Abstract
A thorough understanding of the developmental signals that direct pluripotent stem cells (PSCs) towards a cardiac fate is essential for translational applications in disease modeling and therapy. We screened a panel of 44 cytokines/signaling molecules for their ability to enhance Nkx2.5+ cardiac progenitor cell (CPC) formation during in vitro embryonic stem cell (ESC) differentiation. Treatment of murine ESCs with insulin or insulin-like growth factors (IGF1/2) during early differentiation increased mesodermal cell proliferation and, consequently, CPC formation. Furthermore, we show that downstream mediators of IGF signaling (e.g. phospho-Akt and mTOR) are required for this effect. These data support a novel role for IGF family ligands to expand the developing mesoderm and promote cardiac differentiation. Insulin or IGF treatment could provide an effective strategy to increase the PSC-based generation of CPCs and cardiomyocytes for applications in regenerative medicine.
Keywords: embryonic stem cell, development, mesoderm, cardiac progenitor cell, cardiomyocyte, cardiac differentiation, in vitro screening, insulin, insulin-like growth factor, Akt
Introduction
Despite the availability of many treatment options, heart disease remains the leading cause of death worldwide, prompting the need for more innovative therapeutic strategies such as cell-based therapy [1]. The ability to produce patient-specific induced pluripotent stem cells (iPSCs) holds great promise for such regenerative applications [2-3]. A pivotal challenge in translating the potential of iPSCs into effective cardiac therapy is to generate sufficient quantities of functional cardiomyocytes to replace the large numbers of cells that are lost after myocardial injury [4]. Although in vitro cardiac differentiation protocols for PSCs are readily available, the yield for most PSC lines remains modest and highly variable [5,6]. To improve the efficiency of cardiac differentiation, it is critical to understand the molecular mechanism of pluripotent cell commitment towards mesoderm during early development [7].
In vitro differentiation of ESCs has been used to model early cardiac development due to the limited number of cells available when working with early stage embryos. When provided with appropriate cues, ESCs have been shown to faithfully recapitulate developmental gene expression patterns [8]. During ESC differentiation, a gastrulation-like step takes place resulting in the commitment of some cells into ectodermal lineage and another set of cells into the mesendodermal lineage. A portion of the latter cells gives rise to the Brachyury+ mesodermal cell population. Some of these Brachyury+ cells become the first committed cardiac progenitor cells (CPCs) as defined by their expression of two key cardiac transcription factors, Isl-1 and Nkx2.5 [9-10]. CPCs are multipotent at this stage and can give rise to cardiomyocytes, smooth muscle cells and endothelial cells [11-12].
Cardiogenic commitment is driven by the activation of a number of highly conserved signaling pathways. For example, the transforming growth factor β (TGF-β) superfamily members Activin A, bone morphogenetic protein 4 (BMP4) and Nodal, as well as members of the fibroblast growth factor (i.e. FGF2) and Wnt (i.e. Wnt3a) families of signaling molecules have been shown to enhance or inhibit cardiac differentiation in a spatial- and temporal-specific fashion [5, 13-19]. To comprehensively evaluate signaling pathway activation during early cardiac lineage induction, we systematically screened a panel of 44 candidate cytokines/signaling molecules for their ability to enhance CPC formation. Consistent with previous findings, Wnt3a treatment during early differentiation enhanced mesodermal commitment leading to increased Nkx2.5+ CPC formation [20-21]. Surprisingly, treatment with insulin and insulin-like growth factors (IGFs) positively regulated selective expansion of the mesendodermal cell population resulting in greater CPC formation. These ligands act through phosphorylation and activation of downstream targets such as Akt and mTOR and synergize with Wnt3a and FGF2. Mechanistically, IGF induces selective expansion of the mesodermal cell population through increased proliferation. This study reveals a role for IGFs and insulin as regulators of in vitro mesodermal expansion and provides a strategy to significantly enhance the generation of pluripotent stem cell-derived cardiac progenitor cells.
Materials and Methods
Growth Factor Screening
A previously described ESC line in which a cardiac-specific enhancer and base promoter of the murine Nkx2.5 locus drive enhanced green fluorescent protein (eGFP) gene expression was used for all experiments [11]. Nkx2.5-eGFP ESCs were cultured as previously described [11]. For the in vitro screening studies, cells were cultured in differentiation media containing 2% fetal bovine serum (FBS lot 894969; Life Technologies, Grand Island, NY, USA) and seeded at 4,000 cells/well in gelatin-coated 96-well plates (Corning Life Sciences, Tewksbury, MA, USA). Growth factors and signaling molecules were obtained from R&D Systems (Minneapolis, MN, USA) as lyophilized powder and reconstituted as recommended by the manufacturer. A complete list of the compounds used for screening with the screening concentration range can be found in Supplemental Table 1. Growth factors/signaling molecules were added to the cell culture at day 3 of differentiation at 1:2 dilutions in a dose range 4-1000 μg/mL (i.e. 3.9, 7.8, 15.6, 31.3, 62.5, 125, 250, 500, 1000 μg/mL). The concentration with the greatest effect on GFP+ signal was considered for our screening results. Cells were assayed on day 6 of differentiation using a FACSCalibur high-throughput screening platform for 96-well plates (BD Biosciences, San Jose, CA, USA). Data were analyzed with FlowJo software (Tree Star, Ashland, OR, USA). The percentage of Nkx2.5-eGFP+ cells was assessed for each treatment group and compared to that of solvent-exposed control cells. Each experiment was performed in triplicate and three independent experiments were performed for each condition. A hit was determined as having a P value<0.05 using two-tailed Student’s t-test. For all subsequent experiments, optimized treatment timing and cytokine concentrations were used (Fig. 1D and Supplemental Fig. 1).
Figure 1.
A murine ESC-based high throughput screen to identify growth factors/signaling molecules regulating Nkx2.5+ CPC formation. (A) Schematic diagram of the high-throughput growth factor screen. Candidate growth factors/signaling molecules were added on day 3 of differentiation and cells were harvested on day 6 for flow cytometric analysis. PS, primitive streak; CMC, cardiomyocyte. (B) Quantification of eGFP+ CPCs following growth factor treatment of Nkx2.5-eGFP ESCs. Values represent fold change in eGFP+ cell count by growth factor treatment. **, p<0.01 vs. untreated control cells; NS, non-significant difference with untreated control cells. (C) Representative flow cytometry dot plots of day 6 Nkx2.5-eGFP ESCs after treatment with IGF1, IGF2 or insulin on day 3. (D) Dose responses of Nkx2.5-eGFP ESCs to treatment with FGF2, Wnt3a, IGF1, IGF2 or insulin. Values represent fold increase of eGFP+ cells as compared to untreated control cells. Mean ± standard error of the mean (SEM) of triplicate experiments is shown.
Immunofluorescence Microscopy
Nkx2.5-eGFP ESCs were differentiated as previously described [11] and treated with growth factors at day 1 or 2 of differentiation. Cells were cultured on 0.1%-gelatin-coated coverslips until assay or beating embryoid bodies (EBs) were manually collected and transferred to Nunc Lab-Tek II Chamber Slides (Thermo Fisher Scientific, Waltham, MA, USA). Cells were then fixed with 4% paraformaldehyde solution (Thermo Fisher Scientific) and permeabilized with Triton-X100 (Sigma-Aldrich, St. Louis, MO, USA). After washing 3 times with phosphate-buffered saline (PBS)/10%FBS/0.1% Tween-20 (Sigma-Aldrich), cells were incubated overnight at 4°Celsius (C) with mouse monoclonal antibodies directed against sarcomeric α-actinin (clone EA-53; Sigma-Aldrich; 1:250 dilution), affinity-purified goat-anti-Brachyury IgG (R&D Systems; 1:100 dilution), rabbit anti-Ki67 IgG (clone D3B5; Cell Signaling Technology, Danvers, MA, USA; 1:100 dilution) and rabbit anti-phospho-Akt (Thr308) IgG (clone C31E5E; Cell Signaling Technology, 1:1600 dilution). Next, cells were washed again and incubated for 60 minutes at RT (room temperature) with Alexa Fluor dye-conjugated secondary antibodies (Life Technologies; diluted 1:500 in PBS/10%FBS/0.1% Tween-20). Immunostained samples were mounted using VECTASHIELD mounting medium with 4’,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA, USA). For Draq5 labeling, Draq5 reagent (Cell Signaling Technology, Danvers, MA, USA, 1:5000 dilution) was added to the culture media and incubated for 5 minutes at RT. Live cells were immediately imaged after staining. Images were acquired with an LSM 510 META inverted laser scanning confocal microscope (Carl Zeiss, Oberkochen, Germany) or a digital color camera-equipped fluorescence microscope (Nikon Eclipse 80i; Nikon Instruments Europe, Amstelveen, the Netherlands), and processed using Fiji software (www.fiji.sc) and Adobe Photoshop version 5 (Adobe Systems, San Jose, CA, USA).
Quantitative Reverse Transcription-Polymerase Chain Reaction (RT-qPCR)
Nkx2.5-eGFP ESCs were seeded in 24-well plates (Corning Life Sciences) and treated with growth factors at day 1 or 2 of culture. At different time points after growth factor addition (i.e. day 0, 2, 4, 6 and 8), cells were lysed in TRIzol Reagent (Life Technologies) and total RNA was isolated using the RNeasy Mini Kit (Qiagen, Venlo, the Netherlands). Reverse transcription was done with iScript cDNA Synthesis Kit (Bio-Rad, Hercules CA, USA). Gene expression levels were assayed using HotStart-IT SYBR Green qPCR Master Mix (Affymetrix, Santa Clara, CA, USA) and the primer pairs specified in Supplemental Table 2. PCR amplifications were performed in a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Each condition was tested in three independent experiments using three samples per experiment.
Flow cytometry
Nkx2.5-eGFP ESCs were cultured in 24-well plates and treated at day 1 or 2 with growth factors. At the time point(s) of interest, cells were dissociated using trypsin/EDTA (Life Technologies) and 10 mg/mL collagenase A/B (Roche Applied Science, Indianapolis, IN, USA) solutions in Hank’s buffered salt solution (HBSS;Invitrogen)/20% FBS, fixed in 4% paraformaldehyde solution and permeabilized in ice-cold 100% methanol (Thermo Fisher Scientific). After washing 2 times with PBS, cells were incubated with primary antibodies for 1 hour at RT, washed again and incubated for 1 hour at RT with appropriate Alexa Fluor dye-conjugated secondary antibodies. Following additional washings, cells were suspended in HBSS (Invitrogen) supplemented with 10% FBS and analyzed using a FACSCalibur flow cytometer (BD Biosciences). The following primary antibodies were used: cardiac troponin T (cTnT; clone 13-11; Thermo Fisher Scientific; 1:100 dilution in HBSS/10% FBS) and affinity-purified goat-anti-Brachyury IgG (R&D Systems; 1:100 dilution), mouse anti-GATA4 IgG1 (clone L97-56; BD Biosciences; 1:200 dilution), mouse anti-Nestin IgG2a (clone 307501; R&D Systems; 1:200 dilution) and rabbit anti-Ki67 IgG (clone D3B5; Cell Signaling Technology, Danvers, MA, USA; 1:250 dilution). Appropriate dilutions were determined by comparison with undifferentiated ESCs and growth-inhibited mouse embryonic fibroblasts as negative controls. Data analysis was done using FlowJo software. Each condition was tested in three experiments using three independent samples per experiment.
Flow cytometric analysis of phospho-Akt
Nkx2.5-eGFP ESCs were differentiated as described previously [11]. EBs were collected at day 4. Next, the cells were dissociated, serum-starved for 2 hours and exposed to growth factors for 15 minutes at 37°C in a humidified 5% CO2 atmosphere. Cells were immediately fixed and stained with rabbit anti-phospho-Akt (Thr308) IgG (clone C31E5E; Cell Signaling Technology, diluted as specified by the manufacturer) as described previously [22]. The presence of phospho-Akt was determined using a FACSCalibur flow cytometer and FlowJo software for data analysis.
Small molecule inhibition studies
Validated small-molecule inhibitors of downstream IGF signaling pathway targets were added to Nkx2.5-eGFP ESCs simultaneously with insulin, IGF1 or IGF2 at day 1 or 2. Cells were analyzed by flow cytometry for eGFP expression as previously described. A dose titration was performed to determine the optimal concentration of each compound. The following small molecules were used: MK-2206 dihydrochloride (Selleckchem, Houston, TX, USA), PI 103 hydrochloride (R&D Systems), PP 242 (R&D Systems) and KU-0063794 (Stemgent, Cambridge, MA, USA).
Statistical Methods
A two-tailed Student’s T test was performed using GraphPad Prism software version 6 (GraphPad Software, La Jolla, CA, USA) or Microsoft Excel (Microsoft, Redmond, WA, USA). Results were considered significant at P values <0.05.
Results
IGFs and insulin promote cardiac differentiation of murine ESCs
To identify novel signaling pathways involved in early cardiac lineage commitment, murine Nkx2.5-eGFP ESCs were differentiated in vitro by LIF withdrawal and treated with cytokines/signaling molecules on day 3 of differentiation. On day 6 of differentiation, which coincides with the onset of Nkx2.5+ CPC formation [11], the frequency of eGFP+ cells was quantified by automated flow cytometry (Fig. 1A). Four of the 44 factors that were screened (i.e. IGF1, IGF2, insulin and Wnt3a) significantly increased CPC formation (Fig. 1B-C). Interestingly, treatment with Activin A, BMP2 or BMP4 at day 3 of differentiation decreased CPC formation. The remaining 37 factors showed no significant effect. We validated the effects of the positive hits by demonstrating a dose-dependent increase in eGFP+ cell formation following treatment with IGF1, IGF2, insulin or, as a positive control, Wnt3a, which had been previously implicated in early cardiogenesis [21] (Fig. 1D). Although FGF2 treatment did not reach statistical significance in our initial screening assay, it showed a clear dose-dependent increase in the number of eGFP+ cells when the timing of the treatment was optimized (Supplemental Fig. S1). Also the effect of IGFs and insulin on CPC formation is time/differentiation stage-specific (Supplemental Fig. S1). Accordingly, no increase in cell quantity was observed after treatment of FACS-sorted Nkx2.5-eGFP+ cells (CPCs) with IGF/insulin compared to untreated controls (Supplemental Fig. S2). Furthermore, a synergistic effect on CPC formation was observed between IGF1, IGF2 or insulin and either Wnt3a or FGF2. No synergy was seen between FGF2 and Wnt3a or between members of the IGF family of ligands (Supplemental Fig. S3).
Confirmation of IGF/insulin-mediated increase in CPC formation in differentiating murine ESCs
To validate the ability of IGFs and insulin to enhance cardiogenesis, we measured the expression of cardiac lineage genes in growth factor-treated differentiating cells. Treatment with IGF1, IGF2 or insulin increased the expression of cardiac mesoderm- and CPC-specific genes such as Mesp1, Isl-1, Nkx2.5, GATA4 and MLC2a compared with that in untreated control cells (Fig. 2A-E). The ability of IGF/insulin-induced CPCs to produce cardiomyocyte-like cells with sarcomeric architecture was demonstrated by immunocytological staining for and sarcomeric α-actinin (Fig. 2F). Cells treated with IGF1 also were able to differentiate more readily into cardiomyocytes compared to untreated controls (Fig 2G). Accordingly, IGF-stimulated cells were able to differentiate into Nkx2.5-eGFP+ cells (Supplemental Fig. S4) and showed an increased frequency of beating EBs (Fig. 2H). These findings were supported by flow cytometric quantification of the number of cTnT+ cells (Fig. 2I).
Figure 2.
IGF/insulin treatment of murine ESCs enhances Nkx2.5+ CPC formation. Comparison by RT-qPCR of the expression levels of CPC genes in cells treated with 125 μg/mL Wnt3a, 125 μg/mL IGF1, 300 μg/mL IGF2 or 10 mg/mL insulin (A) Mesp1 (B) Isl1 (C) Nkx2.5 (D) GATA4 and (E) MLC2a. (F) Quantification of sarcomeric α-actinin+ cells of untreated controls and IGF1-treated cells. (G) Representative microscopic images of untreated controls and of IGF1-treated cells (day 12) following immunostaining for sarcomeric α-actinin. DAPI-stained DNA (DAPI; blue), sarcomeric α-actinin (α-Actinin; red). (H) Quantification of beating EBs as % of total EBs (days 10 and 12 of differentiation). (I) Quantification of cTnT+ cells by flow cytometry (day 12). Mean ± SEM of triplicate experiments is shown. NS, non-significant; *, p<0.05; **, p<0.01; ***, p<0.001; ψ, p<0.0001.
IGF/insulin treatment induces mesendoderm formation
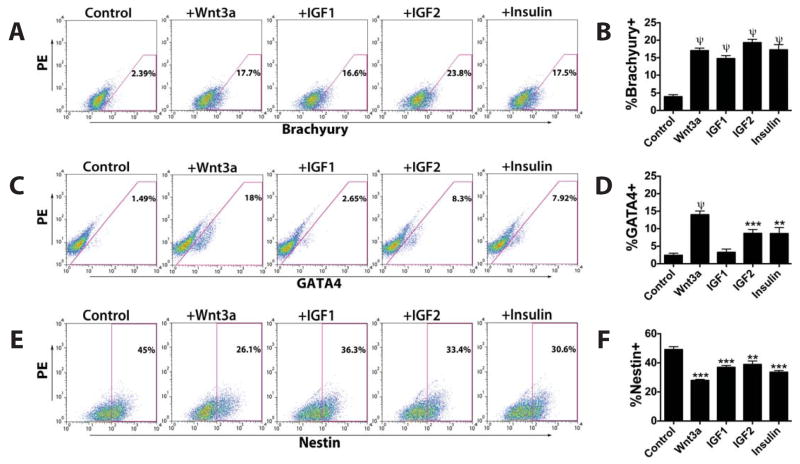
To investigate the mechanism involved in the stimulation of CPC formation by IGFs and insulin, we examined the expression of developmental stage- and lineage-specific genes. No significant difference in the expression of pluripotency markers such as Pou5f1 (Oct 4) (Fig. 3A) and Nanog (Fig. 3B) was found between control and IGF/insulin-treated cells. This suggests that IGF/insulin treatment does not elicit a greater degree of overall differentiation from a pluripotent state. The expression levels of mesoderm- and endoderm-specific genes such as Eomes, Brachyury, GATA4, Goosecoid, AFP, HNF1b, HNF3b and SOX17 were all significantly increased by IGF/insulin treatment (Fig. 3C-D). On the other hand, the expression of ectoderm-specific genes, such as Nestin, GBX2, FGF5 and Pax6 were unchanged or slightly decreased (Fig. 3E). These results were corroborated by flow cytometric analysis of Brachyury (Fig. 4A-B) and GATA4 expression (Fig. 4C-D). Consistently, the number of Nestin+ ectodermal cells decreased after IGF/insulin treatment (Fig. 4E-F). Taken together, these data support the ability of IGF1, IGF2 and insulin to stimulate the formation of Brachyury+ mesoderm or GATA4+ mesendoderm at the expense of ectoderm.
Figure 3.
IGF/insulin stimulation of murine ESCs induces expression of mesendodermal genes. Comparison by RT-qPCR of the expression levels at day 4 of (A) Oct4, (B) Nanog, (C) mesodermal genes, (D) endodermal genes and (E) ectodermal genes in untreated cells and in cells exposed to Wnt3a (125 μg/mL), IGF1 (125 μg/mL), IGF2 (300 μg/mL) or insulin (10 mg/mL). Mean ± SEM of triplicate experiments is shown. NS, non-significant; *, p<0.05; **, p<0.01; ***, p<0.001; ψ, p<0.0001.
Figure 4.
IGF/insulin treatment of murine ESCs gives rise to early mesodermal cells. Flow cytometric analysis of the expression at day 4 of the germ layer markers (A,B) Brachyury, (C,D) GATA4 and (E,F) Nestin in untreated cells and in cells exposed to Wnt3a (125 µg/mL), IGF1 (125 µg/mL), IGF2 (300 µg/mL) or insulin (10 mg/mL). Mean ± SEM of triplicate experiments is shown. **, p<0.01; ***, p<0.001; ψ, p<0.0001.
IGFs and insulin stimulate proliferation of Brachyury+ mesodermal cells
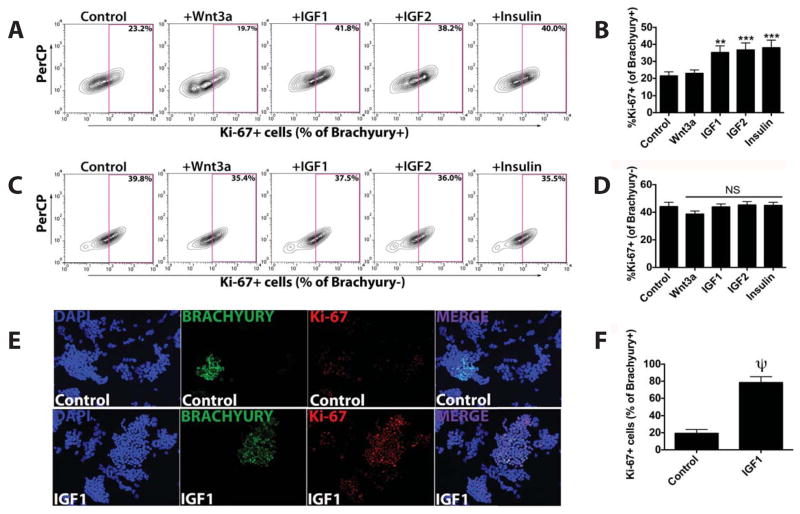
Since IGFs have been implicated in cardiomyocyte proliferation [23-26], we studied the ability of IGFs and insulin to induce proliferation within the mesodermal Brachyury+ cell population. Co-immunostaining for Brachyury and the proliferation marker Ki-67 revealed a significant increase in the percentage of Brachyury+/Ki-67+-double-positive cells after IGF1/2 or insulin treatment, but not after Wnt3a or FGF2 treatment (Brachyury+/Ki-67+: 22.6±4.4%) (Fig. 5A-B,E-F). Interestingly, there was no significant increase in Ki-67 staining within the Brachyury-negative cell population after IGF/insulin treatment (Fig. 5C-D). These data demonstrate that IGFs and insulin selectively promote mesodermal cell proliferation.
Figure 5.
IGF/insulin treatment selectively increases proliferation within the Brachyury+ cell population during murine ESC differentiation. Flow cytometry analysis of the expression at day 4 of the proliferation marker Ki-67 in Brachyury+ or Brachyury- cells in untreated controls and in cells exposed to Wnt3a (125 μg/mL), IGF1 (125 μg/mL), IGF2 (300 μg/mL) or insulin (10 mg/mL). (A,B) Ki-67 expression in Brachyury+ cells. (C,D) Ki-67 expression in Brachyury- cells. Mean ± SEM of triplicate experiments is shown. NS, non-significant; **, p<0.01; ***; p<0.001. (E) Representative microscopy images (200×) of untreated controls and IGF1-treated cells after immunostaining for DAPI (blue), Brachyury (green), Ki-67 (red). (F) Quantification of Ki-67+ cells in Brachyury+ cells of untreated controls and IGF1-treated cells.
IGF/insulin treatment causes rapid Akt phosphorylation in Brachyury+ mesodermal cells
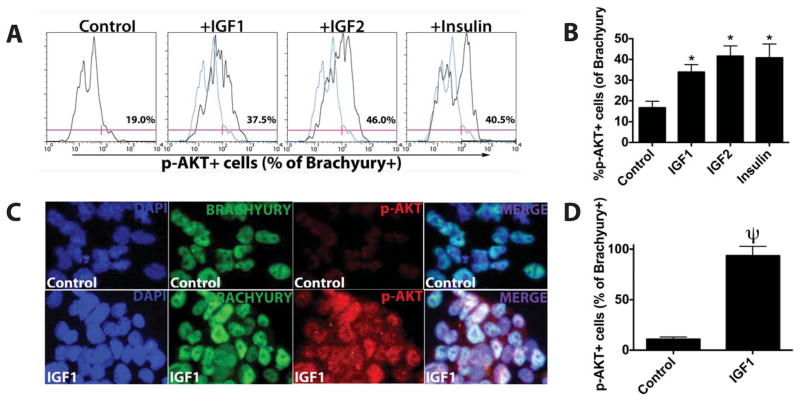
It has been established that Akt phosphorylation is one of the mediators of IGF signaling [27]. To examine whether Akt phosphorylation in mesodermal cells is involved in IGF/insulin-mediated induction of CPC formation, differentiating Nkx2.5-eGFP ESCs were treated with IGF1, IGF2 or insulin and subjected to flow cytometric analysis to investigate the occurrence of Akt phosphorylation in Brachyury+ cells. We found a significant increase in the percentage of Brachyury+/phospho-Akt+ cells following IGF/insulin treatment (Fig. 6A-D). The effect of IGFs and insulin on Akt phosphorylation appears specific since treatment with FGF2 or Wnt3a showed no such effect (Supplemental Fig. S5). These data indicate that IGFs and insulin indeed signal through downstream Akt phosphorylation in the Brachyury+ cell population.
Figure 6.
IGF/insulin treatment promotes Akt phosphorylation in murine ESC-derived Brachyury+ cells. (A) Flow cytometric analysis of phospho-AKT (p-AKT) expression in Brachyury+ cells following treatment of Nkx2.5-eGFP ESCs with IGF1 (125 μg/mL), IGF2 (300 μg/mL) or insulin (10 mg/mL) and untreated control Nkx2.5-eGFP ESCs. (B) Quantification of p-AKT expression in Brachyury+ cells. Mean ± SEM of triplicate experiments is shown. *, p<0.05; ψ, p<0.0001. (C) Representative microscopic images (1000×) of untreated controls and IGF1-treated cells after immunostaining for DAPI (blue), Brachyury (green) and p-AKT (red). (D) Quantification of p-AKT+ cells in Brachyury+ cells of untreated controls and IGF1-treated cells.
Akt and mTOR signaling are required for IGF/insulin-mediated expansion of the CPC pool
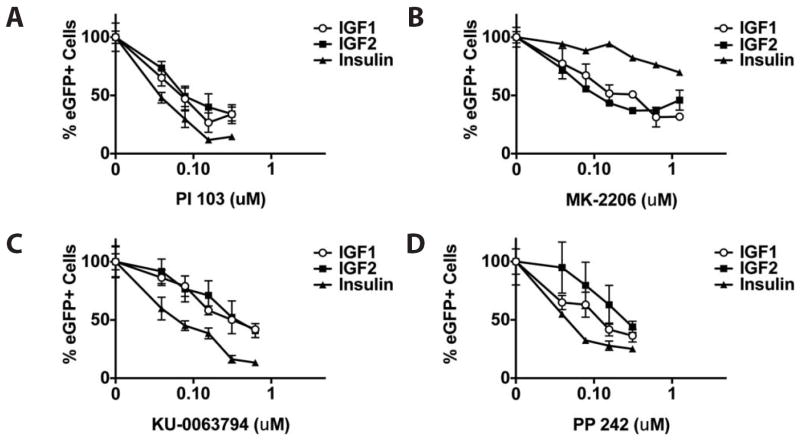
To further examine downstream mediators of IGF signaling responsible for its enhancement of CPC formation, differentiating Nkx2.5-eGFP ESCs were co-incubated with IGF1, IGF2 or insulin and selective small molecule inhibitors of PI3K, Akt and mTOR. IGF/insulin-induced formation of eGFP+ CPCs was dose-dependently reduced when differentiating Nkx2.5-eGFP ESCs were treated with specific inhibitors of PI3K (PI 103), Akt (MK-2206) and mTOR (KU-0063794 and PP 242) (Fig. 7A-D). Furthermore, after treatment of IGF/insulin-stimulated Nkx2.5-eGFP ESCs with MK-2206, the frequency of Brachyury+/Ki-67+-double-positive cells was significantly lower compared to mock-treated cells (Supplemental Fig. S6). These data demonstrate that IGF signaling through PI3K, Akt and mTOR enhances proliferation of mesodermal cells resulting, ultimately, in increased CPC formation.
Figure 7.
Selective inhibition of PI3K, Akt or mTOR abolishes IGF-induced cardiac differentiation of murine ESCs. Flow cytometric analysis of eGFP expression in IGF1- (125 μg/mL), IGF2- (125 μg/mL) or insulin (10 mg/mL)-treated Nkx2.5-eGFP ESCs in the presence of selective small molecule inhibitors of the PI3K/Akt/mTOR signaling pathway. Dose responses were determined for (A) PI 103 (inhibitor of PI3K), (B) MK-2206 (inhibitor of Akt), (C) KU-0063794 (inhibitor of mTOR) and (D) PP 242 (inhibitor of mTOR).
Discussion
In this study, we performed a growth factor screen to identify mediators of early mesoderm induction that can give rise to cardiac lineage induction. The key findings of this study are: 1) IGF1, IGF2, insulin, Wnt3a and FGF2 treatment of murine ESCs significantly increases Nkx2.5+ CPC formation, 2) IGF and insulin selectively promote Brachyury+ mesodermal cell proliferation, 3) IGF treatment leads to an increase in Akt phosphorylation within the Brachyury+ cell population, and 4) Activation of PI3K, Akt and mTOR signaling is required for the ability of IGFs and insulin to enhance CPC formation. These data demonstrate a role for IGFs and insulin in early germ layer development in vitro, by their ability to selectively enhance the proliferation of mesodermal cells, leading to increased cardiac lineage differentiation.
Signaling pathways involved in cardiac differentiation
While our screen identified a novel role for IGFs and insulin to regulate early in vitro cardiogenesis, Activin A, BMP2 and BMP4 treatment at this stage of development inhibited cardiac differentiation. The latter ligands have been shown to play a context-dependent role in cardiac development in previous studies [28-30]. For example, a recent small molecule screening study revealed the ability of a BMP inhibitor, dorsomorphin, to activate cardiogenic differentiation [30]. Other studies have shown both a dose- and stage-specific requirement for BMP signaling to enhance cardiac differentiation [5]. This context-dependent effect of BMP treatment has been previously reported [31]. Consistent with this, the addition of Activin A or BMPs alone to differentiating ESC cultures has been shown to preferentially induce endodermal lineage development while combined Activin A and BMP treatment was shown to induce mesodermal cardiac lineage formation [32].
Role of IGF/insulin in early cardiomyogenesis
Some studies have revealed a negative modulatory effect of IGF and insulin expression on cardiomyocyte differentiation in human ESCs [33-35]. Similarly, a recent study found differential effects of FGF signaling pathway activation in mouse and human ESCs [36]. The apparent discrepancy between these and our findings are most likely due to differences in the developmental stage in which IGF/insulin was added to cells. In this study, we examined the effect of IGF/insulin on mesodermal cells, whereas prior human ESC studies examined the effect of IGF/insulin on cardiomyocyte differentiation from post-mesodermal cells. It has been well recognized that developmental regulators such as Wnt and Notch exert a biphasic role on cardiac lineage differentiation. We believe this is likely to be the case for IGF/insulin as well. Indeed, our preliminary data showed that treatment of mouse cardiac progenitor cells with IGF results in decreased cardiomyocyte differentiation. This suggests that our findings in mouse ESC here are largely consistent with those from prior human ESC studies.
Our data showed that IGF signaling results in selective expansion of Brachyury+ mesodermal cells through induction of proliferation. As such, the mechanism by which IGFs and insulin stimulate CPC formation appears different from that of Wnt3a or FGF2, which showed no obvious proliferation rate changes in Brachyury+ mesodermal cells (Fig. 5). Furthermore, combining either Wnt3a or FGF2 with IGF/insulin resulted in additive increases in Nkx2.5-eGFP+ cells, which are no greater than the increases when each factor was added singularly (Supplementary Fig. S3). These data further support that IGF and insulin act through a different downstream signaling pathway than FGF2 or Wnt3a.
In support of our findings, Morali et al. found that IGF2-/- ESC lines exhibit an impaired ability to differentiate into cardiogenic and myogenic lineages when differentiated in vitro. This phenotype could be partially rescued when differentiating cells were treated with exogenous IGF2 [37]. A more recent study describes the ability of IGF to increase ESC-derived vascular cells as well, a finding that may also be related to the upregulation of Brachyury+ mesodermal cells [38]. Additionally, IGF and insulin treatment showed a marked induction of endodermal markers (Fig. 3D), suggesting this pathway could also be active during endodermal lineage formation. A recent study showed IGF treatment directed hepatocyte differentiation from definitive endoderm [39].
Conclusions
To our knowledge, this is the first study demonstrating a direct role for IGFs and insulin to promote the in vitro differentiation of pluripotent stem cells into mesoderm. We showed that IGF/insulin treatment of murine ESCs selectively expands the Brachyury+ mesodermal cell pool via activation of PI3K, Akt and mTOR, leading to increased cell proliferation. These findings contribute to our understanding of early developmental events and provide a new strategy to enhance directed cardiac differentiation for regenerative applications.
Supplementary Material
Figure S1. Flow cytometric assessment of the optimal timing of IGF/insulin stimulation-induced formation of CPCs from murine ESCs. The growth factors were added on the indicated day after LIF removal. Bars represent fold change in eGFP+ cell count by growth factor treatment. Mean ± SEM of triplicate experiments is shown.
Figure S2. Representative microscopic images of Draq5-labeled untreated controls and IGF1-treated cells (day 11) after fluorescence-activated cell sorting of Nkx2.5-eGFP+ cells at day 6. Quantification of Draq5+ cells after treatment with IGF1 (125 μg/mL), IGF2 (300 μg/mL), insulin (10mg/mL) or untreated controls of sorted Nkx2.5-eGFP+ cells. NS, not significant.
Figure S3. IGFs and insulin synergize with Wnt3a and FGF2 in stimulating the formation of CPCs from murine ESCs. Nkx2.5-eGFP ESCs were exposed to IGF1 (125 μg/mL), IGF2 (300 μg/mL), insulin (10 mg/mL), FGF2 (200 μg/mL) or Wnt3a (125 μg/mL) alone or in pairs as indicated below the graphs and the resulting number of eGFP+ CPCs was determined by flow cytometry. Bars represent mean of SEM in triplicate experiments. NS, non-significant; *, p<0.05; ψ, p<0.0001.
Figure S4. Representative phase contrast photomicroscopic images of differentiating murine Nkx2.5-eGFP ESCs as untreated control or following treatment with IGF1, IGF2 or insulin.
Figure S5. Treatment of Nkx2.5-eGFP ESCs with Wnt3a (125 μg/mL) or FGF2 (200 μg/mL) does not increase Akt phosphorylation in Brachyury+ cells. Flow cytometric analysis of phospho-AKT (pAkt) expression in Brachyury+ cells following treatment of Nkx2.5-eGFP ESCs with Wnt3a or FGF2. Control, untreated Nkx2.5-eGFP ESCs. Bars represent mean ± SEM of triplicate experiments. NS, non-significant.
Figure S6. Selective inhibition of Akt signaling abolishes IGF/insulin-dependent proliferation of murine ESC-derived Brachyury+ cells. Flow cytometric quantification of Ki67 expression in Brachyury+ cells following treatment of differentiating murine ESCs with IGF1, IGF2 or insulin in the presence of the Akt inhibitor ML-2206 or its solvent, DMSO. Mean ± SEM of triplicate experiments is shown. *, p<0.05.
Acknowledgments
Laura Prickett-Rice and Katherine Folz-Donahue at the Center for Regenerative Medicine Harvard Stem Cell Institute Flow Cytometry Core provided assistance with flow cytometry experiments. Andrew Olson at the Stanford Neuroscience Microscopy Service (supported by NIH NS069375) at Stanford School of Medicine provided technical assistance with confocal microscopy imaging. M.C.E. was supported by a Mosaic grant from the Netherlands Organization for Scientific Research (NWO 017.007.064). D.A.P. was supported by NWO (VENI grant 91611070) and S.M.W. was supported by NIH/Officer of the Director’s New Innovator Award (OD004411), NIH/NHLBI U01 (HL099776) and K08 (HL081086) and by the Harvard Stem Cell Institute.
Footnotes
Author Contributions
M.C.E.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, and final approval of manuscript; K.R., R.F. and A.S.: collection and/or assembly of data, data analysis and interpretation, and final approval of manuscript; U.B.N.: provision of study material, financial support, and final approval of manuscript; M.J.S.: financial support, data analysis and interpretation, and final approval of manuscript; A.A.F.V.: data analysis and interpretation, manuscript writing, and final approval of manuscript; D.A.P.: conception and design, data analysis and interpretation, manuscript writing, and final approval of manuscript; S.M.W.: conception and design, financial support, data analysis and interpretation, manuscript writing, and final approval of manuscript.
Disclosures
U.B. Nielsen is the chief executive officer of Silver Creek Pharmaceuticals, Inc.
References
- 1.Fuster V, Voute J, Hunn M, Smith SC., Jr Low priority of cardiovascular and chronic diseases on the global agenda: a cause for concern. Circulation. 2007;23:1966–1970. doi: 10.1161/CIRCULATIONAHA.107.733444. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Wu SM, Hochedlinger K. Harnessing the potential of induced pluripotent stem cells for regenerative medicine. Nat Cell Biol. 2011;13:497–505. doi: 10.1038/ncb0511-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burridge PW, Keller G, Gold JD, Wu JC. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10:16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Burridge PW, Thompson S, Millrod MA, Weinberg S, Yuan X, Peters A, Mahairaki V, Koliatsos VE, Tung L, Zambidis ET. A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PLoS One. 2011;6:e18293. doi: 10.1371/journal.pone.0018293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Vliet P, Wu SM, Zaffran S, Pucéat M. Early cardiac development: a view from stem cells to embryos. Cardiovasc Res. 2012;96:352–362. doi: 10.1093/cvr/cvs270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 9.Cai CL, Cai X, Liang Y, Shi PH, Chu SL, Pfaff J, Chen S, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stanley EG, Biben A, Elefanty L, Barnett F, Koentgen L, Robb L, Harvey RP. Efficient cre-mediated deletion in cardiac progenitor cells conferred by a 3’UTR-ires-cre allele of the homeobox gene Nkx2-5+ Int J Dev Biol. 2002;46:431–439. [PubMed] [Google Scholar]
- 11.Wu SM, Fujiwara Y, Cibulsky SM, Clapham DE, Lien CL, Schultheiss TM, Orkin SH. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell. 2006;127:1137–1150. doi: 10.1016/j.cell.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 12.Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, Sun Y, Evans SM, Laugwitz KL, Chien KR. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 13.Willems E, Spiering, Davidovics H, Lanier M, Xia Z, Dawson M, Cashman J, Mercola M. Small-molecule inhibitors of the Wnt pathway potently promote cardiomyocytes from human embryonic stem cell-derived mesoderm. Circ Res. 2011;109:360–364. doi: 10.1161/CIRCRESAHA.111.249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Klos M, Wilson GF, Herman AM, Lian X, Raval KK, Barron MR, Hou L, Soerens AG, Yu J, Palecek SP, Lyons GE, Thomson JA, Herron TJ, Jalife J, Kamp TJ. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method. Circ Res. 2012;111:1125–1136. doi: 10.1161/CIRCRESAHA.112.273144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tran TH, Wang X, Browne C, Zhang Y, Schinke M, Izumo S, Burcin M. Wnt3a-induced mesoderm formation and cardiomyogenesis in human embryonic stem cells. Stem Cells. 2009;27:1869–1878. doi: 10.1002/stem.95. [DOI] [PubMed] [Google Scholar]
- 16.Hao J, Daleo MA, Murphy CK, Yu PB, Ho JN, Hu J, Peterson RT, Hatzopoulos AK, Hong CC. Dorsomorphin, a selective small molecule inhibitor of BMP signaling, promotes cardiomyogenesis in embryonic stem cells. PLoS One. 2008;3:e2904. doi: 10.1371/journal.pone.0002904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimoji K, Yuasa S, Onizuka T, Hattori F, Tanaka T, Hara M, Ohno Y, Chen H, Egasgira T, Seki T, Yae K, Koshimizu U, Ogawa S, Fukuda K. G-CSF promotes the proliferation of developing cardiomyocytes in vivo and in derivation from ESCs and iPSCs. Cell Stem Cell. 2010;6:227–237. doi: 10.1016/j.stem.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Willems E, Cabral-Teixeira J, Schade D, Cai W, Reeves P, Bushway PJ, Lanier M, Walsh C, Kirchhausen T, Izpisua Belmonte JC, Cashman J, Mercola M. Small molecule-mediated TGF-β type II receptor degradation promotes cardiomyogenesis in embryonic stem cells. Cell Stem Cell. 2012;11:242–252. doi: 10.1016/j.stem.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe Y, Miyagawa-Tomita S, Vincent SD, Kelly RG, Moon AM, Buckingham ME. Role of mesodermal FGF8 and FGF10 overlaps in the development of the arterial pole of the heart and pharyngeal arch arteries. Circ Res. 2010;106:495–503. doi: 10.1161/CIRCRESAHA.109.201665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takada S, Stark KL, Shea MJ, Vassileva G, McMahon JA, McMahon AP. Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev. 1994;8:174–189. doi: 10.1101/gad.8.2.174. [DOI] [PubMed] [Google Scholar]
- 21.Ueno S, Weidinger G, Osugi T, Kohn AD, Golob JL, Pabon L, Reinecke H, Moon RT, Murry CE. Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc Natl Acad Sci USA. 2007;104:9685–9690. doi: 10.1073/pnas.0702859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schulz KR, Danna EA, Krutzik PO, Nolan GP. Single-cell phospho-protein analysis by flow cytometry. Curr Protoc Immunol. 2012;Chapter 8(Unit 8.17):1–20. doi: 10.1002/0471142735.im0817s96. [DOI] [PubMed] [Google Scholar]
- 23.Li P, Cavallero S, Gu Y, Chen TH, Hughes J, Hassan AB, Brüning JC, Pashmforoush M, Sucov HM. IGF signaling directs ventricular cardiomyocyte proliferation during embryonic heart development. Development. 2011;138:1795–1805. doi: 10.1242/dev.054338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oikonomopoulos A, Sereti KI, Conyers F, Bauer M, Liao A, Guan J, Crapps D, Han JK, Dong H, Bayomy AF, Fine GC, Westerman K, Biechele TL, Moon RT, Force T, Liao R. Wnt signaling exerts an antiproliferative effect on adult cardiac progenitor cells through IGFBP3. Circ Res. 2011;109:1363–1374. doi: 10.1161/CIRCRESAHA.111.250282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xin M, Kim Y, Sutherland LB, et al. Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci Signal. 2011;4:ra70. doi: 10.1126/scisignal.2002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDevitt TC, Laflamme MA, Murry CE. Proliferation of cardiomyocytes derived from human embryonic stem cells is mediated via the IGF/PI 3-kinase/Akt signaling pathway. J Mol Cell Cardiol. 2005;39:865–873. doi: 10.1016/j.yjmcc.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings BA. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 28.Sugi Y, Lough J. Activin-A and FGF-2 mimic the inductive effects of anterior endoderm on terminal cardiac myogenesis in vitro. Dev Biol. 1995;168:567–574. doi: 10.1006/dbio.1995.1102. [DOI] [PubMed] [Google Scholar]
- 29.Kruithof BP, van Wijk B, Somi S, Kruithof-de Julio M, PérezPomares JM, Weesie F, Wessels A, Moorman AF, van den Hoff MJ. BMP and FGF regulate the differentiation of multipotential pericardial mesoderm into the myocardial or epicardial lineage. Dev Biol. 2006;295:507–522. doi: 10.1016/j.ydbio.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 30.Ao A, Hao J, Hopkins CR, Hong CC. DMH1, a novel BMP small molecule inhibitor, increases cardiomyocyte progenitors and promotes cardiac differentiation in mouse embryonic stem cells. PLoSOne. 2012;7:e41627. doi: 10.1371/journal.pone.0041627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Pater E, Ciampricotti M, Priller F, Veerkamp J, Strate I, Smith K, Lagendijk AK, Schilling TF, Herzog W, Abdelilah-Seyfried S, Hammerschmidt M, Bakkers J. Bmp signaling exerts opposite effects on cardiac differentiation. Circ Res. 2012;110:578–587. doi: 10.1161/CIRCRESAHA.111.261172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao S, Chen S, Clark J, Hao E, Beattie GM, Hayek A, Ding S. Long-term self-renewal and directed differentiation of human embryonic stem cells in chemically defined conditions. Proc Natl Acad Sci USA. 2006;103:6907–6912. doi: 10.1073/pnas.0602280103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freund C, Ward-van Oostwaard D, Monshouwer-Kloots J, van den Brink S, van Rooijen M, Xu X, Zweigerdt R, Mummery C, Passier R. Insulin redirects differentiation from cardiogenic mesoderm and endoderm to neuroectoderm in differentiating human embryonic stem cells. Stem Cells. 2008;26:724–733. doi: 10.1634/stemcells.2007-0617. [DOI] [PubMed] [Google Scholar]
- 34.Xu XQ, Graichen R, Soo SY, et al. Chemically defined medium supporting cardiomyocyte differentiation of human embryonic stem cells. Differentiation. 2008;76:958–970. doi: 10.1111/j.1432-0436.2008.00284.x. [DOI] [PubMed] [Google Scholar]
- 35.Tran TH, Wang X, Browne C, et al. Wnt3a-induced mesoderm formation and cardiomyogenesis in human embryonic stem cells. Stem Cells. 2009;27:1869–1878. doi: 10.1002/stem.95. [DOI] [PubMed] [Google Scholar]
- 36.Greber B, Wu G, Bernemann C, Joo JY, Han DW, Ko K, Tapia N, Sabour D, Sterneckert J, Tesar P, Schöler HR. Conserved and Divergent Roles of FGF Signaling in Mouse Epiblast Stem Cells and Human Embryonic Stem Cells. Cell Stem Cell. 2010;6:215–226. doi: 10.1016/j.stem.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 37.Morali OG, Jouneau A, McLaughlin KJ, Thiery JP, Larue L. IGF-II promotes mesoderm formation. Dev Biol. 2000;227:133–145. doi: 10.1006/dbio.2000.9875. [DOI] [PubMed] [Google Scholar]
- 38.Piecewicz SM, Pandey A, Roy B, Xiang SH, Zetter BR, Sengupta S. Insulin-like growth factors promote vasculogenesis in embryonic stem cells. PLoS One. 2012;7:e32191. doi: 10.1371/journal.pone.0032191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magner NL, Jung Y, Wu J, Nolta JA, Zern MA, Zhou P. Insulin and IGFs Enhance Hepatocyte Differentiation from Human Embryonic Stem Cells via the PI3K/AKT Pathway. Stem Cells. 2013 Jul 8; doi: 10.1002/stem.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Flow cytometric assessment of the optimal timing of IGF/insulin stimulation-induced formation of CPCs from murine ESCs. The growth factors were added on the indicated day after LIF removal. Bars represent fold change in eGFP+ cell count by growth factor treatment. Mean ± SEM of triplicate experiments is shown.
Figure S2. Representative microscopic images of Draq5-labeled untreated controls and IGF1-treated cells (day 11) after fluorescence-activated cell sorting of Nkx2.5-eGFP+ cells at day 6. Quantification of Draq5+ cells after treatment with IGF1 (125 μg/mL), IGF2 (300 μg/mL), insulin (10mg/mL) or untreated controls of sorted Nkx2.5-eGFP+ cells. NS, not significant.
Figure S3. IGFs and insulin synergize with Wnt3a and FGF2 in stimulating the formation of CPCs from murine ESCs. Nkx2.5-eGFP ESCs were exposed to IGF1 (125 μg/mL), IGF2 (300 μg/mL), insulin (10 mg/mL), FGF2 (200 μg/mL) or Wnt3a (125 μg/mL) alone or in pairs as indicated below the graphs and the resulting number of eGFP+ CPCs was determined by flow cytometry. Bars represent mean of SEM in triplicate experiments. NS, non-significant; *, p<0.05; ψ, p<0.0001.
Figure S4. Representative phase contrast photomicroscopic images of differentiating murine Nkx2.5-eGFP ESCs as untreated control or following treatment with IGF1, IGF2 or insulin.
Figure S5. Treatment of Nkx2.5-eGFP ESCs with Wnt3a (125 μg/mL) or FGF2 (200 μg/mL) does not increase Akt phosphorylation in Brachyury+ cells. Flow cytometric analysis of phospho-AKT (pAkt) expression in Brachyury+ cells following treatment of Nkx2.5-eGFP ESCs with Wnt3a or FGF2. Control, untreated Nkx2.5-eGFP ESCs. Bars represent mean ± SEM of triplicate experiments. NS, non-significant.
Figure S6. Selective inhibition of Akt signaling abolishes IGF/insulin-dependent proliferation of murine ESC-derived Brachyury+ cells. Flow cytometric quantification of Ki67 expression in Brachyury+ cells following treatment of differentiating murine ESCs with IGF1, IGF2 or insulin in the presence of the Akt inhibitor ML-2206 or its solvent, DMSO. Mean ± SEM of triplicate experiments is shown. *, p<0.05.