Abstract
Comparison of the sequences of mouse and human genomes revealed a surprising number of nonexonic, nonexpressed conserved sequences, for which no function could be assigned. To study the possible correlation between these conserved intronic sequences and alternative splicing regulation, we developed a method to identify exons that are alternatively spliced in both human and mouse. We compiled two exon sets: one of alternatively spliced conserved exons and another of constitutively spliced conserved exons. We found that 77% of the conserved alternatively spliced exons were flanked on both sides by long conserved intronic sequences. In comparison, only 17% of the conserved constitutively spliced exons were flanked by suchconserved intronic sequences. The average length of the conserved intronic sequences was 103 bases in the upstream intron and 94 bases in the downstream intron. The average identity levels in the immediately flanking intronic sequences were 88% and 80% for the upstream and downstream introns, respectively, higher than the conservation levels of 77% that were measured in promoter regions. Our results suggest that the function of many of the intronic sequence blocks that are conserved between human and mouse is the regulation of alternative splicing.
The recently published draft sequence of the mouse genome (Waterston et al. 2002) facilitates a great advance in searching for cis-regulatory sequence elements. The 75 million years that have passed since the divergence of the ancestor of the human and mouse lineages allowed a substantial divergence in neutral DNA; the constraint on functional elements has kept them conserved. Indeed, homologous human and mouse exons are, on the average, 85% identical in their sequences, but introns are more poorly conserved: 60% of the nonexonic sequences are nonalignable, and in the alignable regions the average identity level is 69% (Waterston et al. 2002).
However, numerous regions that are conserved between human and mouse are also found in introns (Hardison et al. 1997). Comparison between the human chromosome 21 and the corresponding genomic sequences in mouse revealed that only one-third of the conserved blocks are exons (Dermitzakis et al. 2002). The other two-thirds of highly conserved sequences are intronic and intergenic. These conserved elements were found to be unexpressed in microarray experiments. Thus, the conclusion was that they are probably cis-regulatory sequence elements, but no function could be assigned to most of them (Dermitzakis et al. 2002). We decided to check the possible correlation between the conserved intronic sequences and alternative splicing regulation.
Alternative splicing, a process by which several mRNA isoforms can be generated from a single gene, has received a great deal of attention recently. The current estimates are that 35%–59% of all human genes undergo alternative splicing (Mironov et al. 1999; Brett et al. 2000; International Human Genome Sequencing Consortium 2001). Despite the thousands of alternative splicing events identified to date, very little is known about the regulation of this process (Maniatis and Tasic 2002). Cis-acting sequence elements found within exons, called exonic splicing enhancers (ESEs), show the ability to regulate alternative splicing (Tacke and Manley 1999; Blencowe 2000). These sequences interact with proteins of the SR family, which recruit the splicing machinery toward weak, flanking splice sites and enable the inclusion of the alternatively spliced exon in the mature mRNA (Blencowe 2000). Other cis-regulatory elements for splicing, such as exonic splicing silencers (ESSs) and intronic splicing enhancers and silencers (ISEs and ISSs, respectively), were also shown to regulate individual cases of alternative splicing (Maniatis and Tasic 2002). Sometimes, multiple cis-acting elements cooperatively function to regulate the same alternatively spliced exon (Cartegni et al. 2002). Alternative splicing was also found to be affected by other factors, including the phosphorylation state of SR proteins, the migration of SR and other proteins between the nucleus and the cytoplasm (Chabot 1996), and the promoter of the gene (Cramer et al. 1999).
Although several cis-regulatory elements have been characterized for individual cases, it is still unclear how the majority of alternative splicing events are regulated (Maniatis and Tasic 2002; Modrek and Lee 2002). In addition, intronic elements that regulate splicing have received little attention compared to exonic regulatory elements. We used the sequence of the mouse genome to locate homologous exons that are alternatively spliced in both human and mouse. We found that most of these exons are flanked by long conserved intronic sequences, whereas constitutively spliced exons usually are not flanked by such sequences. Our results suggest that many of the previously uncharacterized intronic sequences conserved between human and mouse are involved in the regulation of alternative splicing.
RESULTS
To investigate the correlation between conserved intronic sequences and alternatively spliced exons, we began by obtaining the intron–exon structures of human genes by using the output of the LEADS software platform (Shoshan et al. 2001; Sorek et al. 2002), run on the draft human genome build 30 and the cDNAs and ESTs from GenBank version 131. This software aligns expressed sequences to the genome, taking alternative splicing into account (the LEADS process is described in detail in [Sorek et al. 2002] and [Sorek and Safer 2003]). Using the same methods described in Sorek et al. (2002), we utilized the LEADS output to collect reliable data sets of 3583 human alternatively spliced internal exons and 7557 human constitutively spliced internal exons.
To identify exons that are conserved in mouse, we first aligned the mouse ESTs (from GenBank version 131) to the human genome, as described in the Methods section. A constitutively spliced exon was deemed “conserved” if there were mouse ESTs aligned to the constitutively spliced exon, as well as to the exons immediately adjacent to it. We were able to identify a conserved mouse counterpart for 1966 human constitutively spliced internal exons. This is, of course, fewer than the actual fraction of constitutively spliced exons conserved between human and mouse. There are several reasons for not identifying all of the conserved exons. First, our method de pends on the existence of mouse ESTs covering the actual exon and the two flanking exons by at least 25 bp on each side (see Methods). Second, the method requires the borders of the exons to be kept conserved between human and mouse. Third, the current draft sequence of the mouse genome is still incomplete.
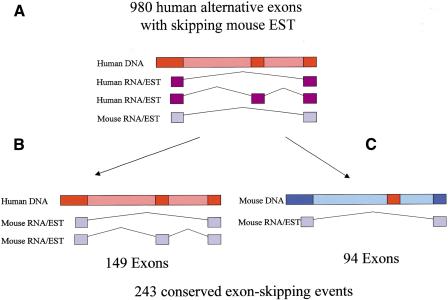
A human exon-skipping was deemed “conserved” in mouse if both splice variants (the variant that skips the exon and the variant that contains the exon) were supported by mouse ESTs. For 149 exon-skipping events, both variants were found in mouse ESTs (Fig. 1B). However, when the variant that contains the alternatively spliced exon is a rare variant, or a variant unique to a tissue that is not represented in mouse EST libraries, there might be no mouse EST covering it. Nevertheless, if the human exon is truly conserved in the mouse transcriptome, we would expect its DNA sequence to be conserved between human and mouse. Here, we rely on the fact that although exons are conserved between the human and mouse genomes at an average level of 85%, introns are conserved at much lower levels (Waterston et al. 2002). Therefore, in cases where there was a skipping variant evident in mouse ESTs, but there was no mouse EST showing the variant that contains the exon, we aligned the sequence of the human exon to the relevant intron in the mouse genome (Fig. 1C). The exon was declared conserved if a significant conservation above 80% identity was found, if the alignment spanned the full length of the human exon, and if the exon was flanked by the canonical AG/GT acceptor and donor sites in the mouse genome. Using this approach we identified 94 additional exon-skipping events conserved between human and mouse. Complete data for the 243 conserved alternatively spliced exons, along with the flanking intronic sequences, are provided as Supplementary Material (available online at www.genome.org).
Figure 1.
Finding exon-skipping events that are conserved between human and mouse: 3583 exon-skipping events were found in the human genome, using the methods described in Sorek et al. (2002). (A) For 980 of these human exons, a mouse EST spanning the intron that represents the exon-skipping variant was found. Human ESTs appear in purple; mouse ESTs are in light blue. (B,C) The two possible ways to identify an exon as conserved in mouse. (B) Identification of mouse ESTs that contain the exon, as well as the two flanking exons. (C) If the exon was not represented in mouse ESTs, the sequence of the human exon was searched against the intron spanned by the skipping mouse EST on the mouse genome. If a significant conservation (above 80%) was found, and the alignment spanned the full length of the human exon, the exon was declared as conserved.
To check the conservation between the intronic sequences immediately flanking alternatively spliced exons, we used Sim4 (Florea et al. 1998) to align the last 100 bases of the intron that is upstream of the human alternatively spliced exon to the last 100 bases of the intron upstream of the respective exon in the mouse genome. We repeated this analysis for the first 100 bases of the intron that is downstream from the alternatively spliced exon. A significant alignment was found for 223/243 (92%) human and mouse 100 bases of upstream introns, and for 199/243 (82%) human and mouse 100 bases of downstream introns. For 188/243 (77%) of the exons, conserved sequences were found in both the upstream and downstream introns. These percentages were similar in both the subset of 149 EST confirmed exons and the subset of the 94 exons supported by the mouse genomic sequence only (76.5% and 78.5%, respectively, P=0.92). This demonstrates the homogeneity of our group of conserved alternatively spliced exons, and further indicates that the 94 genome-supported exons are real events of conserved alternatively spliced exons.
We repeated the same analysis on constitutively spliced exons conserved between human and mouse. For these, a significant alignment was found for 886/1966 (45%) human and mouse last 100 bases of upstream introns, and for 691/1966 (35%) human and mouse first 100 bases of downstream introns. Only 343/1966 (17%) of the constitutively spliced exons had conserved sequences in both the upstream and downstream introns.
We further characterized the alignment between the intronic regions close to conserved alternatively spliced exons. Whenever the conserved intronic region exceeded the 100 bases aligned, we extended the alignment to the end of the conserved region. For alternatively spliced exons, the average length of the conserved intronic sequences immediately flanking the exon was 103 bases in the upstream intron and 94 bases in the downstream intron (medians 72 and 77, respectively). In the 17% of constitutively spliced exons for which a significant conservation was identified, the average length of the conserved intronic sequences was 41 and 38 bases for upstream and downstream introns, respectively (medians 34 and 30).
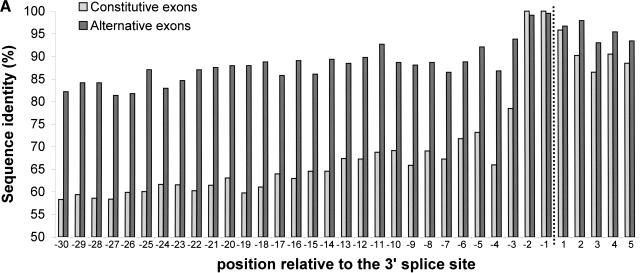
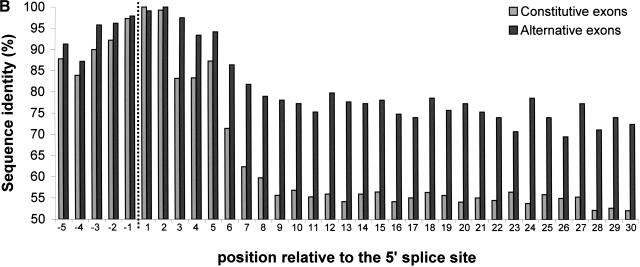
Figure 2 presents the per-position conservation near the splice sites. For alternatively spliced exons, the average identity level in the last 30 bases of the upstream intron was 88%, and 80% for the first 30 bases of the downstream intron. For comparison, promoter regions are conserved between human and mouse at an average level of about 77% (Waterston et al. 2002). The average identity levels gradually decrease with the distance from the splice site, but remain significantly higher than those of constitutively spliced exons (Table 1).
Figure 2.
Per-position conservation near alternatively and constitutively spliced exons. Intronic regions near the splice site were aligned, using GAP (global alignment program) of the GCG package, and identity levels were calculated for each position as described in Methods. All 243 alternative exons and 1966 constitutive exons were used for this analysis. (A) Conservation near the 3′ splice site. Data for the last 30 nt of the intron and the first 5 nt of the exon are shown. Dashed line marks the border between the intron and the exon. (B) Conservation near the 5′ splice site. Data for the last 5 nt of the exon and the first 30 nt of the intron are shown.
Table 1.
Percent Intronic Conservation as a Function of the Distance From the Splice Sitea
|
Upstream introns
|
Downstream introns
|
|||
|---|---|---|---|---|
| Distance from splice site (bp) | Constitutively spliced | Alternatively spliced | Constitutively spliced | Alternatively spliced |
| 1-6b | 81.6 | 93.3 | 87.4 | 95.1 |
| 7-30 | 63.1 | 86.7 | 55.1 | 75.9 |
| 31-60 | 54.9 | 76.0 | 52.0 | 70.1 |
| 61-90 | 50.5 | 65.8 | 49.0 | 65.6 |
Shown are the average identity levels in windows of 30 bases
Conservation for the nucleotides that define the splice site (intronic bases 1-6) was calculated separately
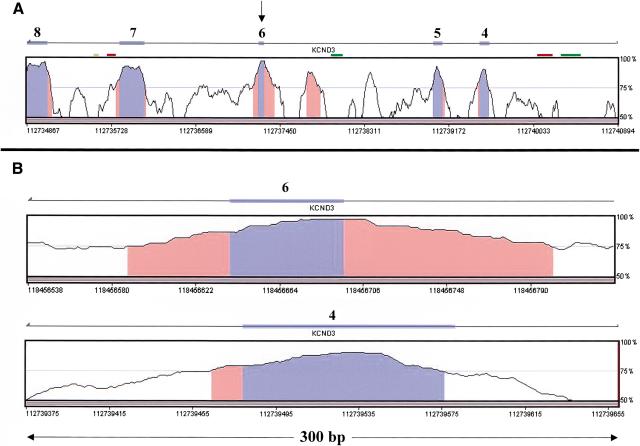
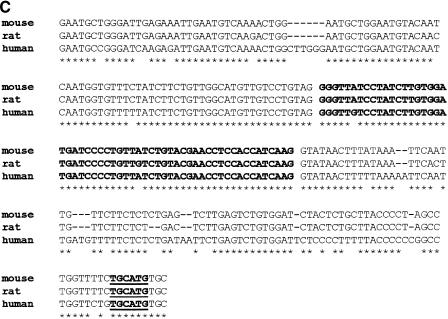
A typical example of conserved intronic elements flanking an alternatively spliced exon is presented in Figure 3. This figure shows a VISTA conservation graph (Mayor et al. 2000) for exons 4–8 of the gene KCND3, corresponding to RefSeq NM_004980. Exon 6 (marked by an arrow) was found, in our analysis, to be an alternatively spliced exon, conserved between human and mouse. The other four exons are constitutively spliced. Long conserved intronic regions (colored orange) are clearly seen flanking the alternatively spliced exon; such long conserved regions are not visible near any of the four constitutively spliced exons. These flanking intronic regions are also highly conserved in rats, a fact that further supports their functionality (Fig. 3C). Another conserved region is visible near (∼400 bp upstream), but not adjacent to, the alternatively spliced exon. This region might be an alternatively spliced exon not yet identified or might serve as another cis-regulatory element.
Figure 3.
Human–mouse alignment of the KCND3 gene, corresponding to RefSeq NM_004980 (from VISTA browser, http://pipeline.lbl.gov/vistabrowser/). x-axis: The nucleotide coordinates on human chromosome 1, according to the assembly version of the human genome from June 2002. y-axis: The level of conservation between the human genome and the corresponding mouse genome, according to the MGSCv3 assembly version of the mouse genome. (A) Blue bars above the conservation area correspond to annotated exons 4–8 of KCND3. Blue areas within the conservation graph mark exons; orange areas mark conserved nonexonic sequences. The exon marked with an arrow (exon 6) is an alternatively spliced one; the others are constitutively spliced exons. (B) Enlarged view of the conservation graphs of the alternatively spliced exon (exon 6), and one of the constitutively spliced exons (exon 4) is presented to show the relative lengths of the conserved areas near the exons. (C) Human, mouse, and rat alignment of exon 6, as well as the 100 bases upstream and downstream of the exon. Exon sequence is bold; asterisks mark identity in all three organisms. Bold and underline mark the hexamer TGCATG, which previously showed the ability to regulate alternative splicing when found in introns downstream to alternatively spliced exons (Lim and Sharp 1998; Deguillien et al. 2001).
The sequences we identified in this study could serve as templates for the identification of regulatory sequences for alternative splicing. To demonstrate this, we conducted a hexamer count in the first and last 100 bases of the downstream and upstream introns, respectively, that flank the human alternatively spliced exons. For this, we took only the 132 cases where the length of the conserved stretch was more than 50 bases. The most abundant hexamer in the conserved intronic sequences downstream of the alternatively spliced exons was TGCATG (excluding TTTTTT, which was also the most abundant in nonconserved introns). This hexamer appeared in 24 (18%) of the examined sequences (appearing twice in five of the sequences), ninefold over the expected frequency. In 93% of the cases, the TGCATG hexamer was conserved in mouse. This hexamer was over-abundant neither in the 100 bases downstream of constitutively spliced exons, nor in intronic sequences located upstream from the alternatively spliced exons. The TGCATG hexamer was previously shown to regulate alternative splicing of several exons, specifically when found in the downstream intron (Lim and Sharp 1998; Deguillien et al. 2001).
DISCUSSION
Our finding that long conserved intronic elements are found near alternatively spliced exons is intriguing, especially because the average level of conservation between human and mouse intronic sequences is relatively low(Waterston et al. 2002). These findings suggest that there might be a regulatory mechanism common to many alternatively spliced exons that involves the intronic sequences immediately flanking these exons. Because typical binding sites for RNA-splicing regulatory proteins are relatively short (4–10 nucleotides; Tacke and Manley 1999; Blencowe 2000; Cartegni et al. 2002; Fairbrother et al. 2002; Maniatis and Tasic 2002), the fact that the length of the conserved regions usually exceeds 50 bases implies the involvement of multiple factors in this regulation. It is also possible that some of the intronic sequences create a secondary structure, or are involved in interactions with the transcriptional machinery, or in chromatin remodeling. All of these processes were shown to regulate alternative splicing (Caceres and Kornblihtt 2002; Damgaard et al. 2002; Manley 2002).
It was recently reported that the frequency of SINEs (short interspersed repetitive elements) insertion into the 150 intronic bases that flank exons tends to be lower than the frequency of their insertion into other parts of introns (Majewski and Ott 2002). Those authors, therefore, concluded that the first and last 150 bp of introns are likely to contain elements required for the splicing process. Our findings fit well with the distance reported by Majewski and Ott (2002) and indicate that most of this tendency might be contributed by introns flanking alternatively spliced, rather than constitutively spliced exons.
We found that 17% of the constitutively spliced exons conserved between human and mouse were flanked on both sides by conserved intronic sequences. These exons were declared constitutively spliced, because many expressed sequences showed their existence, but no expressed sequence skipped them. However, it is possible that these exons are actually alternatively spliced, and the splice isoform skipping the exon is rare, or condition-specific, and therefore not represented in the EST database.
A recent study found that 41% of all mouse genes undergo alternative splicing, with about two alternatively spliced exons observed per alternatively spliced gene (Okazaki et al. 2002). Our results suggest that most of the alternatively spliced exons are flanked on both sides by conserved intronic sequences, averaging about 100 bases in length; these conserved intronic elements possibly function in alternative splicing regulation. This implies that a few million intronic nucleotides in the sequence of the mouse and human genomes are involved in the regulation of alternative splicing. How this regulation occurs remains to be determined.
METHODS
Human ESTs and cDNAs were obtained from NCBI GenBank version 131 (August 2002; www.ncbi.nlm.nih.gov/dbEST) and aligned to the human genome build 30 (August 2002; www.ncbi.nlm.nih.gov/genome/guide/human) using the LEADS clustering and assembly system as described in Sorek et al. (2002). Briefly, the software cleans the expressed sequences from vectors and immunoglobulins, masking them for repeats and low-complexity sequences. The software then aligns the expressed sequences to the genome, taking alternative splicing into account, and clusters overlapping expressed sequences into “clusters” that represent genes or partial genes.
Alternatively spliced internal exons and constitutively spliced internal exons were identified using the same methods described in Sorek et al. (2002). In short, these methods screen for reliable exons requiring canonical splice sites and discarding possible genomic contamination events. “Constitutively spliced internal exon” was defined as an internal exon supported by at least four sequences, for which no alternative splicing was observed. “Alternatively spliced internal exon” was defined as such if there was at least one sequence that contained both the internal exon and the two flanking exons (exon inclusion), and one sequence that contained the two flanking exons, but skipped the middle one (exon skipping).
Mouse ESTs and cDNAs from GenBank version 131 were aligned to the UCSC mouse genome, assembly version 3 (ftp.ensembl.org/pub/assembly/mouse/mgsc_assembly_3/) as follows. Mouse ESTs and cDNAs were cleaned from terminal vector sequences, and low-complexity stretches and repeats in the expressed sequences were masked. Sequences with internal vector contamination were discarded, as were sequences identified as immunoglobulins or T-cell receptors. In the next stage, expressed sequences were heuristically compared with the genome to find likely high-quality hits. They were then aligned to the genome using a spliced alignment model that allows long gaps. Single hits of mouse expressed sequences to the human genome shorter than 20 bases, or having less than 75% identity to the human genome, were discarded. Using these parameters, 1,341,274 mouse ESTs were mapped to the human genome, 511,381 of them having all their introns obeying the GT/AG or GC/AG rules.
To determine whether the borders of a human intron (which define the borders of the flanking exons) were conserved in mouse, a mouse EST spanning the same intron-borders, while aligned to the human genome, was required (with alignment of at least 25 bp on each side of the exon–exon junction). In addition, this mouse EST was required to span an intron (i.e., open a long gap) at the same position along the EST, when aligned to the mouse genome.
Alignment of intronic regions was performed using the local alignment program Sim4 (Florea et al. 1998). An alignment was considered significant according to Sim4 default parameters. This program detects exact matches of length 12 and extends them in both directions with a score of 1 for a match and -5 for a mismatch, stopping when extensions no longer increase the score (Florea et al. 1998). The end of the Sim4 alignment was considered the end of the conserved region. In cases in which the alignment spanned the entire 100 bases, the next 100 intronic bases were aligned, and so forth, until the alignment stopped. A minimal significant alignment was, therefore, of a length of at least 12 exactly matching bases. Lengths of alignments and identity levels were parsed from Sim4 standard output.
For per-position conservation calculation, the first and last 100 bases of the downstream and upstream introns, respectively, that flank the alternatively spliced exons were aligned to their mouse counterparts using the GCG global alignment program GAP (default parameters). For each position, a parameter `N' was assigned, such as N= the number of cases (out of 243 exons) in which this position was conserved. A per-position conservation value `C' was calculated such as C= (N/243) × 100, that is, the percentage of cases in which this position was conserved. This procedure was repeated for the 1966 constitutively spliced exons.
Overrepresentation of TGCATG hexamer was calculated as follows. We analyzed 132 downstream intronic sequences; each of these sequences was 100 bases long. In each such sequence, 95 hexamers are possible. As expected by chance, the probability for a specific hexamer is 1/4096, so that in each 100-base sequence the probability of appearance of a specific hexamer was 95/4096 = 0.023. The mean expected frequency for a specific hexamer in 132 sequences was, therefore, 0.023 × 132 = 3.06. Hexamer counts for alternatively spliced and constitutively spliced exons are provided as Supplementary Material.
Acknowledgments
We thank Eyal Fink and Guy Kol for enabling the study with the mouse LEADS productions; Erez Levanon and Dr. Zurit Levine for fruitful discussions; Han Xie, Pini Akiva, Yossef Kliger, Gad Cojocaru, Dvir Dahary, Kinneret Savitsky, Erez Levanon, and Galit Rotman for helpful comments. This work was partially supported by a grant from the Israel Science Foundation to G.A.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1208803.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Blencowe, B.J. 2000. Exonic splicing enhancers: Mechanism of action, diversity and role in human genetic diseases. Trends Biochem Sci. 25: 106-110. [DOI] [PubMed] [Google Scholar]
- Brett, D., Hanke, J., Lehmann, G., Haase, S., Delbruck, S., Krueger, S., Reich, J., and Bork, P. 2000. EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett. 474: 83-86. [DOI] [PubMed] [Google Scholar]
- Caceres, J.F. and Kornblihtt, A.R. 2002. Alternative splicing: Multiple control mechanisms and involvement in human disease. Trends Genet. 18: 186-193. [DOI] [PubMed] [Google Scholar]
- Cartegni, L., Chew, S.L., and Krainer, A.R. 2002. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 3: 285-298. [DOI] [PubMed] [Google Scholar]
- Chabot, B. 1996. Directing alternative splicing: Cast and scenarios. Trends Genet. 12: 472-478. [DOI] [PubMed] [Google Scholar]
- Cramer, P., Caceres, J.F., Cazalla, D., Kadener, S., Muro, A.F., Baralle, F.E., and Kornblihtt, A.R. 1999. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol. Cell 4: 251-258. [DOI] [PubMed] [Google Scholar]
- Damgaard, C.K., Tange, T.O., and Kjems, J. 2002. hnRNP A1 controls HIV-1 mRNA splicing through cooperative binding to intron and exon splicing silencers in the context of a conserved secondary structure. RNA 8: 1401-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deguillien, M., Huang, S.C., Moriniere, M., Dreumont, N., Benz Jr., E.J., and Baklouti, F. 2001. Multiple cis elements regulate an alternative splicing event at 4.1R pre-mRNA during erythroid differentiation. Blood 98: 3809-3816. [DOI] [PubMed] [Google Scholar]
- Dermitzakis, E.T., Reymond, A., Lyle, R., Scamuffa, N., Ucla, C., Deutsch, S., Stevenson, B.J., Flegel, V., Bucher, P., Jongeneel, C.V., et al. 2002. Numerous potentially functional but non-genic conserved sequences on human chromosome 21. Nature 420: 578-582. [DOI] [PubMed] [Google Scholar]
- Fairbrother, W.G., Yeh, R.F., Sharp, P.A., and Burge, C.B. 2002. Predictive identification of exonic splicing enhancers in human genes. Science 297: 1007-1013. [DOI] [PubMed] [Google Scholar]
- Florea, L., Hartzell, G., Zhang, Z., Rubin, G.M., and Miller, W. 1998. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 8: 967-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison, R.C., Oeltjen, J., and Miller, W. 1997. Long human–mouse sequence alignments reveal novel regulatory elements: A reason to sequence the mouse genome. Genome Res. 7: 959-966. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Lim, L.P. and Sharp, P.A. 1998. Alternative splicing of the fibronectin EIIIB exon depends on specific TGCATG repeats. Mol. Cell. Biol. 18: 3900-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski, J. and Ott, J. 2002. Distribution and characterization of regulatory elements in the human genome. Genome Res. 12: 1827-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T. and Tasic, B. 2002. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 418: 236-243. [DOI] [PubMed] [Google Scholar]
- Manley, J.L. 2002. Nuclear coupling: RNA processing reaches back to transcription. Nat. Struct. Biol. 9: 790-791. [DOI] [PubMed] [Google Scholar]
- Mayor, C., Brudno, M., Schwartz, J.R., Poliakov, A., Rubin, E.M., Frazer, K.A., Pachter, L.S., and Dubchak, I. 2000. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16: 1046-1047. [DOI] [PubMed] [Google Scholar]
- Mironov, A.A., Fickett, J.W., and Gelfand, M.S. 1999. Frequent alternative splicing of human genes. Genome Res. 9: 1288-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek, B. and Lee, C. 2002. A genomic viewof alternative splicing. Nat. Genet. 30: 13-19. [DOI] [PubMed] [Google Scholar]
- Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., Nikaido, I., Osato, N., Saito, R., Suzuki, H., et al. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563-573. [DOI] [PubMed] [Google Scholar]
- Shoshan, A., Grebinskiy, V., Magen, A., Scolnicov, A., Fink, E., Lehavi, D., and Wasserman, A. 2001. Designing oligo libraries taking alternative splicing into account. In Proceedings of SPIE: Microarrays: Optical technologies and informatics (eds. M.L. Bittner, Y. Chen, A.N. Dorsel, and E.R. Dougherty), pp. 86-95. E.R. Vol 4266. SPIE, Bellingham, WA. [Google Scholar]
- Sorek, R. and Safer, H.M. 2003. A novel algorithm for computational identification of contaminated EST libraries. Nucleic Acids Res. 31: 1067-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek, R., Ast, G., and Graur, D. 2002. Alu-containing exons are alternatively spliced. Genome Res. 12: 1060-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke, R. and Manley, J.L. 1999. Determinants of SR protein specificity. Curr. Opin. Cell. Biol. 11: 358-362. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- www.ncbi.nlm.nih.gov/dbEST; Database of expressed sequence tags.
- www.ncbi.nlm.nih.gov/genome/guide/human; Human genomic sequence.
- ftp.ensembl.org/pub/assembly/mouse/mgsc_assembly_3/; Mouse genomic sequence.
- http://pipeline.lbl.gov/vistabrowser/; VISTA Genome Browser.