Abstract
Mapping quantitative trait loci (QTLs)with high resolution facilitates identification and positional cloning of the underlying genes. The novel approach of advanced intercross lines (AILs) generates many more recombination events and thus can potentially narrow QTLs significantly more than do conventional backcrosses and F2 intercrosses. In this study, we carried out QTL analyses in (C57BL/6J × NZB/BlNJ)× C57BL/6J backcross progeny fed either chow or an atherogenic diet to detect QTLs that regulate high-density lipoprotein cholesterol (HDL)concentrations, and in (C57BL/6J × NZB/BlNJ)F11 AIL progeny to confirm and narrow those QTLs. QTLs for HDL concentrations were found on chromosomes 1, 5, and 16. AIL not only narrowed the QTLs significantly more than did a conventional backcross but also resolved a chromosome 5 QTL identified in the backcross into two QTLs, the peaks of both being outside the backcross QTL region. We tested 27 candidate genes and found significant mRNA expression differences for 12 (Nr1i3, Apoa2, Sap, Tgfb2, Fgfbp1, Prom, Ppargc1, Tcf1, Ncor2, Srb1, App, and Ifnar). Some of these underlay the same QTL, indicating that expression differences are common and not sufficient to identify QTL genes. All the major HDL QTLs in our study had homologous counterparts in humans, implying that their underlying genes regulate HDL in humans.
Many cardiovascular diseases result from cholesterol imbalances. In the past several years, prospective, randomized, controlled clinical trials have demonstrated that lowering low-density lipoprotein cholesterol (LDL) can significantly reduce the incidence of cardiovascular diseases. Still, 60% to 70% of treated patients develop adverse cardiovascular events (Shah et al. 2001). Substantial evidence from major epidemiological studies indicates that the level of plasma high-density lipoprotein cholesterol (HDL), especially at average to slightly above average concentrations, is inversely related to coronary artery disease incidence in both women and men. Furthermore, these studies show that coronary artery disease risk associated with HDL is independent of plasma LDL, other lipid parameters (triglycerides, total cholesterol), and other non-lipid risk factors (Franceschini 2001). Many studies indicate that therapies that raise HDL may significantly reduce the incidence of cardiovascular diseases (Boden and Pearson 2000). Although its beneficial properties are not clearly understood, HDL may protect against atherosclerosis in several ways, including stimulating reverse cholesterol transport, inhibiting oxidation of LDL, inhibiting inflammation by suppressing the formation of adhesion molecules and macrophage chemotactic proteins, reducing lipoprotein retention, and attenuating endothelial dysfunction (Libby 2001; Nofer et al. 2002). Thus, it is very important to find the genes that regulate HDL levels: They may provide new therapeutic targets for preventing and curing atherosclerosis.
Plasma HDL levels are controlled by both genes and environment, and up to 70% of the variation of HDL levels in humans is genetically determined (Rader and Maugeais 2000). Some of the responsible genes have been found by searching for polymorphisms in genes that affect HDL. Examples are Abca1 (Bodzioch et al. 1999; Rust et al. 1999) and Lipc (Jansen et al. 1999). Quantitative trait loci (QTL) mapping has been used to identify genomic regions that contain genes regulating HDL. In fact, at least 27 mouse QTLs and 22 human QTLs for HDL levels have been found (Wang and Paigen 2002). Those QTLs often have large confidence intervals (CIs) because they were detected from limited recombination events. Until these QTLs are narrowed, identifying their underlying genes by positional cloning will be challenging.
At least eight different experimental strategies or populations can be used to finely resolve QTLs (Darvasi 1998; McPeek 2000). These are selective phenotyping, selective genotyping, recombinant progeny testing, interval-specific congenic lines (Darvasi 1998), advanced intercross lines (AILs; Darvasi and Soller 1995), recombinant inbred segregation test (Darvasi 1998), recombinant inbred intercross test (D.W. Threadgill, pers. comm.), and genetically heterogeneous stocks (Talbot et al. 1999; Mott et al. 2000). The advantages and disadvantages of each depend on the mapping resolution desired; the size, effect (dominant or additive) and expected number of QTLs; the difficulty of phenotyping the animal models; and the available resources (time, animal space, money).
Regardless of the strategy used, narrowing a QTL interval in a mouse cross depends on generating many recombinations and detecting them by typing the cross progeny with closely spaced polymorphic markers. The large number of recombinations required is usually not generated in F2 and backcross populations, even large ones. On the other hand, repeated intercrossing does generate them, often eliminates linkage disequilibrium, and may resolve a QTL to one-fifth the size possible in backcross and F2 populations (Darvasi and Soller 1995). Such repeated intercrossing is the basis of AILs. To produce such lines, mice of two inbred strains assumed homozygous for alternative alleles at a series of QTLs are used to produce F2, F3, F4, F5, through F10 or more generations, by randomly intercrossing mice in each generation to produce the next, avoiding sibling or cousin pairs. Thus, instead of producing a few recombinations in either a single large F2 or backcross population, the many recombinations required to finely resolve QTLs are accumulated in a single relatively small population produced over the course of many generations. Since AIL were first proposed in 1995 (Darvasi and Soller 1995), their use has been reported only once, to narrow a QTL interval for trypanosomiasis resistance (Iraqi et al. 2000).
C57BL/6J (B6) mice have relatively low plasma HDL levels and are susceptible to atherosclerosis, whereas NZB/BlNJ (NZB) mice have relatively high plasma HDL levels and are resistant to atherosclerosis. In this study, we used a (B6 × NZB)F1 × B6 backcross to detect significant QTLs that either individually or interactively regulate HDL levels. We then used a (B6 × NZB)F11 AIL to finely resolve those QTLs and statistically show that many QTLs exist on the same chromosome. Each QTL was examined for candidate genes, which were tested for mRNA expression differences between the two parental strains.
RESULTS
Plasma Cholesterol Concentrations and Distribution in the Parental Strains, F1, N2, and F11 Progeny
Plasma HDL concentration was twice as high in chow-fed NZB mice than it was in B6 mice (Table 1). Whereas 6 weeks of eating a high-fat diet raised the HDL concentration of the NZB mice by 60%, it did not affect HDL concentrations in B6 mice. These results were consistent with previous research showing that the high-fat diet can increase HDL levels in female NZB mice (Pitman et al. 1998, 2002). The age of the mice had nothing to do with the change of plasma HDL levels: 12-week-old B6 and NZB mice fed chow had the same HDL levels as when they were 8 weeks old (data not shown).
Table 1.
Plasma Lipid Concentrations in the Female Mice From Various B6 × NZB Crosses and Their Parental Strains Before and After High-Fat Diet
|
Total cholesterol
|
HDL
|
Non-HDL
|
||||
|---|---|---|---|---|---|---|
| Before | After | Before | After | Before | After | |
| C57BL/6J (n = 5) | 73±4 | 234±10 | 61±4 | 64±7 | 13±3 | 169±13 |
| NZB/B1NJ (n = 5) | 145±12a | 333±30b | 114±8a | 182±17a | 31±17 | 151±25 |
| F1 (n = 5) | 97±6 | 273±11 | 76±6 | 100±2 | 21±3 | 173±10 |
| N2 (n = 104) | 83±1 | 247±4 | 63±1 | 56±1 | 21±1 | 190±4 |
| F11 (n = 349) | 102±2 | 370±7 | 81±1 | 84±1 | 24±0 | 276±8 |
Plasma from female B6, NZB, (B6 × NZB) F1 mice, and (B6 × NZB) × B6 N2 mice (12 weeks old and fed high-fat diet for 4 weeks) was measured for total cholesterol concentrations with a commercial colorimetric enzymatic assay, and HDL concentrations were measured after apolipoprotein B—containing lipoproteins were selectively precipitated with polyethylene glycol. Non-HDL represents the difference of total cholesterol and HDL. Plasma from (B6 × NZB) F11 mice (10 weeks old and on high-fat diet for 6 weeks) was measured for LDL, VLDL, and HDL concentrations with an enzymatic assay after gel filtration using a Superose 6 column. Non-HDL represents the total of LDL and VLDL concentrations. Values represent mean ± SEM (mg/dL) aP<0.01, bP<0.05, compared with B6 and F1
Non-HDL levels did not differ between NZB and B6 mice fed either diet. The difference in total cholesterol levels between these strains, before and after they were fed a high-fat diet, was due to the difference in HDL levels between the two strains (Table 1). Although the total and HDL cholesterol levels of F1 mice were intermediate between those of the parental strains, they were closer to those of the B6 parents, whether they ate either a chow or a high-fat diet (Table 1). The distributions of HDL levels among the F11 AIL progeny were unimodal, implying that many genes affect the phenotypes (Fig. 1).
Figure 1.
Distribution of HDL concentrations and HDL inducibility in the 345 female (B6 × NZB)F11 AIL progeny. HDL concentrations were determined before (A) and after (B) the mice consumed the high-fat diet for 6 weeks. (C) HDL inducibility was calculated as follows: ([HDL concentrations of mice fed a high-fat diet - HDL concentrations of mice fed chow]/HDL concentrations of mice fed chow) × 100%.
Correlation of Plasma Cholesterol Levels and Atherosclerotic Lesions in the Backcross
The atherosclerotic lesions of all 104 female backcross progeny were measured after the progeny had consumed the high-fat diet for 15 weeks. The total plasma cholesterol and the HDL of each progeny were determined three times: immediately before and 4 and 15 weeks after the progeny began eating the atherogenic diet. The result of a hierarchical clustering analysis is shown in Figure 2. HDL levels and atherosclerosis in mice fed a high-fat diet for 15 weeks were most closely clustered, indicating they were very closely related. Preatherogenic diet cholesterol levels formed the next most closely related cluster to atherosclerosis (75% of total cholesterol in chow-fed mice is HDL; data not shown). Thus, the three cholesterol parameters most closely correlated with atherosclerosis were HDL levels induced by the 15-week atherogenic diet and the basal cholesterol levels before the mice were fed atherogenic diet (total cholesterol and HDL at the start of high-fat diet). In contrast, the four parameters least correlated with atherosclerosis were non-HDL levels induced by the high-fat diet (the bottom four parameters in Fig. 2). Thus, atherogenesis was more closely related to HDL levels than to non-HDL levels. However, we did not find any significant QTL for atherosclerotic lesions in the backcross progeny after they consumed high-fat diet for 15 weeks, because the majority of the mice did not develop significant aortic atherosclerosis.
Figure 2.
Hierachical clustering analysis for atherosclerosis and plasma lipid levels in the 104 (B6 × NZB)F1 × B6 backcross progeny. Total and HDL cholesterol were measured in mice fed chow (0 w), mice fed a high-fat diet for 4 weeks (4 w), and mice fed a high-fat diet for 15 weeks (15 w). Non-HDL was calculated by subtracting HDL from total cholesterol. Atherosclerotic lesions were determined by averaging the lesion sizes of five aortic root cross sections from each mouse. The length along the X-axis represents the distance between two clusters, expressed in semipartial R2 distance. The more similar the traits are, the closer they will appear in the cluster tree.
QTL Analysis of Genetic Factors Affecting Plasma HDL Concentrations
Backcross
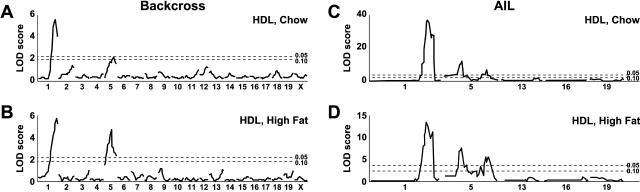
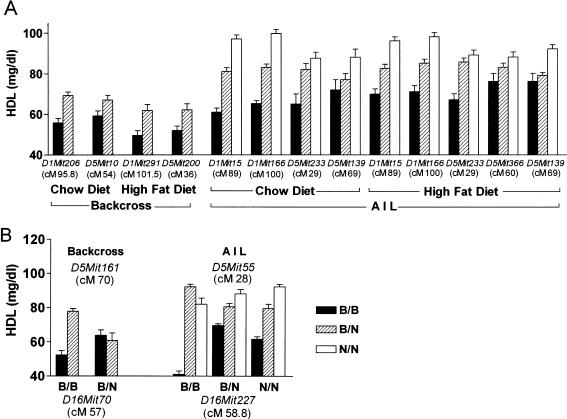
Genome scans in the backcross progeny fed chow and in those fed a high-fat diet revealed QTLs for HDL levels on both Chrs 1 and 5 (Fig. 3A,B). The positions, 95% CIs, LOD scores, and the nearest markers for these QTLs are shown in Table 2. The Chr 1 QTL explained more of the HDL variation than did the Chr 5 QTL (22.3% compared with 9% for chow-fed mice, and 23.3% compared with 19.2% for fat-fed mice; Table 2). Homozygous B6 genotypes at Chrs 1 and 5 QTLs were associated with lower HDL concentrations, and heterozygous genotypes were associated with higher HDL levels (Fig. 4A).
Figure 3.
Genome scans for HDL concentrations in (B6 × NZB)F1 × B6 backcross progeny fed chow (A) and high-fat diet for 4 weeks (B), and HDL concentrations in the (B6 × NZB)F11 AIL progeny fed chow (C) and a high-fat diet for 6 weeks (D). The Y-axis indicates LOD ratio scores, and the X-axis indicates chromosome positions. Horizontal lines indicate suggestive (P < 0.10) and significant (P < 0.05) thresholds, calculated by permutation tests.
Table 2.
HDL QTLs in Female Mice From (C57BL/6J×NZB)×C57BL/6J Backcross and C57BL/6J×NZB/BINJ AIL
| Cross | Phenotype | Chr | cM | 95% CI (cM)a | Nearest marker | LODb | % variance explainedc | Locus name | Homologous human QTL |
|---|---|---|---|---|---|---|---|---|---|
| Backcross | 0 w HDL | 1 | 96 | 84-100 | D1Mit206 | 5.5 | 22.3% | Hdlq5 | |
| 5 | 54 | 40-60 | D5Mit10 | 2.1 | 9.0% | ||||
| 4 w HDL | 1 | 102 | 87-110 | D1Mit291 | 5.8 | 23.3% | Hdlq6 | ||
| 5 | 38 | 34-42 | D5Mit200 | 4.7 | 19.2% | ||||
| AIL | 0 w HDL | 1 | 91 | 87-93 | D1Mit15 | 37.0 | 39.0% | Hdlq5 | 1q21-23,d 1q23-25e |
| 5 | 29 | 25-32 | D5Mit233 | 12.0 | 14.8% | Hdlq7 | 4p16-15.3e | ||
| 5 | 69 | 66-70 | D5Mit242 | 7.1 | 9.0% | Hdlq1 | 12q24.2,g, 13q12-13d,f | ||
| 16 | 59 | 50-60 | D16Mit227 | 1.3 | 0.9% | Hdlq9 | |||
| 6 w HDL | 1 | 89 | 87-93 | D1Mit15 | 13.6 | 16.6% | Hdlq5 | 1q21-23,d 1q23-25e | |
| 1 | 100 | 98-102 | D1Mit166 | 11.4 | 14.1% | Hdlq6 | |||
| 5 | 29 | 26-32 | D5Mit233 | 7.8 | 9.9% | Hdlq7 | 4p16-15.3e | ||
| 5 | 60 | 58-63 | D5Mit155 | 3.6 | 4.7% | Hdlq8 | 1p22f | ||
| 5 | 69 | 66-77 | D5Mit371 | 5.8 | 7.4% | Hdlq1 | 12q24.2,g 13q12-13d,f |
Confidence interval (CI) was calculated according to the posterior probability densities of the QTL locations
Significant thresholds of LOD score were determined by permutation tests. In the backcross, the threshold of suggestive LOD score is 2.0, significant one is 2.3; in AIL, the threshold of suggestive LOD score is 2.5, significant one is 3.0
Phenotypic variance explained by each QTL is based on a single-QTL model
Figure 4.
Allele effect and interaction plots for the QTLs found in the (B6 × NZB)F1 × B6 backcross and in the (B6 × NZB)F11 AIL. (A) Genotypes at marker alleles nearest each QTL identified in the backcross and AIL-affected HDL concentrations in chow-fed and high-fat diet–fed progeny (4 weeks for backcross progeny and 6 weeks for AIL progeny). (B) Genotypes at marker alleles closest to the Chr 5 and 16 QTLs identified in the backcross and AIL interacted to affect HDL concentrations of chow-fed backcross AIL progeny. All HDL concentrations are plotted as mean ± SEM (mg/dL).
A pair-wise genome scan for HDL in chow-fed backcross mice revealed that a Chr 5 QTL near marker D5Mit161 (70 cM) interacted with a Chr 16 QTL near marker D16Mit70 (57 cM). When the D16Mit70 genotype was B/B, HDL levels were higher with a B/N than with a B/B genotype at D5Mit161; when the genotype at D16Mit70 was B/N, HDL levels were the same for either a B/B or B/N genotype at D5Mit161 (Fig. 4B).
Advanced Intercross Lines
Because significant QTLs for HDL levels were found on Chrs 1 and 5 and an interacting locus on Chr 16 in the backcross, we genotyped the AIL progeny with 23 Chr 1 markers, 30 Chr 5 markers, and eight Chr 16 markers. The HDL QTLs subsequently found are shown in Figure 3, C and D, and their positions, 95% CIs, LOD scores, and nearest markers are shown in Table 2. The QTLs were named Hdlq (HDL QTLs) followed by a number. The Chr 1 QTLs for HDL levels in AIL progeny fed chow and in those fed a high-fat diet confirmed and significantly narrowed Hdlq5 found in the backcross progeny (both in those fed chow and in those fed a high-fat diet). In addition to Hdlq5, a second QTL for HDL levels was found on Chr 1 and was given a locus name Hdlq6.
Whereas analysis of AIL chow-fed mice resolved the backcross Chr 5 HDL QTL into two separate ones, it resolved that in AIL mice fed the high-fat diet into three distinct ones. The first, at 29 cM in both chow- and fat-fed mice, was given a locus name Hdlq7; the second, at 60 cM in fat-fed mice, was given a locus name Hdlq8; and the third, at 69 cM in both chow- and fat-fed mice, had already been named Hdlq1 (Pitman et al. 2002).
The Chr 1 QTLs explained more of the variation than did any of the Chr 5 QTLs (39.0% compared with 14.8% and 9.0% for chow-fed mice, and 30.7% [16.6% + 14.1%] compared with 9.9%, 4.7%, and 7.4% for mice fed the high-fat diet; Table 2). Homozygous B6 genotypes at all these QTLs were associated with the lowest HDL concentrations, homozygous NZB genotypes were associated with the highest HDL concentrations, and heterozygosity at these loci was associated with intermediate HDL levels (Fig. 4A).
The LOD score plot patterns in Figure 5, A and B, were similar. Thus, although Hdlq6 was statistically significant only in high fat-fed mice, we still think it exists in the chow-fed mice. Similarly, although Hdlq8 was found only in high fat-fed mice in AIL and appeared as a peak proximal to Hdlq1 in the LOD score plot (Fig. 5D), it may also exist in chow-fed mice, as indicated by a shoulder proximal to Hdlq1 (Fig. 5C).
Figure 5.
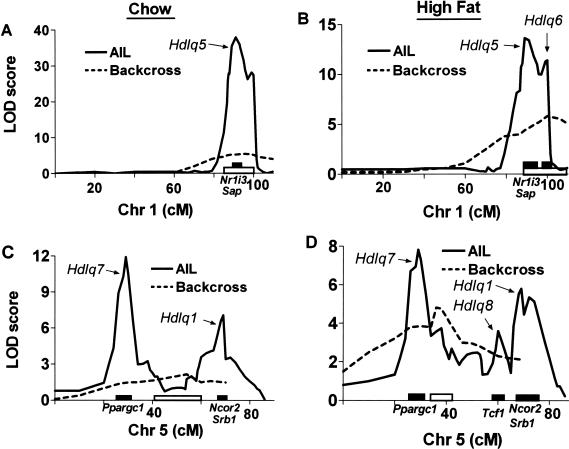
Comparison of QTLs detected in the (B6 × NZB)F1 × B6 backcross and (B6 × NZB)F11 AIL. The fine scans compare the Chr 1 HDL QTLs (A, in chow-fed progeny; B in high fat-fed progeny) and Chr 5 HDL QTLs (C in chow-fed progeny; D in high fat-fed progeny) identified in the backcross (dashed line) and AIL (solid line) progeny. The Y-axis indicates LOD ratio scores, and the X-axis indicates chromosome positions. Solid bars along the X-axis represent 95% confidence intervals (CIs) for the AIL QTLs, and the open bars represent the 95% CI for the backcross QTLs, calculated according to the posterior probability densities of the QTL locations. Some candidate genes are shown along the X-axis.
Because the single marker genome scan showed multiple QTLs on Chr 1 and Chr 5, we carried out a statistical analysis to determine if they were separate QTLs. Simulation tests declaring either one versus two or two versus three QTLs on the same chromosome were carried out 1000 times by using MAKEF2 function in Pseudomarker (release version 0.9; http://aretha.jax.org/jax-cgi/churchill/index.cgi). Maximum LOD scores were calculated based on 1-QTL, 2-QTL, and 3-QTL models. The LOD score difference between 2-QTL and 1-QTL models (ΔLOD2-1) was used to judge whether there were two separate QTLs on the same chromosome; the LOD score difference between 3-QTL and 2-QTL models (ΔLOD3-2) was used to judge whether there were three separate QTLs on the same chromosome. Threshold LOD differences (ΔLOD) were computed by permutation tests, and they were ΔLOD of 1.58 (P = 0.20), ΔLOD of 1.83 (P = 0.10), and ΔLOD of 1.99 (P = 0.05). This means that the LOD score for the 2-QTL model must be at least 1.99 higher than the LOD for the 1-QTL model to accept that a chromosome contains two separate QTLs, and the LOD score for the 3-QTL model must be at least 1.99 higher than the LOD for the 2-QTL model to accept that a chromosome contains three separate QTLs. As shown in Table 3, the LOD score difference for one versus two QTL models (ΔLOD2-1) for the Chr 1 QTLs obtained by analyzing the high-fat diet–fed AIL progeny was 1.5 (not significant); for the Chr 5 QTLs obtained by analyzing the chow-fed progeny, the LOD score difference for one versus two QTL models (ΔLOD2-1) was 2.2 (P < 0.05); for the Chr 5 QTLs obtained by analyzing the high fat-fed progeny, the LOD score difference for one versus two QTL models (ΔLOD2-1) was 2.4 (P < 0.05), and for two versus three QTL models (ΔLOD3-2) was 1.9 (0.10 > P > 0.05; Table 3). Therefore, there were at least two Chr 5 QTLs for HDL levels in both chow and high fat-fed progeny, and possibly three in high fat-fed progeny. Likewise, there is an 80% chance (P 0.2) that there were two distal Chr 1 HDL QTLs in high fat-fed progeny.
Table 3.
Statistical Test for the Existence of Multiple QTLs on the Same Chromosome
|
Chr 1
|
Chr 5
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HDL QTL | LOD1 | LOD2 | ΔLOD2-1 | HDL QTL | LOD1 | LOD2 | LOD3 | ΔLOD2-1 | ΔLOD3-2 | |
| Chow | Hdlq5 | — | — | — | Hdlq7, Hdlq1 | 10.3 | 12.5 | — | 2.2 | — |
| High fat | Hdlq5, Hdlq6 | 13.4 | 14.9 | 1.5 | Hdlq7, Hdlq8, Hdlq1 | 7.0 | 9.4 | 11.3 | 2.4 | 1.9 |
HDL QTL refers to the significant QTLs found in single-QTL model. LOD1, LOD2, and LOD3 are LOD scores for one-QTL, two-QTL, and three-QTL models, respectively. ΔLOD2-1 = LOD2 - LOD1; ΔLOD3-2 = LOD3 - LOD2
Threshold LOD differences were computed by permutation tests: ΔLOD = 1.99 (P = 0.05), ΔLOD = 1.83 (P = 0.10), and ΔLOD = 1.58 (P = 0.20)
A pair-wise scan for Chrs 1, 5, and 16 in the AIL confirmed the interaction between a Chr 16 and a Chr 5 QTL revealed in the backcross: Loci on Chrs 16 (near marker D16Mit227, 58.8 cM) and 5 (Hdlq7, near marker D5Mit55, 28 cM) were found to interactively affect HDL levels (Fig. 4B). However, although the AIL analysis revealed that the position of the Chr 5 interacting locus was ∼30 cM, the backcross analysis revealed it to be ∼70 cM, perhaps because QTLs are more finely resolved and accurately mapped in AIL than in backcrosses. The B allele was associated with lower HDL levels than was the N allele, and the lowest HDL level was found when both the Chr 5 and Chr 16 loci were homozygous for B alleles (Fig. 4B).
Finally, all main effect QTLs detected by genome scans were entered into a multiple regression model to assess their combined effect on HDL levels. To assess those effects, we used F statistics based on adjusted (type III) sum of squares (Table 4). In chow-fed progeny, Hdlq5, Hdlq7, Hdlq1, and Hdlq9 significantly (P < 0.05) regulated HDL concentrations, with Hdlq5 explaining more variance (28.7%) than Hdlq7 and Hdlq1 combined (6.8%). In high fat-fed progeny, Hdlq5, Hdlq6, Hdlq7, Hdlq8, and Hdlq1 each significantly (P < 0.05) affected HDL levels. The multiple regression model indicated that the Chr 1 QTLs affected HDL levels less in high fat-fed progeny than in chow-fed progeny.
Table 4.
Multiple Regression Model for Loci Affecting Plasma HDL Concentrations in Female Mice From C57BL/6J×NZB AIL
| Diet | Chr | cM | Source (locus) | df | Adjusted SS | % variance explained | F | P |
|---|---|---|---|---|---|---|---|---|
| Chow | 1 | 91 | Hdlq5 | 2 | 4.24 | 28.7 | 92.75 | 2.0 × 10-16 |
| 5 | 29 | Hdlq7 | 2 | 0.32 | 6.8 | 7.09 | 7.0 × 10-4 | |
| 5 | 69 | Hdlq1 | 2 | 0.48 | 10.61 | 3.4 × 10-5 | ||
| 16 | 59 | Hdlq9 | 2 | 0.17 | 1.2 | 3.80 | 0.023 | |
| Error | 333 | |||||||
| Total | 341 | |||||||
| High fat | 1 | 89 | Hdlq5 | 2 | 0.40 | 14.1 | 4.69 | 1.0 × 10-3 |
| 1 | 100 | Hdlq6 | 2 | 0.21 | 2.42 | 0.090 | ||
| 5 | 29 | Hdlq7 | 2 | 0.43 | 9.0 | 5.00 | 7.2 × 10-3 | |
| 5 | 60 | Hdlq8 | 2 | 0.33 | 3.82 | 0.023 | ||
| 5 | 69 | Hdlq1 | 2 | 0.26 | 3.03 | 0.049 | ||
| Error | 334 | |||||||
| Total | 344 |
P values are based on the F distribution with 2 and 333 degrees of freedom (df) in chow diet mice and with 2 and 334 degrees of df in high fat-fed mice. F statistics from the multiple regression model are based on adjusted sums of squares
Comparison of the QTLs for HDL Levels in Backcross and AIL
The 95% CIs computed for the AIL QTLs were considerably narrower than those computed for the QTLs identified in the backcross (Table 2; Fig. 5). Whereas the backcross CI for Hdlq5 in chow-fed mice was 16 cM, it was 6 cM in AIL; whereas the backcross CI for the same QTL in high fat-fed mice was 23 cM, it was 6 cM in AIL. Likewise, the Chr 5 QTLs identified in the AIL were considerably narrower than the Chr 5 QTL identified in the backcross. Furthermore, the AIL analysis resolved the broad Chr 5 QTL identified in the backcross into separate loci (Fig. 5C,D; Table 2). The backcross Chr 5 QTL (whether found in the mice fed chow or the high-fat diet) was between the first and second main effect QTLs found in comparable AIL mice (Fig. 5C,D).
Real-Time PCR Quantification of mRNA Expression of Candidate Genes for Plasma HDL Concentrations in B6 and NZB Mice
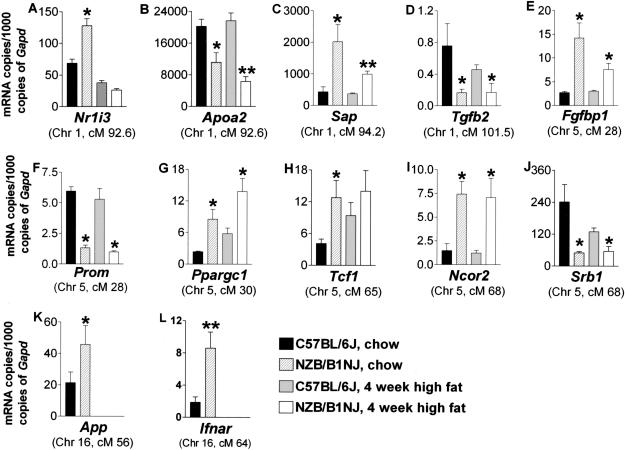
Candidate genes for the HDL QTLs detected in the AIL were scrutinized in the Ensembl Mouse Genome Server (http://www.ensembl.org/Mus_musculus/). Real-time PCR with mRNA extracted from B6 and NZB mouse liver tissue was used to quantify expression levels of 27 candidate genes (a list of the candidate genes and the primer sequences for these genes are listed in Supplemental Table 1 available at www.genome.org). We normalized these expression levels to those of glyceraldehyde-3-phosphate dehydrogenase (Gapd), the expression of which does not differ between chow- and high fat-fed mice of the same strain (data not shown). Among the six Hdlq5 candidate genes tested, mRNA expression levels of Nr1i3 (in chow-fed mice only), Apoa2, and Sap were significantly (P < 0.05) different between B6 and NZB mice. Transforming growth factor (β2 Tgfb2), a candidate gene for Hdlq6, was expressed significantly more (P < 0.05) in B6 than in NZB mice. Among the 13 Chr 5 candidate genes tested, mRNA expression levels of Fgfbp1, Prom, Ppargc1, Tcf1, Ncor2, and Srb1 were significantly (P < 0.05) different between B6 and NZB mice. Among the seven Chr 16 candidates tested, mRNA expression levels of App and Ifnar were significantly (P < 0.05) different between B6 and NZB mice (Fig. 6). In summary, NZB mice expressed more mRNA copies of Nr1i3, Sap, Fgfbp1, Ppargc1, Tcf1, Ncor2, App, and Ifnar than did B6 mice, and B6 expressed more mRNA copies of Apoa2, Tgfb2, Prom, and Srb1 than did NZB mice.
Figure 6.
Real-time PCR quantification of mRNA expression levels of candidate genes for plasma HDL concentrations in B6 and NZB mice. Total RNA was extracted from the livers of female mice-fed chow and from mice fed a high-fat diet for 4 weeks. It was then transcribed into cDNA. mRNA expression levels for each candidate gene was quantified with real time PCR by using fluorescent SYBR. Results were normalized to Gapd and expressed as mRNA copies of candidates per 1000 copies of Gapd. (*P < 0.05, and **P < 0.01, compared with B6 mice on the same diet).
DISCUSSION
Atherosclerosis is a progressive process contributing to the pathogenesis of coronary artery and cerebral vascular diseases. Its very complex etiology involves many genetic and environmental risk factors. A great deal of evidence indicates that it is attenuated by relatively high HDL concentrations (Gordon and Rifkind 1989). To find the genes that control the HDL concentrations, many QTL studies in mice and humans have been carried out. In all, these studies have identified 27 mouse and 22 human HDL-regulating QTLs (Wang and Paigen 2002).
In this study, we used a backcross to detect HDL QTLs and AIL to confirm and finely resolve them. AIL proved to be a very powerful tool for narrowing QTL: The 16 cM and 23 cM CI for Hdlq5 identified in the chow- and fat-fed backcross progeny, respectively, were each narrowed to 6 cM by using AIL. Not only did AIL significantly narrow Hdlq5, it also detected a second HDL QTL (Hdlq6) on Chr 1 in high fat-fed mice.
Most interestingly, the AIL approach resolved the backcross Chr 5 QTL into three separate ones, each with a higher LOD score than that of the backcross. However, none of the three localized to the backcross QTL position. Thus, which are the real QTLs? Haley and Knott (1992) analyzed a simulated regression in which a presumed single QTL may localize in the middle of what are actually two linked QTLs. This appears to be the situation in our study: The Chr 5 QTL (54 cM) identified in the chow-fed backcross progeny was between the first (Hdlq7, 29 cM) and the second (Hdlq1, 69 cM) QTL identified in the AIL; the Chr 5 QTL (38 cM) identified in the high fat-fed backcross progeny was between the first (Hdlq7, 29 cM) and the second (Hdlq8, 60 cM) QTLs identified in the AIL. Perhaps the backcross Chr 5 QTL is a “ghost” QTL, and the flanking AIL Chr 5 QTLs are the real ones. A ghost QTL is the result of model mis-specification (Martinez and Curnow 1992). In a genome scan, we assume that there is only one QTL. When there are in fact two (or more) QTLs and they are moderately tightly linked, the LOD curve can present a peak that does not correspond to either of the two QTLs. Such a ghost peak is most likely to occur in the middle, and as we have demonstrated, the CI based on the ghost peak may exclude both of the real QTLs. It is interesting that QTLs for HDL levels with similar positions to our backcross Chr 5 QTL have been found before: once in a backcross (Pitman et al. 2002) and three times in three different intercrosses (Machleder et al. 1997; Mehrabian et al. 2000; Pitman et al. 2002). Thus, AIL, with many more recombination events, and thus more resolving power than conventional backcrosses and intercrosses, revealed the true and much narrower QTLs. Without a refined mapping strategy such as AIL, pursuing genes underlying ghost QTLs would be fruitless.
By analyzing our AIL with a recently developed statistical approach to detect gene interactions (Sen and Churchill 2001), we detected a gene interaction between Hdlq7 on Chr 5 and Hdlq9 on Chr 16. Finding gene interactions has some practical implications. First, they can help determine which genetic background is best for producing overlapping congenic lines. Such lines, which are used to finely resolve QTL and confirm their association with a phenotype, are constructed by introgressing a locus into a nonnative background. Interactions between this locus and loci in the new background may dramatically alter and lead to a misinterpretation of its effects. Second, genes can interact in many ways: They may either co-activate or co-repress the same target gene(s) in the same pathway, they may work antagonistically, and their ligands and receptors may physically interact. Understanding such dynamics might help an investigator choose the right candidates for testing. Third, identifying interactions between mouse QTL genes may help identify interacting genes involved in human disease (Glorioso et al. 2001; Herrera et al. 2001; Korstanje and Paigen 2002).
Once the QTLs identified in the backcross were more finely resolved with AIL, we searched for candidate genes in the Mouse Genome Server (http://www.ensembl.org/Mus_musculus/) and tested several of them. We were surprised to find that as many as 12 out of the 27 candidates, some for the same QTLs, had twofold or more differences in expression levels between B6 and NZB mice. This implies that interstrain gene expression differences detected by the sensitive real-time PCR method may be common and insufficient evidence that a gene underlies a QTL. It is noteworthy that the mRNA expression level of a gene is only a crude predictor of its functions. Proving that a gene underlies a QTL requires additional evidence, such as either testing its expression in the F2 progeny or showing that a polymorphism in its regulatory region affects transcription. Of course, a mutation in a gene underlying a QTL may not result in an expression difference between two strains; it may only affect the function of a protein.
Many QTLs that regulate HDL concentrations in humans have been reported (for review, see Wang and Paigen 2002), and some are homologous to the QTLs we found in mice: Two QTLs on human Chr 1 (Coon et al. 2001; Elbein and Hasstedt 2002) are homologous to Hdlq5, a human QTL on 4p16–15.3 (Coon et al. 2001) is homologous to Hdlq7, a human QTL on 1p22 (Peacock et al. 2001) is homologous to Hdlq8, and two human QTLs on chromosomes 12q24.2, (Almasy et al. 1999) and 13q12–13,(Peacock et al. 2001; Elbein and Hasstedt 2002) are homologous to Hdlq1 (Table 2). This implies that the human and mouse homologous HDL QTLs may have the same candidate genes.
In summary, we used a (B6 × NZB)F1 × B6 backcross to detect significant QTLs that either individually or interactively regulate HDL levels, and we used (B6 × NZB)F11 AIL to confirm and resolve those QTLs finely enough for us to test their candidate genes. We have thus shown that the AIL approach has the potential to finely resolve large QTLs (some perhaps being ghost QTLs) and eliminate false ones. Future studies could compare the sequences of the promoter and coding regions of promising candidate genes between NZB and B6 mice, compare the protein levels of these candidate genes between the two strains, and explore the functions of the candidate genes in gene-targeted and transgenic mice. Once the genes underlying these HDL QTL are found in mice, they may be directly tested in humans.
METHODS
Mice and Diets
Backcross Progeny
We obtained NZB/BlNJ (NZB) and C57BL/6J (B6) mice from The Jackson Laboratory, Bar Harbor, Maine, and mated them to produce 104 (B6 × NZB)F1 × B6 backcross females. Mice were caged with pine shavings at The Jackson Laboratory in a climate-controlled room (22°C to 23°C) with a 14-h/10-h light/dark cycle (lights on at 6:00 a.m.), and they had free access to acidified water and a chow diet (Old Guilford 234A, CT) until they were 8 weeks old. At that time, some mice were fed an atherogenic diet containing 15% dairy fat, 50% sucrose, 20% casein, 0.5% cholic acid, 1.0% cholesterol, cellulose, vitamins, and minerals (Nishina et al. 1990) for 15 weeks. All experiments were approved by the Animal Care and Use Committee at The Jackson Laboratory.
AIL Progeny
The AIL progeny were produced by Pierre L. Roubertoux at the University Paris-René Descartes, and then maintained at the Centre National De La Recherche Scientifique (CNRS) in Orléans, France. B6 and NZB mice were chosen to construct the AIL because they met two critical criteria for successful AIL analysis: simple sequence length polymorphisms (SSLP) are common between the two strains, and the lipid levels in NZB mice are considerably higher than those of B6 mice (see Mouse Phenome Database in MGI). The mice were caged with dust-free sawdust in a climate-controlled room (23.5 ± 0.5°C) with a 12-h/12-h light/dark cycle (lights on at 7:00 a.m.), and they had free access to water and chow (Old Guilford 234A, CT). B6 × NZB AIL (F2 through F11) were constructed by randomly intercrossing mice (avoiding sibling and cousin pairs) in each generation to produce the next (Darvasi and Soller 1995). The first F11 litters of the F10 females were discarded. When the F10 females were visibly close to parturition for their second litters, they were isolated and, ∼1 d after they gave birth, their litters were culled to five to seven pups to reduce postnatal effects of large litters. Litters smaller than five were discarded. A total of 345 F11 females were produced. When they were 28 ± 2 d old, they were weaned and fed the same atherogenic diet as the backcross progeny for 6 weeks.
Lipid Measurements
Plasma lipid levels were measured three times in the same backcross progeny: (1) when the progeny were 8 weeks old, immediately before they were fed the atherogenic diet, (2) when they had been fed the atherogenic diet for 4 weeks, and (3) when they had been fed the atherogenic diet for 15 weeks. Plasma lipid levels were measured twice in the same AIL progeny: (1) when the progeny were 4 weeks old, immediately before they were fed the atherogenic diet; and (2) when they had been fed the atherogenic diet for 6 weeks. Mice were fasted for 4 h, and blood from their periorbital sinuses was collected in EDTA-containing tubes. The plasma was separated by centrifuging the samples for 5 min at 1500 rpm at 4°C. Plasma total cholesterol concentrations of backcross progeny were measured with a commercial colorimetric enzymatic assay (Pitman et al. 1998). HDL concentrations were measured after apolipoprotein B-containing lipoproteins were selectively precipitated with polyethylene glycol (Pitman et al. 1998). We measured very low density lipoprotein (VLDL), LDL, and HDL from the AIL progeny by gel filtration by using a Superose 6 column. Briefly, plasma was diluted 1/5 in 10 mM Tris (pH 7.4) containing 1 mM EDTA and 150 mM NaCl. Twenty microliters of the sample were injected into the Superose 6 column (preequilibrated in the same buffer), and lipoproteins were separated during a 56-min run. After the cholesterol detection enzymatic reagent (Biomerieux-France) was added, the OD was measured at 500 nm after a 10-min reaction at 37°C. Cholesterol concentration was then calculated using a standard serum from Biomerieux, with the same protocol. Results are expressed as mean ± standard error (SE) in milligrams per decaliter.
Evaluation of Aortic Atherosclerotic Lesions
After the 104 female progeny from the backcross had been on the atherogenic diet for 15 weeks, they were killed by cervical dislocation. Their hearts and ascending aortas were removed and fixed in 4% formaldehyde. Atherosclerotic lesions on the aortic root of each mouse were measured as described previously (Paigen et al. 1987). Lesions were not measured in AIL progeny because 6 weeks of exposure to the atherogenic diet is not sufficient to produce lesions.
DNA Isolation and Genotyping
DNA was isolated from either tail tips or spleens. Approximately either 1 cm of tail tip or one third of a spleen from each mouse was digested overnight in 500 μL of 1× digestion buffer (50 mM Tris-Cl, 100 mM EDTA, 100 mM NaCl, and 1% SDS at pH 8.0) containing 1mg/mL proteinase K in a 55°C water bath. The digested products were mixed with one volume of 25:24:1 phenol:chloroform:isoamyl alcohol and centrifuged for 5 min at 1350 rpm at room temperature. The aqueous phase was isolated, and DNA was precipitated from it with two volumes of 100% ethanol. Strands of DNA were wound around a glass capillary pipette and air-dried. The dried DNA pellets were resuspended in 1 ml TE (10 mM Tris-HCl and 1 mM EDTA at pH 7.5–8.0).
We genotyped each backcross mouse with 97 SSLP markers (MIT MapPairs primers; Research Genetics) spaced 15 to 20 cM apart throughout the mouse genome (a density of three to seven markers per chromosome). Because most of the genetic information for a quantitative trait can be determined from the population members exhibiting the phenotypic extremes of the trait, we first genotyped only the backcross progeny with HDL concentrations that were in the upper and lower 30% of the HDL concentrations of the entire population, a total of 61 of the 104 progeny. After the initial genome-wide scan identified QTLs for HDL levels, we genotyped the remaining 43 mice for those chromosome markers and genotyped all the mice for 11 more markers within the QTL regions identified.
Because AIL are best suited for confirming and mapping QTLs initially detected in either F2 or backcross progeny (Darvasi and Soller 1995), after the backcross genome scan detected main effect QTLs for HDL levels on Chrs 1 and 5, and an interaction between QTLs on Chrs 16 and 5, we genotyped the 345 AIL progeny with 23 polymorphic markers on Chr 1, 29 on Chr 5, and eight on Chr 16.
PCR genotyping for both the backcross and the AIL was carried out for 35 cycles under the following conditions: 30 sec at 94°C, 30 sec at 55°C, and 1 min at 72°C. Polymorphisms were detected by electrophoresing the PCR products on 4% Nusieve® 3:1 agarose gels in 1× Tris-borate-EDTA running buffer for 2 h at 190 V. Gels were then stained with ethidium bromide and photographed under UV light. All SSLP markers used in this study were ordered according to their physical position retrieved from either Ensembl Mouse Genome Server (http://www.ensembl.org/Mus_musculus/) or Celera Discovery System (http://www.celera.com).
Real-Time PCR Analysis
Total RNA was extracted by using RNeasy Mini Kit (Qiagen), following the manufacturer's instructions. For each RNA sample, spectrophotometric absorption at 260 nm was measured, and RNA concentration was calculated as A260 × 40 (μg/mL) × dilution factor. cDNA was synthesized by reverse-transcribing 2 μg of total RNA with Omniscript RT Kit (Qiagen), using oligo(dT)15 primer (Promega). Quantitative real-time PCR was performed by using the ABI Prism 7700 sequence detection system (PE Applied Biosystems). Primers were designed by using the primer design software Primer Express 2.0 (PE Applied Biosystems). The forward and reverse primers for each pair of primers are located on different exons. To be sure the primers amplified a unique and desired cDNA segment, each potential pair of primers was checked in the BLAST program in Ensembl Mouse Genome Server (http://www.ensembl.org/Mus_musculus/) and Celera database (http://www.celera.com). The primers used are shown in Supplemental Table 1.
cDNA samples were mixed with primers and SYBR Master Mix (PE Applied Biosystems) in a total volume of 15 μL. PCR was conducted using the following parameters: 2 min at 50°C, 10 min at 95°C, and 40 cycles of 15 s at 95°C and 1 min at 60°C. PCR reactions were performed in 96-well optical reaction plates (Applied Biosystems). Quantitative real-time PCR was normalized to the copies of Gapd mRNA from the same sample. Acquired data were analyzed by Sequence Detector software (PE Applied Biosystems). All PCR assays were performed in triplicate.
Statistical Analysis
Single marker genome scans to detect main effect QTLs were performed by using the method of Sen and Churchill (2001). We performed marker regression-based genome scans using Matlab Software (Mathworks, Inc.) and computed the oneway ANOVA F statistic at each marker in the genotyping array. LOD ratio scores were computed at 2-cM intervals for the whole genome, and significance thresholds were assessed by permutation tests (Churchill and Doerge 1994). QTLs were deemed significant if they either met or exceeded the 95% genome-wide threshold; they were deemed suggestive if they met the 90% genome-wide threshold but were not significant. We used the more conservative P = 0.1 instead of P = 0.37 suggested by Lander and Kruglyak (1995) as a threshold P value for suggestive QTLs so that fewer false positives would be found. CIs of QTLs were calculated according to the posterior probability density of the QTL locations, as described previously (Sen and Churchill 2001).
Some loci may affect HDL levels through epistatic interactions, so we also performed simultaneous pair-wise genome scans to search for pairs of interacting loci (Sen and Churchill 2001). This method examines all pairs of marker loci for association with HDL levels in a two-dimensional genome scan. The detailed procedure has previously been described (Sugiyama et al. 2001). We chose a nominal level of 0.05 as a significance threshold for interacting QTLs.
Finally, all main effect and interacting QTLs for HDL levels that had been detected by genome scans (single marker and pair-wise) were entered into a multiple regression model by using Minitab software, procedure GLM (Minitab Inc.). We used F statistics based on adjusted (type III) sum of squares to make final determinations on the contribution of a given QTL (main and interacting effects) in combination with all other QTLs.
Hierarchical clustering analysis was performed by using Ward's minimum-variance method in the procedure of Proc Cluster in SAS Program (SAS Institute, Inc., version 8.1). In Ward's minimum-variance method, the distance between two clusters is the ANOVA sum of squares between the two clusters added up over all the variables. At each generation, the within-cluster sum of squares is minimized over all partitions obtainable by merging two clusters from the previous generation. The sums of squares are divided by the total sum of squares to give proportions of variance (squared semipartial correlations).
To compare the difference of the mRNA expression levels between B6 and NZB strains, we used Student's t test. All values are expressed as mean ± SE.
Acknowledgments
This work was funded by GlaxoSmithKline, France, by the Centre National de la Recherche Scientifique (CNRS) and Ministere de la Recherche et de la Technology of France, and by the Program for Genomic Applications of the Heart, Lung and Blood Institute, National Institutes of Health (HL66611). We thank Coralie Outreville for technical assistance and Ray Lambert for his assistance in preparing the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1185803.
Footnotes
Article published online before print in June 2003.
[Supplemental material is available online at www.genome.org.]
References
- Almasy, L., Hixson, J.E., Rainwater, D.L., Cole, S., Williams, J.T., Mahaney, M.C., VandeBerg, J.L., Stern, M.P., MacCluer, J.W., and Blangero, J. 1999. Human pedigree-based quantitative-trait-locus mapping: Localization of two genes influencing HDL-cholesterol metabolism. Am. J. Hum. Genet. 64: 1686-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden, W.E. and Pearson, T.A. 2000. Raising low levels of high-density lipoprotein cholesterol is an important target of therapy. Am. J. Cardiol. 85: 645-650, A610. [DOI] [PubMed] [Google Scholar]
- Bodzioch, M., Orso, E., Klucken, J., Langmann, T., Bottcher, A., Diederich, W., Drobnik, W., Barlage, S., Buchler, C., Porsch-Ozcurumez, M., et al. 1999. The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat. Genet. 22: 347-351. [DOI] [PubMed] [Google Scholar]
- Churchill, G.A. and Doerge, R.W. 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon, H., Leppert, M.F., Eckfeldt, J.H., Oberman, A., Myers, R.H., Peacock, J.M., Province, M.A., Hopkins, P.N., and Heiss, G. 2001. Genome-wide linkage analysis of lipids in the Hypertension Genetic Epidemiology Network (HyperGEN) Blood Pressure Study. Arterioscler. Thromb. Vasc. Biol. 21: 1969-1976. [DOI] [PubMed] [Google Scholar]
- Darvasi, A. 1998. Experimental strategies for the genetic dissection of complex traits in animal models. Nat. Genet. 18: 19-24. [DOI] [PubMed] [Google Scholar]
- Darvasi, A. and Soller, M. 1995. Advanced intercross lines: An experimental population for fine genetic mapping. Genetics 141: 1199-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbein, S.C. and Hasstedt, S.J. 2002. Quantitative trait linkage analysis of lipid-related traits in familial type 2 diabetes: Evidence for linkage of triglyceride levels to chromosome 19q. Diabetes 51: 528-535. [DOI] [PubMed] [Google Scholar]
- Franceschini, G. 2001. Epidemiologic evidence for high-density lipoprotein cholesterol as a risk factor for coronary artery disease. Am. J. Cardiol. 88: 9N-13N. [DOI] [PubMed] [Google Scholar]
- Glorioso, N., Filigheddu, F., Troffa, C., Soro, A., Parpaglia, P.P., Tsikoudakis, A., Myers, R.H., Herrera, V.L., and Ruiz-Opazo, N. 2001. Interaction of α1-Na,K-ATPase and Na,K,2Cl-cotransporter genes in human essential hypertension. Hypertension 38: 204-209. [DOI] [PubMed] [Google Scholar]
- Gordon, D.J. and Rifkind, B.M. 1989. High-density lipoprotein: The clinical implications of recent studies. N. Engl. J. Med. 321: 1311-1316. [DOI] [PubMed] [Google Scholar]
- Haley, C.S., and Knott, S.A. 1992. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69: 315-324. [DOI] [PubMed] [Google Scholar]
- Herrera, V.L., Lopez, L.V., and Ruiz-Opazo, N. 2001. α1 Na,K-ATPase and Na,K,2Cl-cotransporte/D3Mit3 loci interact to increase susceptibility to salt-sensitive hypertension in Dahl S(HSD) rats. Mol. Med. 7: 125-134. [PMC free article] [PubMed] [Google Scholar]
- Iraqi, F., Clapcott, S.J., Kumari, P., Haley, C.S., Kemp, S.J., and Teale, A.J. 2000. Fine mapping of trypanosomiasis resistance loci in murine advanced intercross lines. Mamm. Genome 11: 645-648. [DOI] [PubMed] [Google Scholar]
- Jansen, H., Chu, G., Ehnholm, C., Dallongeville, J., Nicaud, V., and Talmud, P.J. 1999. The T allele of the hepatic lipase promoter variant C-480T is associated with increased fasting lipids and HDL and increased preprandial and postprandial LpCIII:B: European Atherosclerosis Research Study (EARS) II. Arterioscler. Thromb. Vasc. Biol. 19: 303-308. [DOI] [PubMed] [Google Scholar]
- Korstanje, R. and Paigen, B. 2002. From QTL to gene: The harvest begins. Nat. Genet. 31: 235-236. [DOI] [PubMed] [Google Scholar]
- Lander, E. and Kruglyak, L. 1995. Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nat. Genet. 11: 241-247. [DOI] [PubMed] [Google Scholar]
- Libby, P. 2001. Managing the risk of atherosclerosis: The role of high-density lipoprotein. Am. J. Cardiol. 88: 3N-8N. [DOI] [PubMed] [Google Scholar]
- Machleder, D., Ivandic, B., Welch, C., Castellani, L., Reue, K., and Lusis, A.J. 1997. Complex genetic control of HDL levels in mice in response to an atherogenic diet: Coordinate regulation of HDL levels and bile acid metabolism. J. Clin. Invest. 99: 1406-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, O. and Curnow, R.N. 1992. Estimating the locations and the sizes of the effects of quantitative trait loci using flanking markers. Theor. Appl. Genet. 85: 480-488. [DOI] [PubMed] [Google Scholar]
- McPeek, M.S. 2000. From mouse to human: fine mapping of quantitative trait loci in a model organism. Proc. Natl. Acad. Sci. 97: 12389-12390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabian, M., Castellani, L.W., Wen, P.Z., Wong, J., Rithaporn, T., Hama, S.Y., Hough, G.P., Johnson, D., Albers, J.J., Mottino, G.A., et al. 2000. Genetic control of HDL levels and composition in an interspecific mouse cross (CAST/Ei × C57BL/6J). J. Lipid Res. 41: 1936-1946. [PubMed] [Google Scholar]
- Mott, R., Talbot, C.J., Turri, M.G., Collins, A.C., and Flint, J. 2000. A new method for fine mapping quantitative trait loci in outbred animal stocks. Proc. Natl. Acad. Sci. 97: 12649-12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishina, P.M., Verstuyft, J., and Paigen, B. 1990. Synthetic low and high fat diets for the study of atherosclerosis in the mouse. J. Lipid Res. 31: 859-869. [PubMed] [Google Scholar]
- Nofer, J.R., Kehrel, B., Fobker, M., Levkau, B., Assmann, G., and von Eckardstein, A. 2002. HDL and arteriosclerosis: Beyond reverse cholesterol transport. Atherosclerosis 161: 1-16. [DOI] [PubMed] [Google Scholar]
- Paigen, B., Morrow, A., Holmes, P.A., Mitchell, D., and Williams, R.A. 1987. Quantitative assessment of atherosclerotic lesions in mice. Atherosclerosis 68: 231-240. [DOI] [PubMed] [Google Scholar]
- Peacock, J.M., Arnett, D.K., Atwood, L.D., Myers, R.H., Coon, H., Rich, S.S., Province, M.A., and Heiss, G. 2001. Genome scan for quantitative trait loci linked to high-density lipoprotein cholesterol: The NHLBI Family Heart Study. Arterioscler. Thromb. Vasc. Biol. 21: 1823-1828. [DOI] [PubMed] [Google Scholar]
- Pitman, W.A., Hunt, M.H., McFarland, C., and Paigen, B. 1998. Genetic analysis of the difference in diet-induced atherosclerosis between the inbred mouse strains SM/J and NZB/BINJ. Arterioscler. Thromb. Vasc. Biol. 18: 615-620. [DOI] [PubMed] [Google Scholar]
- Pitman, W.A., Korstanje, R., Churchill, G.A., Nicodeme, E., Albers, J.J., Cheung, M.C., Staton, M.A., Sampson, S.S., Harris, S., and Paigen, B. 2002. Quantitative trait locus mapping of genes that regulate HDL cholesterol in SM/J and NZB/B1NJ inbred mice. Physiol. Genomics 9: 93-102. [DOI] [PubMed] [Google Scholar]
- Rader, D.J. and Maugeais, C. 2000. Genes influencing HDL metabolism: New perspectives and implications for atherosclerosis prevention. Mol. Med. Today 6: 170-175. [DOI] [PubMed] [Google Scholar]
- Rust, S., Rosier, M., Funke, H., Real, J., Amoura, Z., Piette, J.C., Deleuze, J.F., Brewer, H.B., Duverger, N., Denefle, P., et al. 1999. Tangier disease is caused by mutations in the gene encoding ATP-binding cassette transporter 1. Nat. Genet. 22: 352-355. [DOI] [PubMed] [Google Scholar]
- Sen, S. and Churchill, G.A. 2001. A statistical framework for quantitative trait mapping. Genetics 159: 371-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, P.K., Kaul, S., Nilsson, J., and Cercek, B. 2001. Exploiting the vascular protective effects of high-density lipoprotein and its apolipoproteins: An idea whose time for testing is coming, part I. Circulation 104: 2376-2383. [DOI] [PubMed] [Google Scholar]
- Sugiyama, F., Churchill, G.A., Higgins, D.C., Johns, C., Makaritsis, K.P., Gavras, H., and Paigen, B. 2001. Concordance of murine quantitative trait loci for salt-induced hypertension with rat and human loci. Genomics 71: 70-77. [DOI] [PubMed] [Google Scholar]
- Talbot, C.J., Nicod, A., Cherny, S.S., Fulker, D.W., Collins, A.C., and Flint, J. 1999. High-resolution mapping of quantitative trait loci in outbred mice. Nat. Genet. 21: 305-308. [DOI] [PubMed] [Google Scholar]
- Wang, X. and Paigen, B. 2002. Quantitative trait loci and candidate genes regulating HDL cholesterol: A murine chromosome map. Arterioscler. Thromb. Vasc. Biol. 22: 1390-1401. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.celera.com; Celera discovery system.
- http://www.ensembl.org/Mus_musculus/; Ensembl mouse genome server.
- http://aretha.jax.org/jax-cgi/churchill/index.cgi; Pseudomarker, release version 0.9.