 | GSTT2f,i | 156 | 978 | 235 | -4.8 | -8.4 | -1.0 | -2.2 | Stress response |
| 3855 | KRT7 | 218 | 1608 | 479 | -4.7 | -11.8 | -1.2 | -3.7 | Structural |
| 9173 | IL1RL1 | 150 | 1217 | 220 | -4.2 | -12.6 | 1.3 | -2.2 | Signal transduction |
| 6000 | RGS7f | 141 | 763 | 163 | -3.8 | -7.5 | 1.1 | -1.6 | Signaling pathway |
| 5157 | PDGFRLg | 150 | 860 | 514 | -3.6 | -8.8 | -1.7 | -5.7 | Signaling pathway |
| 7784 | ZP3Af,g | 150 | 654 | 109 | -3.4 | -5.8 | 1.0 | 1.8 | Extracellular matrix |
| 51632 | NCUBE1i | 159 | 633 | 675 | -3.3 | -4.7 | -3.5 | -5.1 | Ubiquitin-related |
| 8710 | SERPINB7 | 129 | 665 | 428 | -3.1 | -7.5 | -2.0 | -4.8 | Protease inhibitor |
| 868 | CBLB | 316 | 1453 | 389 | -3.0 | -6.4 | 1.1 | -1.7 | Signal transduction |
| 5055 | SERPINB2 | 2113 | 9784 | 7863 | -3.0 | -8.3 | -2.5 | -6.6 | Protease inhibitor |
| 10397 | NDRG1 | 1431 | 5198 | 834 | -3.0 | -4.4 | 1.4 | 2.2 | Signal tranduction |
| 1746 | DLX2 | 128 | 513 | 146 | -2.8 | -5.4 | 1.3 | -1.5 | Transcription factor |
| 1022 | CDK7 | 289 | 1021 | 479 | -2.7 | -4.7 | -1.2 | -2.3 | Cell cycle |
| 27063 | CARP | 119 | 561 | 171 | -2.6 | -7.0 | 1.3 | -2.1 | DNA-binding protein |
| 6272 | SORT1g | 109 | 360 | 283 | -2.5 | -4.1 | -2.2 | -3.1 | Golgi transmembrane |
| 1012 | CDH13 | 405 | 1477 | 948 | -2.5 | -5.3 | -1.7 | -3.4 | Cell-cell interactions |
| 154 | ADRB2 | 130 | 425 | 117 | -2.5 | -4.1 | -1.1 | 1.4 | Signal transduction |
| 10413 | YAP1h | 165 | 525 | 593 | -2.5 | -4.0 | -2.9 | -4.5 | WW domain protein |
Genes show a lower bound of 95% confidence interval of greater than or equal to 2.5-fold change
Donor species [(Hsa) Homo sapiens, (Ppa) Pan pansicus, (Ggo) Gorilla gorilla] along with gene expression score
95% confidence interval of ratios of Hsa/Ppa or Hsa/Ggo gene expression levels
Lower (LB) and upper (UB) bounds of 95% confidence interval
Confirmed by Northern blot analysis
Greater than or equal to 2.0-fold up-regulated in chimpanzee brain relative to human brain (Enard et al. 2002a)
Greater than or equal to 2.0-fold up-regulated in chimpanzee liver relative to human liver (Enard et al. 2002a)
Greater than or equal to 2.0-fold up-regulated in chimpanzee brain and liver relative to human brain and liver (Enard et al. 2002a)
DISCUSSION
Currently, there is little known about the variation in geneexpression profiles in specific cell types among humans and the African great apes. Here, we examined gene-expression profiles in fibroblasts from humans and two species of African great apes, bonobos, and gorillas. Cultured fibroblasts are frequently used to study properties of connective tissue, inherited metabolic diseases, and cellular aging (Connolly 1998; Cristofalo et al. 1998; Zamir and Geiger 2001). The substantial number of fibroblast cell lines from donors of known age and gender allows us to evaluate and correct for intraspecies variation that mask or falsely indicate the presence of differentially expressed genes. The inclusion of two African great ape species (bonobos and gorillas) enables us to more accurately identify gene-expression changes arising in the human lineage. Therefore, this system provides a means to analyze the effect of genomic organization on gene-expression patterns among these species and to screen for genetic changes that may have given rise to species-specific phenotypes.
Although human and African great ape chromosomal organization is strikingly similar, a chromosomal fusion, several large-scale rearrangements, and other more subtle changes have created sequences unique to humans (Yunis and Prakash 1982; Johnson et al. 2001). We found no evidence of gene clusters with similar expression patterns located near the region involved in the telomere-to-telomere fusion event that lead to the formation of human chromosome 2 (Fan et al. 2002) and the pericentric inversions involving human chromosomes 4, 12, and 17 (Nickerson and Nelson 1998; Kehrer-Sawatzki et al. 2002). Therefore, these rearrangements do not appear to have major long-range influences on geneexpression patterns in this system. Furthermore, there were only a few cases in which the differentially expressed genes were closely spaced together, which may indicate the absence of large chromosomal blocks containing differentially expressed genes in this system. It will be necessary to analyze additional cell types with more inclusive microarrays and also consider African great ape genome map data (Samonte and Eichler 2002) to rigorously rule out the occurrence of such large blocks. Finally, there was no evidence of chromosomes that show an increased or decreased number of such genes than expected by chance. This is in contrast to an observation that protein evolution is more than 2.2 times faster in chromosomes that have undergone structural rearrangements in humans and chimpanzees than in those that are collinear between the species (Navarro and Barton 2003).
It has been reported that the rate acceleration of brain gene-expression differences in the human lineage (Enard et al. 2002a) was due to up-regulation (induction) of genes in the human lineage (Gu and Gu 2003). This analysis is complicated by mismatches of African great ape RNAs with microarray probes designed to interrogate human RNAs that cause the hybridization data to underestimate the abundance of certain transcripts. In cases involving a substantial overabundance of induced human lineage-specific genes, such as found in the human frontal cortex (Enard et al. 2002a; Gu and Gu 2003), the effect of these artifacts will be diminished by the presence of true signal. Here, if we assume that ∼43% of the genes up-regulated in humans are artifacts (on the basis of Northern blot data), there is significantly less evidence of pronounced gene induction in the human relative to the bonobo lineage. Geneexpression experiments from African great apes and other primates using microarrays containing species-specific probe sequences are needed in order to most rigorously address these issues.
It has been reported that the levels of several mitosis-related genes in cultured human fibroblasts are strongly influenced by donor age (Ly et al. 2000). Here, we found that the expression of these genes were among the most sensitive to growth conditions. In fact, we found no correlation between donor age and the expression patterns of these genes within these species. Overall, this suggests that subtle changes in culture conditions can influence gene-expression patterns of mitosis-related genes more strongly than donor age.
Due to potential mismatches between nonhuman primate RNAs and human probes, genes predicted by microarray analysis to be differentially expressed among humans and African great apes should be confirmed by an independent method prior to further studies. Below, we discuss the function and possible significance of differentially expressed genes whose levels have been confirmed by Northern blot analysis.
Several genes whose gene products are part of or interact with the extracellular matrix were differentially expressed across species. The glypican 3 (GPC3) gene is not expressed in human, but is moderately expressed in bonobo and gorilla cell lines (Fig. 2A, Table 3). GPC3 is a cell-surface heparin sulfate-modified proteoglycan that plays a role in controlling cell growth and division and shows differential expression in several cancers (Filmus and Selleck 2001; Toretsky et al. 2001; Xiang et al. 2001; Midorikawa et al. 2003). Mutations in GPC3 cause Simpson-Golabi-Behmel (SGB) syndrome, a recessive familial overgrowth disorder that manifests in craniofacial and skeletal abnormalities and mental retardation (Filmus and Selleck 2001). Altered GPC3 gene regulation during development could provide one mechanism for differences in growth rates, skeletal structure, and cognitive functions among hominoids. Similar patterns of GPC3 expression were also found in human and chimpanzee brains (Enard et al. 2002a). Another differentially expressed gene whose product is involved with the extracellular matrix is the transforming growth factor β-induced (TGFBI) gene. TGFBI is up-regulated in human relative to African great ape cell lines (Fig. 2B; Table 4). TGFBI contains an RGD motif that is found in extracellular matrix proteins and acts as a ligand-recognition sequence for several integrins (Munier et al. 1997). Differential expression of genes involved in the extracellular matrix could influence cell adhesion and signaling in hominoid connective tissue.
The phyantol CoA hydroxylase (PHYH) gene is upregulated in human relative to African great ape cell lines (Fig. 2A; Table 4). PHYH catalyzes the initial α-oxidation step in the degradation of phytanic acid by converting phytanoyl-CoA to 2-hydroxyphytanoyl-CoA (Jansen et al. 1997; Mihalik et al. 1997; Wanders et al. 2001). PHYH deficiency causes elevated phytanic acid levels in blood and other tissues (Wanders et al. 2001). Therefore, these expression patterns may be related to an increased need to metabolize phytanic acid in humans due to dietary changes (Stanford and Bunn 2001; Wanders et al. 200l; Broadhurst et al. 2002; C.E. Finch and C.B. Stanford, in prep.) This can result in Refsum's disease as well as several peroxisomal biogenesis disorders that are characterized by peripheral neuropathy, cerebellar ataxia, and skeletal abnormalities (Powers and Moser 1998; Moser 2000; Wanders et al. 2001). Similar differences in PHYH expression levels were found in human and chimpanzee livers and brains (Enard et al. 2002a). Phytanic acid is only obtained in humans via dietary sources such as dairy products, ruminant fats, and seafood that are not parts of African great ape diets. Therefore, these expression patterns may be related to an increased need to metabolize phytanic acid in humans due to dietary changes (Stanford and Bunn 2001; Wanders et al. 2001; Broadhurst et al. 2002). Other pleiotrophic effects related to brain development and skeletal morphology could result from differential PHYH expression in specific human cells and give rise to species-specific traits.
Several other genes from diverse functional classes were differentially expressed among humans and the African great apes (Fig. 2B,C,D; Tables 3 and 4; Suppl. Fig. 1). The glutathione S-transferase theta 2 (GSTT2) gene, up-regulated in bonobos relative to both humans and gorillas (Table 3; Fig. 2C), catalyzes the conjugation of reduced glutathione to a variety of electrophilic and hydrophobic compounds (Landi 2000). This could reflect the evolutionary history of GST gene family members or adaptations to environmental toxins. Similar patterns of GSTT2 expression were also found in human and chimpanzee livers and brains (Enard et al. 2002a). Genes involved in signal transduction pathways, the regulator of G-protein signaling 7 (RGS7) gene (Rose et al. 2000; Ingi and Aoki 2002) and muscle contraction, myosin light chain kinase (MYLK) gene (Lazar and Garcia 1999; Giorgi et al. 2001), were also differentially expressed among humans and the African great apes (Tables 3 and 4; Fig. 2B; Suppl. Fig. 1). Furthermore, MYLK appears to be regulated in a similar fashion in human and chimpanzee brains (Enard et al. 2002a). In addition, we found that the stromal cell-derived factor 1 (SDF1) is up-regulated in human relative to the African great ape fibroblast cell lines (Fig. 2D; Table 4). SDF1 is the principal ligand for the CXCR4 coreceptor for HIV-1 and is also involved in neuronal cell migration, differentiation, and proliferation (Ma et al. 1998; Barbero et al. 2002).
Here, we have taken a first step in characterizing the heterogeneity of gene-expression patterns among characterized populations of a specific cell type within human and the African great apes. These gene-expression patterns may reflect biochemical differences in these species that could manifest in phenotypic changes. Because phenotypic differences among humans and African great apes are likely caused by variations in protein activities and levels as well as posttranslational modifications during development and adulthood (King and Wilson 1975; Goldman et al. 1987; Gagneux and Varki 2001; Gagneux et al. 2001; Hacia 2001), investigation in tissues and other cell types from a variety of primates is needed to determine their effect on physiological processes (Goodman 1999). Collections of these materials will continue to play a vital role in understanding the genetic and biochemical basis for the recent evolution of complex human phenotypes involved in health and disease.
METHODS
Fibroblast Cell Lines
Primary human fibroblast cell lines were obtained from Coriell Cell Repositories. Primary bonobo (Pan paniscus) and gorilla (Gorilla gorilla) fibroblasts were obtained from the Zoological Society of San Diego and the Center for the Reproduction of Endangered Species (CRES). All primary fibroblast cultures were initiated from explants of 2-mm-punch skin biopsies and cultured in selective medium. All human cultures had normal karyotypes, and the age and gender of all fibroblast donors were documented.
Cell Culture and RNA Isolation
Primary fibroblasts were cultured in rich medium (DMEM supplemented with vitamins, essential and nonessential amino acids, antibiotic and 10% FBS) at 37°C in 5% CO2. Growth rates were measured in a 96-well format using Cell-Titer 96-AQueous One Solution Cell Proliferation Assay (Promega).
RNA was isolated from the cell lines when they reached 80% confluency using the RNA Stat-60 reagent (Tel-Test, Inc.). Gene-expression profiles from one human (AG11745), one bonobo (KB10025), and one gorilla (KB7621) cell line harvested at 50% confluency are available in Supplementary Table 8. Confluency did not have a significant impact on the interspecies gene-expression differences listed in Tables 3 and 4 (Suppl. Table 9).
Oligonucleotide Microarray Experiments
Fibroblast total RNA samples (20 μg/sample) were converted into biotin-labeled cRNA using standard protocols recommended by Affymetrix. For each cell line, 15 μg of fragmented cRNA was applied to a U95Av2 microarray designed to interrogate the relative abundance of >10,000 human transcripts. Microarrays were hybridized 12–16 h at 45°C under standard conditions. Following hybridization, microarrays were washed using the Affymetrix Fluidics Station 400, stained twice with a streptavidin-phycoerythrin conjugate, and then read using a Hewlett Packard GeneArray Scanner.
Microarray Data Analysis
We generated raw gene-expression scores for every gene in every sample using GENECHIP version 5.0 software (Affymetrix). To normalize the relative hybridization signal from each experiment, we calculated multiplicative scaling factors on the basis of the median intensity of the 60th to 95th percentile of gene-expression scores. All gene-expression scores below 100 were set to 100 to minimize noise associated with less robust measurements of rare transcripts. On average, 40% of the human and 37% of the bonobo and gorilla transcripts examined were called present in these cell lines. Geneexpression data from all experiments are available in Affymetrix GENECHIP (.CEL) format on our Web site and Microsoft Excel 2000 (.XLS) format in Supplementary Table 8.
Normalized gene-expression data was imported into dChip software (http://www.biostat.harvard.edu/complab/dchip/) for hierarchical clustering analysis using the average linkage algorithm and for calculating 95% confidence interval (CI) for the fold changes in gene expression among the species. Clusters were based upon gene-expression profiles from >5300 genes whose coefficient of variation (SD/mean) is >0.3 across all samples. This allowed us to focus on the expression profiles of genes showing intra- and/or interspecies variation. We found independently labeled human and African great ape RNA samples to provide highly reproducible microarray results (average correlation coefficient of 0.98).
Because 16 perfect match probes interrogate each gene, the effect of mismatches between African great ape targets and human probes should be minimal (Hacia et al. 1998, 1999; Bigger et al. 2001; Enard et al. 2002a). However, when probes interrogating a given transcript are partially overlapping, a single nucleotide change or small deletion in African great ape mRNA can reduce target affinity to more than one human probe and have a major affect on the analysis. In Table 4, we omitted genes with highly overlapping probes that could be affected by single-nucleotide differences between human and the African great ape genomes. Furthermore, we omitted genes in which different probe tilings gave conflicting results. In Table 3, we omitted the IGFBP2 gene, as it showed greater than 2.5-fold gene-expression differences between the Hsa1 and Hsa2 groups. The elimination of these genes will increase the specificity of identifying differentially expressed genes among humans and African great apes by avoiding possible artifacts.
Northern Blot Analysis
Ten micrograms of total RNA or 200 ng of mRNA from at least two cell lines representing each species were resolved on 1% formaldehyde gels and blotted onto Hybond XL nylon membranes (Amersham Pharmacia Biotech). SuperScript One-Step RT–PCR (Invitrogen) kits were used to amplify the 3′-UTR of genes of interest from human, bonobo, and gorilla fibroblast RNA samples. Human placental RNA (Invitrogen) was used as template for the GPC3 gene, as no RT–PCR product could be generated from human fibroblast RNA. For each gene analyzed, human and African great ape amplicons were mixed in equimolar amounts and radiolabeled using the Rediprime II kit (Amersham Pharmacia Biotech). Therefore, the orthologous human, bonobo, and gorilla mRNAs had equal opportunity to hybridize to radiolabeled gene-specific probes regardless of species-specific sequence differences. Blots were hybridized in ExpressHyb solution (Clontech) and washed according to established protocols. Processed blots were analyzed using the Storm PhosphorImager system (Molecular Dynamics). Band intensities were quantified using ImageQuant v5.2 software (Molecular Dynamics) and corrected for loading differences on the basis of β-actin mRNA or 28S rRNA levels.
DideoxySequencing Analysis
Amplicons generated by RT–PCR from African great ape fibroblast RNA samples were subcloned using the TOPO TA Cloning kit (Invitrogen) and sequenced using M13 forward and reverse primers. Sequencher v3.0 software (Gene Codes Corp.) was used to analyze and compare sequences from different species. African great ape sequences were compared against human probe sequences in the U95Av2 microarray to identify possible mismatches that might have resulted in an underestimation of specific message levels (Suppl. Table 1).
Gene Localization and Statistical Analysis
Chromosomal gene locations were determined using the Human Genome Resources on the NCBI Web site (http://www.ncbi.nlm.nih.gov/genome/guide/human/). We used the Human Genome Browser (June 2002 freeze) at the UCSC Genome Bioinformatics site (http://www.genome.ucsc.edu/) to determine the location of genes that gave ambiguous results in the above method.
We calculated the ratio of the total number of differentially expressed genes to the total number of probe tilings on the U95Av2 microarray. This provides us with the number of differentially expressed genes per probe tiling. Then we determined the expected number of differentially expressed genes on each chromosome on the basis of the number of probe tilings representing that chromosome on the U95Av2 microarray. The 95% confidence intervals for the number of genes predicted to be differentially expressed on each chromosome was calculated on the basis of a binomial distribution using InStat (GraphPad Software)
Acknowledgments
We thank Francis Collins, Jeff Trent, Mike Bittner, Aeryn Mayer, Larry Brody, Suellen Charter, and Elizabeth Novotny for technical assistance and/or discussion. We thank Mario Caceres, Carollee Barlow, and David Lockhart for sharing unpublished data. Partial support for this work was provided by the James Zumberge Award (J.G.H.)
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1289803.
Footnotes
References
- Barbero, S., Bajetto, A., Bonavia, R., Porcile, C., Piccioli, P., Pirani, P., Ravetti, J.L., Zona, G., Spaziante, R., Florio, T., et al. 2002. Expression of the chemokine receptor CXCR4 and its ligand stromal cell-derived factor 1 in human brain tumors and their involvement in glial proliferation in vitro. Ann. NY Acad. Sci. 973: 60-69. [DOI] [PubMed] [Google Scholar]
- Bigger, C.B., Brasky, K.M., and Lanford, R.E. 2001. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J. Virol. 75: 7059-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten, R.J. 2002. Divergence between samples of chimpanzee and human DNA sequences is 5%, counting indels. Proc. Natl. Acad. Sci. 99: 13633-13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadhurst, C.L., Wang, Y., Crawford, M.A., Cunnane, S.C., Parkington, J.E., and Schmidt, W.F. 2002. Brain-specific lipids from marine, lacustrine, or terrestrial food resources: Potential impact on early African Homo sapiens. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 131: 653-673. [DOI] [PubMed] [Google Scholar]
- Chang, H.Y., Chi, J.T., Dudoit, S., Bondre, C., van de Rijn, M., Botstein, D., and Brown, P.O. 2002. Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc. Natl. Acad. Sci. 99: 12877-12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F.C. and Li, W.H. 2001. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 68: 444-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, H.H., Takematsu, H., Diaz, S., Iber, J., Nickerson, E., Wright, K.L., Muchmore, E.A., Nelson, D.L., Warren, S.T., and Varki, A. 1998. A mutation in human CMP-sialic acid hydroxylase occurred after the Homo-Pan divergence. Proc. Natl. Acad. Sci. 95: 11751-11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, H.H., Hayakawa, T., Diaz, S., Krings, M., Indriati, E., Leakey, M., Paabo, S., Satta, Y., Takahata, N., and Varki, A. 2002. Inactivation of CMP-N-acetylneuraminic acid hydroxylase occurred prior to brain expansion during human evolution. Proc. Natl. Acad. Sci. 99: 11736-11741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly, G.P. 1998. Fibroblast models of neurological disorders: Fluorescence measurement studies. Trends Pharmacol. Sci. 19: 171-177. [DOI] [PubMed] [Google Scholar]
- Cristofalo, V.J., Allen, R.G., Pignolo, R.J., Martin, B.G., and Beck, J.C. 1998. Relationship between donor age and the replicative lifespan of human cells in culture: A reevaluation. Proc. Natl. Acad. Sci. 95: 10614-10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler, E.E., Johnson, M.E., Alkan, C., Tuzun, E., Sahinalp, C., Misceo, D., Archidiacono, N., and Rocchi, M. 2001. Divergent origins and concerted expansion of two segmental duplications on chromosome 16. J. Hered. 92: 462-468. [DOI] [PubMed] [Google Scholar]
- Enard, W., Khaitovich, P., Klose, J., Zollner, S., Heissig, F., Giavalisco, P., Nieselt-Struwe, K., Muchmore, E., Varki, A., Ravid, R., et al. 2002a. Intra- and interspecific variation in primate gene expression patterns. Science 296: 340-343. [DOI] [PubMed] [Google Scholar]
- Enard, W., Przeworski, M., Fisher, S.E., Lai, C.S., Wiebe, V., Kitano, T., Monaco, A.P., and Paabo, S. 2002b. Molecular evolution of FOXP2, a gene involved in speech and language. Nature 418: 869-872. [DOI] [PubMed] [Google Scholar]
- Fan, Y., Linardopoulou, E., Friedman, C., Williams, E., and Trask, B.J. 2002. Genomic structure and evolution of the ancestral chromosome fusion site in 2q13-2q14.1 and paralogous regions on other human chromosomes. Genome Res. 12: 1651-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filmus, J. and Selleck, S.B. 2001. Glypicans: Proteoglycans with a surprise. J. Clin. Invest. 108: 497-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneux, P. and Varki, A. 2001. Genetic differences between humans and great apes. Mol. Phylogenet. Evol. 18: 2-13. [DOI] [PubMed] [Google Scholar]
- Gagneux, P., Amess, B., Diaz, S., Moore, S., Patel, T., Dillmann, W., Parekh, R., and Varki, A. 2001. Proteomic comparison of human and great ape blood plasma reveals conserved glycosylation and differences in thyroid hormone metabolism. Am. J. Phys. Anthropol. 115: 99-109. [DOI] [PubMed] [Google Scholar]
- Giorgi, D., Brand-Arpon, V., and Rouquier, S. 2001. The functional myosin light chain kinase (MYLK) gene localizes with marker D3S3552 on human chromosome 3q21 in a >5-Mb yeast artificial chromosome region and is not linked to olfactory receptor genes. Cytogenet. Cell. Genet. 92: 85-88. [DOI] [PubMed] [Google Scholar]
- Goldman, D., Giri, P.R., and O'Brien, S.J. 1987. A molecular phylogeny of the hominoid primates as indicated by two-dimensional protein electrophoresis. Proc. Natl. Acad. Sci. 84: 3307-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman, M. 1999. The genomic record of Humankind's evolutionary roots. Am. J. Hum. Genet. 64: 31-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, J. and Gu, X. 2003. Induced gene expression in human brain after the split from chimpanzee. Trends Genet. 19: 63-65. [DOI] [PubMed] [Google Scholar]
- Gu, Z., Nicolae, D., Lu, H.H., and Li, W.H. 2002. Rapid divergence in expression between duplicate genes inferred from microarray data. Trends Genet. 18: 609-613. [DOI] [PubMed] [Google Scholar]
- Hacia, J.G. 2001. Genome of the apes. Trends Genet. 17: 637-645. [DOI] [PubMed] [Google Scholar]
- Hacia, J.G., Makalowski, W., Edgemon, K., Erdos, M.R., Robbins, C.M., Fodor, S.P., Brody, L.C., and Collins, F.S. 1998. Evolutionary sequence comparisons using high-density oligonucleotide arrays. Nat. Genet. 18: 155-158. [DOI] [PubMed] [Google Scholar]
- Hacia, J.G., Fan, J.B., Ryder, O., Jin, L., Edgemon, K., Ghandour, G., Mayer, R.A., Sun, B., Hsie, L., Robbins, C.M., et al. 1999. Determination of ancestral alleles for human single-nucleotide polymorphisms using high-density oligonucleotide arrays. Nat. Genet. 22: 164-167. [DOI] [PubMed] [Google Scholar]
- Ingi, T. and Aoki, Y. 2002. Expression of RGS2, RGS4 and RGS7 in the developing postnatal brain. Eur. J. Neurosci. 15: 929-936. [DOI] [PubMed] [Google Scholar]
- Irie, A., Koyama, S., Kozutsumi, Y., Kawasaki, T., and Suzuki, A. 1998. The molecular basis for the absence of N-glycolylneuraminic acid in humans. J. Biol. Chem. 273: 15866-15871. [DOI] [PubMed] [Google Scholar]
- Irving, J.A., Pike, R.N., Lesk, A.M., and Whisstock, J.C. 2000. Phylogeny of the serpin superfamily: Implications of patterns of amino acid conservation for structure and function. Genome Res. 10: 1845-1864. [DOI] [PubMed] [Google Scholar]
- Jansen, G.A., Ofman, R., Ferdinandusse, S., Ijlst, L., Muijsers, A.O., Skjeldal, O.H., Stokke, O., Jakobs, C., Besley, G.T., Wraith, J.E., et al. 1997. Refsum disease is caused by mutations in the phytanoyl-CoA hydroxylase gene. Nat. Genet. 17: 190-193. [DOI] [PubMed] [Google Scholar]
- Johnson, M.E., Viggiano, L., Bailey, J.A., Abdul-Rauf, M., Goodwin, G., Rocchi, M., and Eichler, E.E. 2001. Positive selection of a gene family during the emergence of humans and African apes. Nature 413: 514-519. [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki, H., Schreiner, B., Tanzer, S., Platzer, M., Muller, S., and Hameister, H. 2002. Molecular characterization of the pericentric inversion that causes differences between chimpanzee chromosome 19 and human chromosome 17. Am. J. Hum. Genet. 71: 375-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, M.C. and Wilson, A.C. 1975. Evolution at two levels in humans and chimpanzees. Science 188: 107-116. [DOI] [PubMed] [Google Scholar]
- Kreitman, M. 2000. Methods to detect selection in populations with applications to the human. Annu. Rev. Genomics Hum. Genet. 1: 539-559. [DOI] [PubMed] [Google Scholar]
- Landi, S. 2000. Mammalian class θ GST and differential susceptibility to carcinogens: A review. Mutat. Res. 463: 247-283. [DOI] [PubMed] [Google Scholar]
- Lazar, V. and Garcia, J.G. 1999. A single human myosin light chain kinase gene (MLCK; MYLK). Genomics 57: 256-267. [DOI] [PubMed] [Google Scholar]
- Ly, D.H., Lockhart, D.J., Lerner, R.A., and Schultz, P.G. 2000. Mitotic misregulation and human aging. Science 287: 2486-2492. [DOI] [PubMed] [Google Scholar]
- Ma, Q., Jones, D., Borghesani, P.R., Segal, R.A., Nagasawa, T., Kishimoto, T., Bronson, R.T., and Springer, T.A. 1998. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc. Natl. Acad. Sci. 95: 9448-9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midorikawa, Y., Ishikawa, S., Iwanari, H., Imamura, T., Sakamoto, H., Miyazono, K., Kodama, T., Makuuchi, M., and Aburatani, H. 2003. Glypican-3, overexpressed in hepatocellular carcinoma, modulates FGF2 and BMP-7 signaling. Int. J. Cancer 103: 455-465. [DOI] [PubMed] [Google Scholar]
- Mihalik, S.J., Morrell, J.C., Kim, D., Sacksteder, K.A., Watkins, P.A., and Gould, S.J. 1997. Identification of PAHX, a Refsum disease gene. Nat. Genet. 17: 185-189. [DOI] [PubMed] [Google Scholar]
- Moser, H.W. 2000. Molecular genetics of peroxisomal disorders. Front. Biosci. 5: D298-306. [DOI] [PubMed] [Google Scholar]
- Munier, F.L., Korvatska, E., Djemai, A., Le Paslier, D., Zografos, L., Pescia, G., and Schorderet, D.F. 1997. Kerato-epithelin mutations in four 5q31-linked corneal dystrophies. Nat. Genet. 15: 247-251. [DOI] [PubMed] [Google Scholar]
- Navarro, A. and Barton, N.H. 2003. Chromosomal speciation and molecular divergence—accelerated evolution in rearranged chromosomes. Science 300: 321-324. [DOI] [PubMed] [Google Scholar]
- Nickerson, E. and Nelson, D.L. 1998. Molecular definition of pericentric inversion breakpoints occurring during the evolution of humans and chimpanzees. Genomics 50: 368-372. [DOI] [PubMed] [Google Scholar]
- Nimchinsky, E.A., Gilissen, E., Allman, J.M., Perl, D.P., Erwin, J.M., and Hof, P.R. 1999. A neuronal morphologic type unique to humans and great apes. Proc. Natl. Acad. Sci. 96: 5268-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, M.V. 1999. When less is more: Gene loss as an engine of evolutionary change. Am. J. Hum. Genet. 64: 18-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, M.V. and Varki, A. 2003. Sequencing the chimpanzee genome: Insights into human evolution and disease. Nat. Rev. Genet. 4: 20-28. [DOI] [PubMed] [Google Scholar]
- Powers, J.M. and Moser, H.W. 1998. Peroxisomal disorders: Genotype, phenotype, major neuropathologic lesions, and pathogenesis. Brain Pathol. 8: 101-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, J.J., Taylor, J.B., Shi, J., Cockett, M.I., Jones, P.G., and Hepler, J.R. 2000. RGS7 is palmitoylated and exists as biochemically distinct forms. J. Neurochem. 75: 2103-2112. [DOI] [PubMed] [Google Scholar]
- Samonte, R.V. and Eichler, E.E. 2002. Segmental duplications and the evolution of the primate genome. Nat. Rev. Genet. 3: 65-72. [DOI] [PubMed] [Google Scholar]
- Schneider, E.L., Braunschweiger, K., and Mitsui, Y. 1978. The effect of serum batch on the in vitro lifespans of cell cultures derived from old and young human donors. Exp. Cell. Res. 115: 47-52. [DOI] [PubMed] [Google Scholar]
- Stanford, C.B. and Bunn, H.T. 2001. Meat-eating & human evolution. Oxford University Press, Oxford, UK; New York.
- Stankiewicz, P., Park, S.S., Inoue, K., and Lupski, J.R. 2001. The evolutionary chromosome translocation 4;19 in Gorilla gorilla is associated with microduplication of the chromosome fragment syntenic to sequences surrounding the human proximal CMT1A-REP. Genome Res. 11: 1205-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer, C. 2002. Modern human origins: Progress and prospects. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 357: 563-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toretsky, J.A., Zitomersky, N.L., Eskenazi, A.E., Voigt, R.W., Strauch, E.D., Sun, C.C., Huber, R., Meltzer, S.J., and Schlessinger, D. 2001. Glypican-3 expression in Wilms tumor and hepatoblastoma. J. Pediatr. Hematol. Oncol. 23: 496-499. [DOI] [PubMed] [Google Scholar]
- Wanders, R.J., Jansen, G.A., and Skjeldal, O.H. 2001. Refsum disease, peroxisomes and phytanic acid oxidation: A review. J. Neuropathol. Exp. Neurol. 60: 1021-1031. [DOI] [PubMed] [Google Scholar]
- Xiang, Y.Y., Ladeda, V., and Filmus, J. 2001. Glypican-3 expression is silenced in human breast cancer. Oncogene 20: 7408-7412. [DOI] [PubMed] [Google Scholar]
- Yunis, J.J. and Prakash, O. 1982. The origin of man: A chromosomal pictorial legacy. Science 215: 1525-1530. [DOI] [PubMed] [Google Scholar]
- Zamir, E. and Geiger, B. 2001. Molecular complexity and dynamics of cell-matrix adhesions. J. Cell. Sci. 114: 3583-3590. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Webb, D.M., and Podlaha, O. 2002. Accelerated protein evolution and origins of human-specific features. Foxp2 as an example. Genetics 162: 1825-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.biostat.harvard.edu/complab/dchip/; dChip software.
- http://www.genome.ucsc.edu/; UCSC Genome Bioinformatics site.
- http://www.ncbi.nlm.nih.gov/genome/guide/human/; Human Genome Resources on the NCBI Web site.
- http://www.ncbi.nlm.nih.gov/LocusLink/; gene annotation.