Abstract
Microarray gene expression data provide a wealth of information for elucidating the mode and tempo of molecular evolution. In the present study,we analyze the spatial expression pattern of human duplicate gene pairs by using oligonucleotide microarray data,and study the relationship between coding sequence divergence and expression divergence. First,we find a strong positive correlation between the proportion of duplicate gene pairs with divergent expression (as presence or absence of expression in a tissue) and both synonymous (KS) and nonsynonymous divergence (KA). The divergence of gene expression between human duplicate genes is rapid, probably faster than that between yeast duplicates in terms of generations. Second,we compute the correlation coefficient (R) between the expression levels of duplicate genes in different tissues and find a significant negative correlation between R and KS. There is also a negative correlation between R and KA,when KA ≤ 0.2. These results indicate that protein sequence divergence and divergence of spatial expression pattern are initially coupled. Finally,we compare the functions of those duplicate genes that show rapid divergence in spatial expression pattern with the functions of those duplicate genes that show no or little divergence in spatial expression.
Ever since Ohno (1970), the evolution of duplicate genes has been a subject of extensive theoretical modeling and empirical research. Lately, there has been much interest in whether a positive correlation exists between coding region divergence and gene expression divergence. In particular, two recent studies (Wagner 2000; Gu et al. 2002b) used yeast microarray data to test the presence of such correlation on a genome-wide scale. Wagner (2000) explored the relationship between protein sequence divergence and mRNA expression divergence among 144 yeast duplicate genes. The expression was measured at multiple time points in four physiological processes. No significant correlation was observed, implying decoupling of coding sequence (CDS) divergence and expression divergence. Gu et al. (2002b) investigated expression divergence in a larger sample of yeast duplicate genes (400 pairs) and used the microarray expression data from 14 processes. The expression divergence between duplicate genes was significantly correlated with their synonymous divergence (KS) and with their nonsynonymous divergence (KA) when KA ≤ 0.30, contrary to the conclusion of Wagner (2000).
In the present study, we investigate the relationship between CDS divergence and spatial expression divergence among human duplicate genes (paralogs). To our knowledge, this is the first study that uses microarray data to analyze the evolution of human gene expression on a genome-wide scale. Specifically, we focus on the following questions: (1) how quickly do human paralogs diverge in their expression; (2) does expression divergence increase with gene sequence divergence, that is, evolutionary time; (3) what are the functions of gene pairs with rapid divergence in expression; and (4) does the present study of spatial expression of human paralogs support the conclusion drawn from the study of temporal expression of yeast paralogs (Gu et al. 2002b)? It is believed that transcription regulation is more complex in mammals than in lower eukaryotes, for example, in yeast (Huang et al. 1999). We intend to explore whether this has any implications for the tempo of gene expression evolution. The studies on yeast investigated temporal expression only, as it is difficult to study spatial expression in a single cell organism. Because there are no comprehensive data on temporal gene expression in humans (Ly et al. 2000; Cho et al. 2001), we used the data of Su et al. (2002), who generated a spatial gene expression profile for human genes by using the U95A oligonucleotide array (Affymetrix). It is the largest study of spatial (tissue) expression of human genes available to date.
RESULTS
Identification of Duplicate Genes
Human U95A oligonucleotide array contains 12,387 probes. These include probes from 7565 human genes with annotated CDSs in GenBank. The other probes predominantly correspond to ESTs, which were not used in this study. Duplicate genes with annotated CDSs were identified and grouped into multigene families by using a rigorous method developed by Gu et al. (2002a; see Methods). From this analysis, we estimated that the U95A array contains 875 multiple gene families.
A total of 1404 independent duplicate gene pairs were selected for further analysis. KS and KA divergences between duplicate genes were calculated. The expression data for these gene pairs studied in 25 independent and nonredundant tissues were retrieved from Su et al. (2002).
Proportion of Gene Pairs With Diverged Expression Increases With Time
To study the dynamics of spatial expression divergence, we calculated the proportion of gene pairs with diverged expression among all pairs duplicated at approximately the same time, that is, having the same KS value. This analysis was limited to 1230 gene pairs for which at least one member of a pair is expressed in at least one tissue (for the definition of a gene being expressed, see Methods). Two duplicate genes are said to have diverged expression in a particular tissue if one gene is expressed in that tissue and the other is not. We used two definitions of gene expression divergence. In the first one, a gene pair is said to have diverged in expression if it shows diverged expression in at least one of the tissues studied. In the second definition, a gene pair is said to have diverged in expression if it shows diverged expression in at least two of the tissues studied. The latter definition is more robust against errors in microarray typing. Both definitions are conservative because they exclude cases in which both genes are expressed, in which both genes are not expressed, or in which one is expressed (or not expressed) and the other is marginally expressed. These definitions are also conservative in a sense that they do not take into account quantitative differences in expression. Thus, they underestimate the divergence in expression. However, they highlight the evolution of tissue-specific expression. The measure that takes into account the quantitative differences in expression is described in the next section.
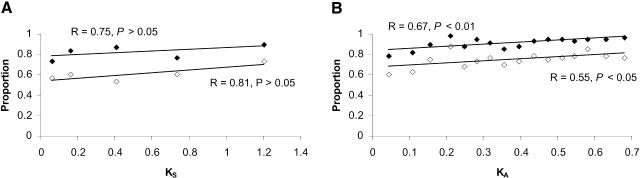
First, we used KS as a proxy of divergence time. A high positive correlation (although not significant) is observed between the proportion of gene pairs with diverged expression and KS (Fig. 1A). This is true for the proportion of genes with diverged expression in at least one tissue and in at least two tissues. Strikingly, 73.3% of the gene pairs with an average KS of only 0.064 already have diverged in expression in at least one tissue, whereas 56.7% of these genes have diverged in expression in at least two tissues. These percentages increase to 90.0% and 73.3%, respectively, for gene pairs with an average KS of 1.2. Thus, rapid divergence in spatial expression pattern is observed between duplicate genes. The relationship between divergence time (measured by KS) and the proportion of gene pairs with diverged expression is approximately linear.
Figure 1.
The relationship between sequence divergence and the proportion of human gene pairs with diverged expression. (A) Synonymous divergence (KS) is used to represent sequence divergence. Each point represents 30 gene pairs. (B) Nonsynonymous divergence (KA) is used to represent sequence divergence. Each point represents 60 gene pairs. Solid diamonds represent the proportion of gene pairs with diverged expression in at least one tissue, and open diamonds represent proportion of gene pairs with diverged expression in at least two tissues. Solid and punctured lines are the corresponding linear regressions.
A statistically significant positive correlation is observed between KA and the proportion of gene pairs with diverged expression in either at least one or in at least two tissues (Fig. 1B). However, the correlation coefficient is smaller than the one observed when KS is used because KS is a better proxy of evolutionary time (see Discussion). Again, divergence in gene expression occurs very rapidly. Indeed, at an average KA of 0.044, 78.3% of gene pairs have diverged in expression in at least one tissue, and 60% of them have diverged in expression in at least two tissues. The proportion of genes with diverged expression increases rapidly and reaches a plateau at KA = 0.2. At an average KA of 0.212, almost all gene pairs (98.3%) have diverged in expression in at least one tissue, and 88.3% of gene pairs have diverged in expression in at least two tissues. Thus, even when we used KA as a proxy of evolutionary time, we observed rapid divergence in gene expression among duplicate genes and a significant correlation between KA and the proportion of gene pairs with diverged expression.
Correlation Between CDS Divergence and Expression Divergence
Another way of measuring similarity in expression pattern between two genes is to compute the Pearson correlation coefficient (R) between the expression levels of two genes over the tissues studied. As will be explained in the Discussion, this measure is less desirable than that described above, but it can also give insights into the dynamics of gene expression divergence. First, we considered cases in which both copies of a gene pair were expressed in at least five of the tissues studied. Only 269 gene pairs (group A) satisfied this criterion. Next, we considered cases in which at least one of the two copies was expressed in at least five of the tissues studied. This adds some noise to the calculation of R; however, it allows us to increase the sample size. A total of 895 gene pairs were selected originally, but later only 841 gene pairs (group B) were retained for the final analysis because in the other 54 gene pairs only one gene of the pair was expressed, resulting in R = 0. We used the transformation ln[(1 + R)/(1 - R)] and then carried out the normal linear regression between each pair of KS (or KA) and the transformed R.
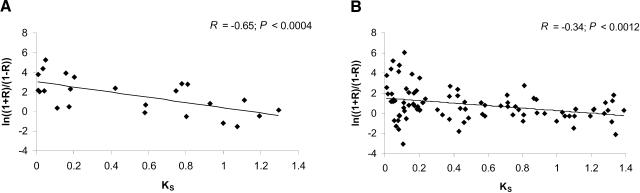
A significant negative correlation was found between ln[(1 + R)/(1 - R)] and KS for genes in group A (R = -0.65, P < 0.0004; Fig. 2A) and in group B (R = -0.34, P < 0.0012; Fig. 2B). To test whether the transformation changed our conclusion, we also carried out the linear regression between KS and R (data not shown). This again resulted in a significant negative correlation for both group A (R = -0.63, P < 0.0005) and group B (R = -0.31, P < 0.0164). Thus, the correlation coefficient of gene expression between duplicate genes decreases approximately linearly with divergence time as measured by KS.
Figure 2.
The relationship between synonymous rate (KS) and the transformed correlation coefficient of gene expression values between duplicate genes: in which both genes are expressed in at least five tissues (24 gene pairs, group A; A) and in which at least one gene is expressed in at least five tissues (94 gene pairs, group B; B). Only gene pairs with KS < 1.4 were included.
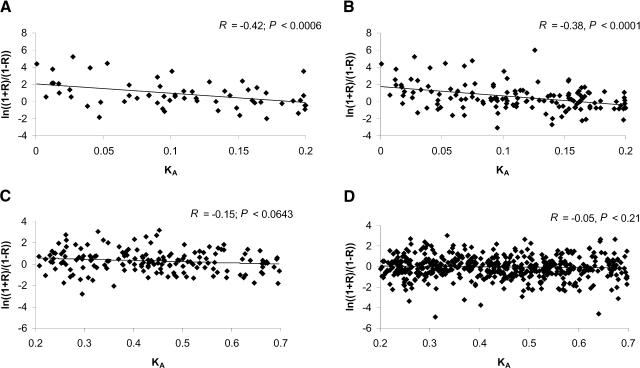
A weak negative correlation (data not shown) was observed between KA (KA < 0.70) and ln[(1 + R)/(1 - R)] for group A (R = -0.26, P < 0.0001) and group B (R = -0.19, P < 0.0001). However, this correlation becomes stronger for both groups (R = -0.42, P < 0.0006 for group A and R = -0.38, P < 0.0001 for group B) when only gene pairs with KA < 0.2 are examined (Fig. 3A,B). With KA > 0.2 (Fig. 3C,D), the correlation is considerably weaker and no longer statistically significant (R = -0.15, P < 0.0643 for group A and R = -0.05, P < 0.21). The choice of KA < 0.2 as a dividing point is arbitrary; however, the correlation coefficient changes only slightly from R = -0.41 (R = -0.36 for group B) for KA < 0.15 to R = -0.36 (R = -0.37 for group B) for KA < 0.25. Therefore, initially there is a coupling between gene expression divergence and KA.
Figure 3.
The relationship between nonsynonymous rate (KA) and the transformed correlation coefficient of gene expression values between duplicate genes: 60 gene pairs with KA < 0.2 from group A (A); 153 gene pairs with KA < 0.2 from group B (B); 165 gene pairs with 0.2 < KA < 0.7 from group A (C); and 609 gene pairs with 0.2 < KA < 0.7 from group B (D).
Functions of Gene Pairs With Rapid Divergence or No Divergence in Expression
It is interesting to look into the functions of duplicate genes that show rapid divergence in expression. Thus, we investigated the functions of the duplicate gene pairs with KS < 0.3 and with diverged expression (as presence or absence of expression in a tissue) in at least 50% of the tissues studied (we considered only the tissues in which at least one gene of a pair is expressed). There were 38 such gene pairs (Table 1). Also, we examined duplicate gene pairs with KS < 0.3 and a correlation coefficient of gene expression (R) < 0.5. There were 18 gene pairs in this group (Table 1). Interestingly, most of the gene pairs in these two groups overlapped. Thus, the results from the two measures concur. The functions of these genes were retrieved from LocusLink (http://www.ncbi.nlm.nih.gov/LocusLink/) manually. The gene pairs in these two groups encode enzymes (oxidoreductases, hydrolases, transferases, and an isomerase), proteins of the immune system (e.g., lymphocyte antigen, cytokine gro-beta, MHC proteins, and immunoglobulins), transcription factors, structural proteins (e.g., amelogenin, keratin, and skeletal muscle protein), and receptors (Table 1). To determine whether any of the functions were overrepresented among genes with rapid divergence in expression, we compared their functions with the functions of the other duplicate genes using the Gene Ontology database (Camon et al. 2003). There was indeed a significantly higher proportion of immune response genes among gene pairs with rapid divergence in expression compared with other gene pairs in our study (P < 0.009 for gene pairs with KS < 0.5 and diverged expression in at least 50% of studied tissues; P < 0.001 for gene pairs with KS < 0.5 and R < 0.5).
Table 1.
Duplicate Genes That Have Rapidly Diverged in Gene Expression
| Protein 1 | Protein 2 | KA | KS | N expra | N divb | Rc | Function of protein 1 | Function of protein 2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene pairs that have diverged in expression (presence or absence) in at least 50% of the tissues studied (in which at least one of the two duplicate genes is expressed) | ||||||||||||||||
| AAA02487 | CAA36842 | 0.116 | 0.217 | 2 | 2 | n/ad | activator protein 2B | transcription factor AP-2 | ||||||||
| AAA02993 | BAA00310 | 0.109 | 0.164 | 1 | 1 | n/a | cytochrome P450 PCN3 | cytochrome P-450 HFLa | ||||||||
| AAA35658 | BAA04619 | 0.093 | 0.201 | 7 | 6 | 0.19 | chlordecone reductase | unknown function | ||||||||
| AAA35781 | AAC50056 | 0.079 | 0.115 | 25 | 25 | 0.13 | DNA-binding protein | RNA-binding protein | ||||||||
| AAA35827 | AAA36051 | 0.165 | 0.188 | 25 | 18 | 0.20 | IgG Fc fragment receptor precursor | IgG Fc receptor β-Fc-γ-RII | ||||||||
| AAA35946 | CAA46096 | 0.077 | 0.141 | 7 | 6 | >0.5 | human complement factor H | serum protein | ||||||||
| AAA36014 | AAA51831 | 0.034 | 0.162 | 2 | 1 | n/a | δ-5-δ-4 isomerase type II | δ-5-δ-4-isomerase | ||||||||
| AAA36236 | AAA59772 | 0.047 | 0.086 | 15 | 15 | -0.15 | lymphocyte antigen | lymphocyte antigen | ||||||||
| AAA36444 | AAA36445 | 0.029 | 0.088 | 4 | 4 | n/a | phospholipase D | phospholipase D | ||||||||
| AAA36511 | AAA52607 | 0.069 | 0.082 | 5 | 3 | >0.5 | β1-glycoprotein | β1-glycoprotein precursor | ||||||||
| AAA36516 | CAA35612 | 0.126 | 0.114 | 5 | 4 | >0.5 | β1-glycoprotein | β1-glycoprotein | ||||||||
| AAA36793 | CAA68415 | 0.078 | 0.189 | 2 | 1 | n/a | UDP-glucuronosyltransferase | precursor | ||||||||
| AAA51718 | AAC21581 | 0.068 | 0.057 | 14 | 7 | -0.34 | amelogenin Y | amelogenin X | ||||||||
| AAA52576 | CAA55364 | 0.007 | 0.073 | 2 | 1 | n/a | glycerol kinase | glycerol kinase | ||||||||
| AAA59755 | AAA59823 | 0.049 | 0.076 | 6 | 6 | -0.34 | HLA DQ-β | MHC DQw1 β surface glycoprotein | ||||||||
| AAA60066 | AAA60067 | 0.066 | 0.096 | 3 | 3 | n/a | platelet factor 4 | platelet factor 4 | ||||||||
| AAA63183 | AAA63184 | 0.063 | 0.059 | 7 | 6 | >0.5 | cytokine gro-β | cytokine gro-β | ||||||||
| AAA75171 | AAA75172 | 0.165 | 0.201 | 9 | 9 | 0.14 | cysteine protease | cysteine protease | ||||||||
| AAA81368 | CAA11262 | 0.075 | 0.111 | 11 | 10 | 0.47 | zinc finger protein | transcriptional repressor | ||||||||
| AAB21124 | CAA43715 | 0.017 | 0.020 | 2 | 1 | n/a | p50-NF-κB homolog | NF-κB subunit | ||||||||
| AAB01380 | AAC50613 | 0.018 | 0.083 | 19 | 19 | -0.10 | NADP-dependent malic enzyme | NADP-dependent malic enzyme | ||||||||
| AAB42011 | CAA63427 | 0.095 | 0.105 | 5 | <5 | -0.91 | MHC class I molecule | MHC class I chain-related protein A | ||||||||
| AAB53424 | CAA69164 | 0.115 | 0.108 | 2 | 2 | n/a | butyrophilin | put. B7,3 molecule of CD80-CD86 family | ||||||||
| AAB53426 | AAB53430 | 0.092 | 0.169 | 24 | 20 | >0.5 | butyrophilin | butyrophilin | ||||||||
| AAB68615 | CAA72414 | 0.045 | 0.086 | 2 | 2 | n/a | 8-hydroxyguanine glycosylase | DNA glycosylase/AP lyase | ||||||||
| AAB86528 | CAA75867 | 0.042 | 0.047 | 25 | 24 | >0.5 | apoptosis inhibitor | apoptosis inhibitor homolog | ||||||||
| AAB87667 | AAC99761 | 0.139 | 0.139 | 14 | 11 | 0.38 | immunoglobulin-like receptor | immunoglobulin-like receptor | ||||||||
| AAC50187 | AAC50189 | 0.042 | 0.173 | 2 | 2 | n/a | α(1,3/1,4) fucosyltransferase | α(1,3) fucosyltransferase | ||||||||
| AAC51146 | AAD03159 | 0.154 | 0.166 | 7 | 6 | 0.44 | NK-receptor | killer cell inhibitory receptor | ||||||||
| AAC52104 | AAD02195 | 0.057 | 0.236 | 25 | 22 | >0.5 | aflatoxin aldehyde reductase AFAR | aflatoxin B1-aldehyde reductase; AFAR | ||||||||
| AAD02203 | AAD03158 | 0.153 | 0.223 | 2 | 2 | n/a | immunoglobulin-like transcript 7 | immunoglobulin-like transcript 10 | ||||||||
| AAD02289 | BAA24506 | 0.075 | 0.085 | 5 | 5 | >0.5 | paired-box transcription factor | Pax-4 | ||||||||
| BAA07512 | BAA07513 | 0.084 | 0.160 | 1 | 1 | n/a | yeast PMS1 homolog | yeast PMS1 homolog | ||||||||
| BAA11829 | CAA43795 | 0.019 | 0.190 | 15 | 15 | 0.32 | collagen binding protein 2 | collagen-binding protein | ||||||||
| CAA04922 | CAA04923 | 0.063 | 0.079 | 1 | 1 | n/a | NKG2C | NKG2E | ||||||||
| CAA39460 | CAA39462 | 0.174 | 0.133 | 3 | 3 | n/a | eosinophil derived neurotoxin | eosinophil cationic protein | ||||||||
| CAA46987 | CAA51545 | 0.033 | 0.052 | 23 | 21 | >0.5 | skeletal muscle C protein | skeletal muscle C protein | ||||||||
| CAA57956 | CAA76384 | 0.027 | 0.221 | 23 | 16 | 0.49 | hair keratin acidic 3-II | keratin, type I | ||||||||
| Gene pairs with R < 0.5e | ||||||||||||||||
| AAA67367 | CAA38201 | 0.008 | 0.174 | 25 | <5 | 0.37 | myosin regulatory light chain | myosin regulatory light chain | ||||||||
| AAA98669 | BAA08533 | 0.030 | 0.067 | 8 | <5 | -0.56 | U2AFBPL | U2AF1-RS2 | ||||||||
| AAC97383 | BAA31626 | 0.075 | 0.111 | 23 | <5 | 0.26 | guanine nucleotide factor | unknown function | ||||||||
| BAA19516 | CAA60130 | 0.059 | 0.082 | 9 | <5 | -0.66 | homolog of the murine Llglh gene | homolog to Drosophila tumor suppressor gene | ||||||||
N expr is the number of tissues studied in which at least one of the two duplicate genes is expressed
N div is the number of tissues in which one gene is expressed and the other is not (i.e., the two genes have diverged in expression)
R is the correlation coefficient between the expression levels of the two genes over the tissues studied
n/a is not applicable because the number of tissues in which at least one of the duplicates is expressed was less than five
Fourteen gene pairs with R < 0.5 have already been mentioned above and so are not included here
It is also interesting to look into the function of duplicate genes that show no or little expression divergence, even though they duplicated a long time ago. Thus, we investigated gene pairs with KS > 3 and with no divergence in tissue expression (a total of 33 gene pairs; Table 2). Interestingly, two thirds of these gene pairs are almost ubiquitously expressed (expressed in 24 to 25 out of the 25 tissues analyzed), and another 15% are expressed in one tissue only (i.e., tissue-specific). Then we added gene pairs with R > 0.8 and KS > 3 (a total of six gene pairs). Only one gene pair is shared between the two groups. The gene pairs that have been well conserved in expression are enzymes (transferases, hydrolases, and helicases), transcription factors, membrane-bound proteins (e.g., adducins and connexins), structural proteins (keratin and tubulin), and proteasome components (Table 2). However, as the number of the proteins in each functional class is small, none of these classes is found to be significantly overrepresented among the gene pairs with slow divergence in expression, when they are analyzed using the Gene Ontology database.
Table 2.
Duplicate Genes With Low Divergence in Gene Expression
| Protein 1 | Protein 2 | KA | KS | N expra | Rb | Function of protein 1 | Function of protein 2 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Duplicates in which both genes are expressed in the same tissues | ||||||||||||||
| BAA06336 | CAA85523 | 0.705 | 3.01 | 25 | <0.8 | eukaryotic initiation factor 4All | nuclear RNA helicase (DEAD family) | |||||||
| AAC50682 | BAA13199 | 0.256 | 3.12 | 24 | <0.8 | cytoplasmic phosphoprotein | similar to mouse dishevelled-3 | |||||||
| AAB53494 | AAC95431 | 0.378 | 3.35 | 22 | <0.8 | tub homolog | tubby-like protein 3 | |||||||
| AAA35720 | BAA07703 | 0.548 | 3.41 | 25 | <0.8 | cysteine-rich protein | ESP1/CRP2 | |||||||
| AAB28361 | CAA64685 | 0.220 | 3.47 | 25 | <0.8 | actin depolymerizing factor | cofilin | |||||||
| AAB06274 | AAC63143 | 0.563 | 3.51 | 24 | <0.8 | vascular endothelial growth factor B precursor | vascular endothelial growth factor | |||||||
| AAA36446 | CAA40621 | 0.092 | 3.71 | 25 | <0.8 | phospholipase A2 | HS1 | |||||||
| AAA51813 | CAA80661 | 0.615 | 3.74 | 1 | n/ac | bcl2-α protein | unknown function | |||||||
| BAA26001 | CAA54793 | 0.581 | 3.77 | 1 | n/a | CDC2δT | CDK-activating kinase | |||||||
| AAC95472 | CAA27856 | 0.450 | 3.79 | 20 | <0.8 | connexin 31.1 | gap junction protein | |||||||
| BAA13327 | CAA42060 | 0.306 | 3.80 | 24 | <0.8 | rhodanese | unknown function | |||||||
| AAA92734 | AAC99402 | 0.728 | 3.83 | 25 | <0.8 | prosomal protein P30-33K | proteasome subunit HSPC | |||||||
| AAA58455 | CAA34651 | 0.484 | 3.90 | 25 | <0.8 | amplaxin | haematopoietic lineage cell protein | |||||||
| AAA60190 | CAA52882 | 0.670 | 3.91 | 25 | <0.8 | peripherin | keratin 8 | |||||||
| AAA52620 | CAA56071 | 0.628 | 3.91 | 25 | <0.8 | γ-tubulin | β-tubulin | |||||||
| AAA36501 | CAA26528 | 0.716 | 4.04 | 2 | n/a | protein Z | protein C precursor | |||||||
| AAC62107 | AAC62108 | 0.403 | 4.11 | 14 | <0.8 | UP50 | UPH1 | |||||||
| AAA74425 | CAA47732 | 0.288 | 4.17 | 1 | n/a | preprocarboxypeptidase A2 | unknown function | |||||||
| BAA76708 | CAB46271 | 0.534 | 4.19 | 25 | <0.8 | RuvB-like DNA helicase TIP49b | erythrocyte cytosolic protein | |||||||
| AAA35563 | AAA35568 | 0.504 | 4.24 | 25 | 0.82 | aspartate aminotransferase | aspartate aminotransferase precursor | |||||||
| AAA70225 | BAA08576 | 0.807 | 4.32 | 1 | n/a | coagulation factor XII precursor | HGF activator-like protein | |||||||
| AAA35705 | AAB23769 | 0.161 | 4.33 | 1 | n/a | calcineurin A1 | calcineurin A catalytic subunit | |||||||
| BAA03095 | CAA55908 | 0.094 | 4.34 | 25 | <0.8 | human rab GDI | GDP-dissociation inhibitor | |||||||
| AAC15920 | BAA75899 | 0.438 | 4.39 | 25 | <0.8 | copine I | N-copine | |||||||
| AAA36025 | AAA87704 | 0.748 | 4.40 | 25 | <0.8 | 90-kD heat-shock protein | TNF1 receptor-associated protein | |||||||
| AAB97309 | CAA88401 | 0.116 | 4.42 | 25 | <0.8 | polyadenylate binding protein | polyadenylate binding protein II | |||||||
| AAA51582 | AAC17470 | 0.110 | 4.48 | 25 | <0.8 | α-actinin | α-actinin | |||||||
| CAA41149 | CAA41176 | 0.456 | 4.60 | 25 | <0.8 | erythrocyte α-adducin | β-adducin | |||||||
| BAA05393 | BAA07641 | 0.319 | 4.68 | 25 | <0.8 | homolog of female sterile homeotic mRNA | female sterile homeotic (fsh) homolog RING3 | |||||||
| AAA64895 | CAA61107 | 0.258 | 4.74 | 25 | <0.8 | plakoglobin | β-catenin | |||||||
| AAA80559 | BAA25173 | 0.103 | 4.77 | 18 | <0.8 | transcription activator | hSNF2H | |||||||
| BAA07238 | CAA50709 | 0.381 | 4.86 | 25 | <0.8 | proteasome subunit Z | proteasome-like subunit MECL-1 | |||||||
| BAA24855 | CAA12204 | 0.507 | 4.86 | 15 | <0.8 | unknown function | ZNF198 protein | |||||||
| Gene pairs with R > 0.8 | ||||||||||||||
| AAA36338 | AAA36458 | 0.291 | 3.74 | >5 | 0.83 | interferon-induced Mx protein | p78 protein | |||||||
| AAC96325 | BAA11179 | 0.154 | 3.94 | >5 | 0.86 | ZIC2 protein | Zic protein | |||||||
| AAA20580 | AAA58248 | 0.674 | 4.11 | >5 | 0.87 | Mu opiate receptor | somatostatin receptor isoform 2 | |||||||
| AAA60316 | AAB48394 | 0.644 | 4.07 | >5 | 0.87 | serotonin 1D receptor | 5-hydroxytryptamine 7 receptor isoform d | |||||||
| AAA52451 | AAA52644 | 0.336 | 4.15 | >5 | 0.91 | p55-c-fgr protein | protein tyrosine kinase | |||||||
N expr is the number of tissues in which both genes of a pair are expressed
R is the correlation coefficient between the expression levels of the two genes over the tissues studied
n/a is not applicable because the number of tissues in which at least one of the duplicates is expressed was less than five
DISCUSSION
We found that a large proportion of human duplicate genes have diverged rapidly in their spatial expression. Assuming that the average synonymous rate in higher primates is 1.5 × 10-9 nucleotide substitutions per site per year (Yi et al. 2002), 75.5% of human paralogs diverge in their expression in at least one tissue after only 25 Myr (KS = 0.068). It is likely that the true proportion of gene pairs with diverged gene expression is even higher than shown here, because only 25 tissues were analyzed and only a single (presumably normal) physiological condition was studied. In addition, the classification of tissues used by Su et al. (2002) does not correspond to the histological classification. For example, such complex organs as pancreas are called tissues in Su et al. (2002), whereas in reality they are composed of multiple tissues. These organs by themselves are likely to exhibit a wealth of differential spatial gene expression. We estimate that the rate of expression divergence in human paralogs is ∼40 times slower than that of yeast paralogs (Gu et al. 2002b), if the absolute time of divergence is considered. However, the generation time is several orders of magnitude shorter in yeast than in humans. Thus, when calculated per generation, expression divergence is more rapid in humans than in yeast. This might be due to a more complex transcription regulation in mammals than in lower eukaryotes (Huang et al. 1999). It could also be because of more possibilities in which such divergence can be manifested; for example, gene expression is regulated in a larger number of tissues in humans than in yeast. Alternatively (or additionally), this could be intrinsic to the spatial pattern of gene expression. A study of temporal gene expression divergence in humans should distinguish between these two possibilities.
Expression divergence increases approximately linearly with KS and, thus, with the evolutionary time. Therefore, similar to that in yeast duplicates (Gu et al. 2002b), gene sequence divergence and expression divergence are coupled for human duplicates. Interestingly, the linear relationship between expression divergence and KS, when extrapolated to time 0, does not pass through the origin. We propose two possible factors for this observation. First, this might reflect that expression divergence is more discrete in nature compared with sequence divergence, which is continuous. Second, this might be partly because a duplication might have not included all the regulatory elements, so that the two duplicates had already differed in expression to some extent right after duplication.
Note that the correlation coefficient (R) was calculated over many tissues (tissues in which at least one of the duplicates is expressed). Such pooling of data will include genes that are not relevant to the experiment under consideration. Such genes may show similar expression patterns, and thus, their inclusion would tend to increase the correlation of expression and underestimate the divergence in expression.
Initially, R and KA are coupled (KA < 0.2). After KA becomes >0.2, R is not correlated with KA. Note that at KA = 0.2, almost all duplicates have already diverged in their expression in at least one tissue.
In this study, KS and KA, but not protein sequence divergence (d), were used as proxies of time since gene duplication. KS is a more appropriate proxy of divergence time compared with the other two measures because KS varies substantially less among genes than does KA or d (Li 1997). Both KA and d are much affected by selection, which may differ greatly among genes. KS, on the other hand, is less affected by selection, particularly in mammals, in which there is no evidence for strong selection on codon bias (Urrutia and Hurst 2001). However, KS is affected by regional variation in mutation rate within a genome (Li 1997; Lercher et al. 2001; Williams and Hurst 2002). As a result, KS is still variable among genes, which may partly explain why we do not observe a strong correlation between KS and expression divergence measured by R.
The expression data obtained by the hybridization of RNA to the oligonucleotide arrays are supposed to be more accurate than is cDNA microarray data (Wodicka et al. 1997). The Affymetrix array probes are designed to represent the unique portions of a gene. Each probe sequence is scanned against the available genomic sequences, minimizing cross-hybridization between duplicate genes. This approach has a drawback of excluding recently duplicated genes from an array, as unique probes cannot be designed for them. The arrays based on cDNAs are more prone to cross-hybridization of duplicate genes to the same probe. Nevertheless, our results based on oligonucleotide array data are in agreement with the results of Gu et al. (2002b), who used mainly cDNA arrays. Still, the microarray data are expected to be quite noisy, decreasing the strengths of correlations inferred in the present study.
It is important to note that cross-hybridization tends to underestimate the degree of expression divergence. Therefore, the presence of cross-hybridization should reinforce rather than contradict our conclusion of rapid expression divergence between duplicate genes.
Nevertheless, to test the ability of the Affymetrix arrays to discriminate between the paralogs under study, we performed two tests. First, we compared the probe sequences between two genes for each duplicate pair. (Each gene was represented by 16 oligonucleotide probes; each probe was 25 nucleotides long.) From the original 1404 independent gene pairs selected, only seven had one or more probes (two to seven probes in each case) with identical (or reverse complement) sequences. Additional four gene pairs had probes with one nucleotide mismatch (one to five probes in each case). Thus, it seems that cross-hybridization between duplicate genes was not a serious problem and did not significantly affect our results.
Second, we considered duplicate pairs that were expressed in multiple tissues and that showed differing expression in at least one tissue. These were the cases in which the probes were apparently able to discriminate between the duplicates to some degree at least. Here the genes were considered diverged in expression in a tissue if their expression values differed as average difference (AD) > 200 (see Methods). Most duplicate gene pairs satisfy this criterion. We tested for a relationship between expression and sequence divergence in the remaining tissues for these genes. The results (data not shown) did not significantly differ from the original results, ensuring that the correlation is real.
Our investigation of gene pairs that have rapidly diverged in their expression indicates that typically spatial expression pattern alters both in terms of presence or absence in particular tissues and in terms of the absolute amounts of mRNA transcripts (Table 1). An interesting observation regarding gene pairs that show no divergence in their expression over extensive evolutionary time is that they are usually either ubiquitously expressed or tissue-specific (Table 2). For these gene duplicates, both copies are preserved in the genome without a change in their spatial expression and are most likely maintained by purifying selection. We speculate that in such cases, it is advantageous to have a higher dosage of the gene transcript in the cell.
We found a large number of proteins involved in the defense system of an organism among the duplicate pairs with rapid divergence in spatial expression. This is in agreement with a strong selective pressure for adaptation in such proteins (Hughes and Nei 1988; for review, see Wolfe and Li 2003).
It is worth noting that only a subset of human duplicate genes has been included in this study. These included largely the well-characterized genes that had been discovered before the completion of the Human Genome Project. This could have introduced a bias, for instance, toward inclusion of duplicates that have differing functions and that, therefore, may be more likely to have differing expression patterns than randomly selected duplicate gene pairs would.
This study examines divergence in one of the phenotypic manifestations of duplicate genes, namely, divergence in the pattern of spatial expression. It would be of great interest to investigate the molecular basis of such divergence, that is, divergence in the regulatory regions of gene expression.
METHODS
Identification of Duplicate Genes
The GenBank accession numbers for the sequences of the U95A array (Affymetrix) were downloaded from the Affymetrix Web site (http://www.affymetrix.com). The corresponding nucleotide sequences were retrieved by using Batch Entrez. Then, GenBank entries were parsed, and only the entries with the annotated CDS (CDS tag) were used in a subsequent analysis.
To identify duplicate gene pairs, we followed the method of Gu et al. (2002a). Briefly, every protein was used as the query to search against all other proteins by using FASTA (E = 10). Two proteins are scored as forming a link if (1) the FASTA-alignable region between them is >80% of the longer protein, and (2) the identity (I) between them I ≥ 30% if the alignable region is longer than 150 aa and I ≥ 0.01n + 4.8L-0.32[1 + exp(-L/1000)] (Rost 1999) for all other protein pairs, in which n = 6 and L is the alignable length between the two proteins. Proteins with the same sequence, but different names, were deleted from the database. Clustering was performed by using the single-linkage clustering algorithm. All protein pairs with identity (excluding gaps) >97% were manually inspected, and isoforms were deleted. Each protein was used as the query to search against the database of human repetitive elements. If the proteins formed a link because of their homology with the same repetitive element, they were deleted. All steps were repeated in the second-round grouping to identify gene families.
The yn00 module (Yang and Nielsen 2000) of PAML (Yang 1997) with default parameters was used to calculate the number of synonymous substitutions per synonymous site (KS) and the number of nonsynonymous substitutions per nonsynonymous site (KA). Independent pairs of duplicate genes were selected by using the following procedure. For each multiple gene family, gene pairs were sorted by KS in ascending order. The pair with the smallest KS was selected first. Later, we proceeded by selecting independent pairs (pairs that do not contain genes already selected) with increasing KS.
All gene pairs were aligned using CLUSTALW (Thompson et al. 1994). Duplicate genes with KS > 1.4 were excluded because of difficulties to obtain reliable estimates. Likewise, gene pairs with KA > 0.7 were also excluded.
Expression Data Analysis
The expression data for the 25 human tissues were retrieved from http://expression.gnf.org (Su et al. 2002). Expression values were averaged among replicas. We followed the method of Su et al. (2002) in defining expressed and not expressed genes. For calculating the proportion of gene pairs with altered expression, an AD value of >200 was used to call a gene expressed in a particular tissue (this corresponds to approximately three to five copies of mRNA per cell). Similarly, a gene was called not expressed if AD was <100. Genes with 100 < AD < 200 were called marginally expressed and were excluded from the analysis. The gene pairs analyzed are given in Supplemental Table 1, available at www.genome.org.
For studying the relationship between KS (or KA) and the correlation coefficient of gene expression, we analyzed only the gene pairs in which either both (group A; Suppl. Table 2) or at least one (group B; Suppl. Table 3) of the genes was expressed in at least five tissues (AD > 200), and only these tissues were considered. The AD values were log2-transformed. The Pearson correlation coefficient R was transformed into ln[(1+R)/(1 - R)] to make the scale more appropriate for the linear regression analysis. The linear regression was carried out between each pair of KS (or KA) and the transformed R.
Acknowledgments
We are grateful to Z. Gu, H. Kaessmann, and T. Oakley for comments on the earlier versions of the manuscript; to the reviewers for many excellent comments improving our manuscript; and to A. Nekrutenko for help in revising this manuscript. This study was supported by NIH grants.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1133803.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Camon, E., Magrane, M., Barrell, D., Binns, D., Fleischmann, W., Kersey, P., Mulder, N., Oinn, T., Maslen, J., Cox, A., et al. 2003. The Gene Ontology Annotation (GOA) Project: Implementation of GO in SWISS-PROT, TrEMBL, and InterPro. Genome Res. 13: 662-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, R.J., Huang, M., Campbell, M.J., Dong, H., Steinmetz, L., Sapinoso, L., Hampton, G., Elledge, S.J., Davis, R.W., and Lockhart, D.J. 2001. Transcriptional regulation and function during the human cell cycle. Nat. Genet. 27: 48-54. [DOI] [PubMed] [Google Scholar]
- Gu, Z., Cavalcanti, A., Chen, F.-C., Bouman, P., and Li, W.-H. 2002a. Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol. Biol. Evol. 19: 256-262. [DOI] [PubMed] [Google Scholar]
- Gu, Z., Nicolae, D., Lu, H.H., and Li, W.-H. 2002b. Rapid divergence in expression between duplicate genes inferred from microarray data. Trends Genet. 18: 609-613. [DOI] [PubMed] [Google Scholar]
- Huang, L., Guan, R.J., and Pardee, A.B. 1999. Evolution of transcriptional control from prokaryotic beginnings to eukaryotic complexities. Crit. Rev. Eukaryot. Gene Expr. 9: 175-182. [DOI] [PubMed] [Google Scholar]
- Hughes, A.L. and Nei, M. 1988. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335: 167-170. [DOI] [PubMed] [Google Scholar]
- Lercher, M.J., Williams, E.J., and Hurst, L.D. 2001. Local similarity in evolutionary rates extends over whole chromosomes in human–rodent and mouse–rat comparisons: Implications for understanding the mechanistic basis of the male mutation bias. Mol. Biol. Evol. 18: 2032-2039. [DOI] [PubMed] [Google Scholar]
- Li, W.-H. 1997. Molecular evolution. Sinauer Associates, Sunderland, MA.
- Ly, D.H., Lockhart, D.J., Lerner, R.A., and Schultz, P.G. 2000. Mitotic misregulation and human aging. Science 287: 2486-2492. [DOI] [PubMed] [Google Scholar]
- Ohno, S. 1970. Evolution by gene duplication. Springer-Verlag, New York.
- Rost, B. 1999. Twilight zone of protein sequence alignments. Protein Eng. 12: 85-94. [DOI] [PubMed] [Google Scholar]
- Su, A.I., Cooke, M.P., Ching, K.A., Hakak, Y., Walker, J.R., Wiltshire, T., Orth, A.P., Vega, R.G., Sapinoso, L.M., Moqrich, A., et al. 2002. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. 99: 4465-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia, A.O. and Hurst, L.D. 2001. Codon usage bias covaries with expression breadth and the rate of synonymous evolution in humans, but this is not evidence for selection. Genetics 159: 1191-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, A. 2000. Decoupled evolution of coding region and mRNA expression patterns after gene duplication: Implications for the neutralist-selectionist debate. Proc. Natl. Acad. Sci. 97: 6579-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, E.J. and Hurst, L.D. 2002. Is the synonymous substitution rate in mammals gene-specific? Mol. Biol. Evol. 19: 1395-1398. [DOI] [PubMed] [Google Scholar]
- Wodicka, L., Dong, H., Mittmann, M., Ho, M.H., and Lockhart, D.J. 1997. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat. Biotechnol. 15: 1359-1367. [DOI] [PubMed] [Google Scholar]
- Wolfe, K.H. and Li, W.-H. 2003. Molecular evolution meets the genomics revolution. Nat. Genet. 33: 255-265. [DOI] [PubMed] [Google Scholar]
- Yang, Z. 1997. PAML: A program package for phylogenetic analysis by maximum likelihood. CABIOS 13: 555-556. [DOI] [PubMed] [Google Scholar]
- Yang, Z. and Nielsen, R. 2000. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 17: 32-43. [DOI] [PubMed] [Google Scholar]
- Yi, S., Ellsworth, D.L., and Li, W.-H. 2002. Slow molecular clocks in old world monkeys, apes, and humans. Mol. Biol. Evol. 19: 2191-2198. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.ncbi.nlm.nih.gov/LocusLink/; LocusLink.
- http://www.affymetrix.com; Affymetrix Web site.
- http://expression.gnf.org; expression data for the 25 human tissues.