Abstract
Mice with maternal duplication of proximal chromosome 6 die in utero at an early embryonic stage. Recently, two imprinted genes, paternally expressed Sgce and maternally expressed Asb4, were identified in this region. This report analyzes the imprinting status of genes within a 1-Mb region containing these two genes. Peg10, which is next to Sgce, shows complete paternal expression, like Sgce. Conversely, Neurabin, Pon2, and Pon3 show preferential maternal expression at embryonic stages, although they all show biallelic expression in neonatal tissues. These results demonstrate that there is a large novel imprinted gene cluster in this region. 5′-RACE (Rapid Amplification of cDNA Ends) analysis of Peg10 revealed the existence of a novel first exon separate from the second exon, which encoded two putative ORFs similar to the viral Gag and Pol proteins. A differentially methylated region established in sperm and eggs is located just within the region containing the two first exons of Peg10 and Sgce, and may play an important role in regulating the two paternally expressed genes: Peg10 and Sgce.
Maternal uniparental duplication in mouse proximal chromosome 6 causes early embryonic lethality (Beechey 2000; also see http://www.mgu.har.mrc.ac.uk/imprinting/imprinting.html). Previously, we identified Peg1/Mest as the first imprinted gene in the mouse proximal chromosome 6 (Kaneko-Ishino et al. 1995). However, Peg1/Mest-deficient mice show perinatal growth retardation and abnormal maternal behavior, but do not show early embryonic lethality (Lefebvre et al. 1998). Recently, the existence of two imprinted regions has been demonstrated in this region; mice with a maternal duplication proximal to T77H die in utero before 11.5 dpc, whereas those with a duplication distal to T77H show growth retardation (Beechey 2000). Peg1/Mest maps to the latter region, and two novel imprinted genes, paternally expressed Sgce and maternally expressed Asb4, have been identified in the former region (Piras et al. 2000; Mizuno et al. 2002). We have also reported that human retrotransposon-derived PEG10, which is adjacent to SGCE, is a paternally expressed imprinted gene (Ono et al. 2001).
Because many imprinted genes form clusters in some chromosome regions, it is very possible that there is a large imprinted gene cluster in the Sgce-Asb4 region. Therefore, we analyzed the imprinting status of six other genes in this region (Cas1, Peg10, Neurabin, Pon1, Pon3, and Pon2) and showed that four genes were imprinted. Peg10 was paternally expressed, whereas the other three genes showed preferential maternal expression in the embryonic stage. In relation to these imprinted genes, we discuss the phenotypes observed in mice with maternal duplication of proximal chromosome 6 (early embryonic lethality and perinatal growth retardation) and in human maternal disomy of chromosome 7, which is associated with the growth retardation phenotype known as Silver-Russell syndrome (SRS; OMIM No.180860).
RESULTS
Gene Alignment and Analysis of CpG Islands of the Col1a2-Asb4 Region in the Mouse
Nine genes in mouse proximal chromosome 6 map to a 1-Mb region between 1.2 and 2.2 Mb of the Mus musculus Whole-Genome Shotgun (WGS) supercontig Mm6_WIFeb01_97 (GenBankaccession no. NW_000272): Col1a2, Cas1, Sgce, Peg10, Neurabin, Pon1, Pon3, Pon2, and Asb4 (from proximal to distal, Fig. 1A). This gene alignment is conserved in the human syntenic 7q21 region (data not shown). In this report, we characterize the imprinting status of six of these genes (Cas1, Peg10, Neurabin, Pon1, Pon3, and Pon2), because Sgce and Asb4 are already known to be imprinted and Col1a2 is reported to show biallelic expression (Piras et al. 2000; Mizuno et al. 2002). This region contained four CpG islands (CGIs) corresponding to the promoter regions of the Cas1, Peg10-Sgce, Neurabin, and Pon2 genes. Bisulfite sequencing analyses of both day 10 embryo and placenta showed that three regions were nonmethylated (Fig. 1B), and that of Peg10-Sgce was differentially methylated (Fig. 2A).
Figure 1.
Genomic structure of the Col1a2-Asb4 region in the mouse. (A) Physical map of the genes identified in a 1-Mb region in the Mus musculus WGS supercontig Mm6_WIFeb01_97 (GenBank accession no. NW_000272). The arrows show the direction of each transcription unit, and imprinted genes are indicated by color: red and pink indicate strong and weak preferential maternal expression, respectively, and blue indicates complete paternal expression. Genes showing no expression biases between parental alleles are shown in black. (B) CpG islands (CGIs) and their DNA methylation states in the domain. There are four CGIs (length over 300 bp) shown in green boxes in the Col1a2-Asb4 region, and the three nonmethylated CGIs determined by bisulfite sequencing of genomic DNA isolated from (B6 × JF1) F1 embryo and placenta (day 10) are shown. DNA polymorphisms were used to determine paternal and maternal alleles of Cas1 CGI and Pon2 CGI, and showed that both alleles were nonmethylated in this region. No available DNA polymorphisms were found in Neurabin CGI, but no methylated CpGs were observed in this region either. Each horizontal line indicates the sequence from a single clone. Each CpG dinucleotide is represented by an oval. White and black ovals indicate nonmethylated and methylated CpGs, respectively. Details of the differentially methylated region of the Peg10-Sgce CGI are shown in Fig. 2A.
Figure 2.
Peg10 imprinted expression associated with primary DMR. (A) The genomic structure of Peg10 and bisulfite sequencing analyses of the Peg10 repeated sequence region. The full-length Peg10 sequence was determined by 5′-RACE analysis. The white boxes are untranslated regions and putative open reading frames are shown with black boxes. Two putative ORFs of Peg10 are shown in gray boxes below; ORF1 shares 30% amino acid identity with the gag protein, and ORF2 predicted from a -1 frameshift of ORF1 shares 25% identity with the pol protein of Sushi-ichi retrotransposon, respectively. The translational frameshifting of ORF1–ORF2 is commonly observed in retroviruses and gypsy-type LTR retrotransposons. The arrows indicate the 5′–3′ direction of Peg10 and Sgce. The DNA methylation status of Peg10 repeats in day 10 embryo, eggs, and sperm are shown. Differential methylation was observed in the day 10 embryo as well as the day 10 placenta (data not shown). DNA polymorphisms between JF1 and B6 were used to distinguish parental alleles. The entire Peg10-Sgce CpG island indicated by gray lollipops shows a differentially methylated status similar to that in the Peg10 repeats (data not shown). (B) Twelve Peg10 intronic 29-bp repeat sequences. Shaded boxes indicate homology with the most frequent sequence, GCGCTTCATGCGCTACAAAATACTCATAG (four times). (C) Northern blot profiles of Peg 10 in mouse adult tissues. Total RNA from mouse adult brain (lane 1), heart (lane 2), lung (lane 3), liver (lane 4), spleen (lane 5), kidney (lane 6), stomach (lane 7), small intestine (lane 8), skeletal muscle (lane 9), skin (lane 10), thymus (lane 11), testis (lane 12), uterus (lane 13), and placenta (lane 14) was analyzed. Ribosomal RNA detected by ethidium bromide (EtBr) staining was used as a marker. Strong Peg10 expression was observed only in placenta, and the major transcript was estimated to be -6.5 kb long. (D) Paternal expression of Peg10. Paternal expression of Peg10 in day 10 embryo, placenta, yolk sac, and neonatal brain is demonstrated by direct sequencing of the RT-PCR products, by comparing the sequence profiles of genomic DNA containing B6 and JF1 alleles equally.
We previously reported that human PEG10 was derived from a Sushi-ichi retrotransposon encoding two ORFs (ORF1 and ORF2) showing similarity to retroviral Gag and Pol proteins, respectively (Fig. 2A; Poulter and Butler 1998; Ono et al. 2001). Peg10 exists in all five mammals that we have examined (human, cat, dog, olive baboon, and chimpanzee), in the same location as in the mouse (Fig. 1A; GenBankaccession no. AC069292, AC108197, AC113572, AC092529, and AC094111, respectively). Expressed sequence tags (ESTs) corresponding to Peg10 are also registered for the cow, rat, mink, and pig (GenBank accession no. AV608102, AI599367, MVU00594, and BF191703, respectively). Therefore, retrotransposon-derived Peg10 is highly conserved in mammals.
Genomic Structure and Verification of Imprinting of Mouse Peg10
We determined the full-length sequence of Peg10, which consists of 6407 bp, by 5′-RACE (Rapid Amplification of cDNA Ends; GenBankaccession no. AB091827). The first exon of Peg10 was identified 6.5 kb upstream of the second exon and was within 250 bp of the first exon of Sgce, which is oriented in a head-to-head manner (Fig. 2A). The genomic structure of these two genes is also conserved in the human genome (data not shown). Therefore, it is not an intron-less gene, as we previously reported in humans (Ono et al. 2001).
A CGI overlaps the first two exons of Sgce and Peg10 (Fig. 2A), and there are 12 direct repeats of a 29-bp GC-rich sequence in the Peg10 intron 1, just downstream of the first Peg10 exon (Fig. 2B). Bisulfite sequencing analyses showed that the whole CGI was differentially methylated in paternal and maternal alleles in both day 10 embryo (Fig. 2A, lower part) and placenta (data not shown). Furthermore, it was revealed that differential methylation was already established in oocytes and sperm. Therefore, this region is the primary differentially methylated region (DMR), which indicates that it has an important function in regulating the paternal expression of both Peg10 and Sgce (see following).
To verify tissue-specific expression, we carried out Northern blot analyses using a Peg10 3′untranslated region (UTR) fragment as a probe. As shown in Figure 2C, a high level of Peg10 expression was observed only in placentas. The major transcript was estimated to be -6.5 kb, which is consistent with the full-length cDNA that we identified. Strong expression of human SGCE and PEG10 in placenta has been reported previously (McNally et al. 1998; Ono et al. 2001).
To verify the imprinting status of Peg10, we examined DNA polymorphisms in the Peg10 3′UTR between JF1 and C57BL/6 (the G and A residues indicated by arrows in Fig. 2D, respectively) by direct sequencing. In day 10 F1 embryos of the crosses (B6 × JF1 and JF1 × B6), only paternal G residues were detected in the placentas, yolksacs, and neonatal brains in the former, and only paternal A residues were detected in the latter samples. Therefore, paternal expression of Peg10 was confirmed in mice, as occurs with human PEG10.
Genes Showing Preferential Maternal Expression
To test the imprinting status of the remaining five genes (Cas1, Neurabin, Pon1, Pon3, and Pon2), we examined DNA polymorphisms between JF1 and C57BL/6. Allele-specific expression analyses were carried out using a restriction fragment length polymorphism (RFLP) method combined with the Hot-stop RT-PCR method (Uejima et al. 2000). To exclude the possible misinterpretation of imprinting status because of the existence of overlapping RNAs, we confirmed that no other cDNA bands of different sizes or genomic DNA bands of the same size were detected in RT-PCR experiments for each gene, when the two primers were located in different exons. In addition, the amplified bands were directly sequenced to confirm that they were derived from the genes in question.
Cas1, which encodes a putative glycosyltransferase, is a conserved gene found in humans, Drosophila, plants, and bacteria (Janbon et al. 2001). In the mouse, RT-PCR experiments with RNA from a range of adult tissues showed that Cas1 was expressed ubiquitously (Fig. 3A). In the RFLP analysis, Cas1 showed complete or equal biallelic expression in neonatal brain. It was also biallelically expressed in day 10 and 13 embryo, placenta, and yolksac samples, but there seemed to be weakmaternal biases, especially in extraembryonic tissues (data not shown). Therefore, an RFLP analysis combined with Hot-stop PCR was performed to quantify the expression of each allele precisely. The levels of JF1 and B6 expression went up and down reciprocally in two reciprocal F1 samples (Fig. 3B), indicating its imprinting. However, the expression ratios of maternal/paternal alleles (M/P values) in these samples were small (no more than twofold) and were not conclusive.
Figure 3.
Characterization of Cas1 and Neurabin. (A) Expression profiles of Cas1. The RT-PCR products (30 cycles for Cas1 and 25 cycles for the control Gapdh) using total RNA from a range of adult tissues (brain, skeletal muscle, spleen, kidney, liver, and lung) are shown. (B) Cas1 expression in the embryonic stage. In the DdeI RFLP experiment combined with Hot-stop PCR using a radioisotope-labeled primer, the B6 and JF1 alleles yielded 102- and 670-bp fragments, respectively. Three experiments were carried out in the same samples and the average ratios between the B6 and JF1 alleles, and also between maternal and paternal alleles (M/P values), are shown below. Similar results were obtained in the tissues from different individuals (data not shown). (C) Northern blot profiles of Neurabin in mouse adult tissues. Strong expression is observed in the brain sample, whereas weak ubiquitous expression is observed in all other tissues. (D) Maternal expression of Neurabin. In the AciI RFLP analysis with Hot-stop PCR, the B6 and JF1 alleles yielded 240- and 100-bp fragments, respectively. Preferential maternal expression was observed in placenta (day 10) and yolk sac (day 10 and 13) samples. The gray boxes indicate clear maternal biases in reciprocal crosses.
Neurabin was first isolated as an actin filament (F-actin)-binding protein in the rat. It regulates synapse formation in vitro and is specifically expressed in neural tissues (Nakanishi et al. 1997). As shown in Figure 3C, Neurabin was strongly expressed in the brain. Allelic expression analysis using RFLP with Hot-stop PCR showed biallelic expression of Neurabin in the neonatal brain, and preferential maternal expression was observed in placenta (day 10) and yolksac (day 10 and 13) samples (Fig. 3D). In these tissues, the ratio of expression of a 240-bp band derived from the B6 allele to that of a 100-bp band from the JF1 allele was constantly higher in (B6 × JF1) F1 samples and lower in reciprocal (JF1 × B6) F1 samples. The M/P values of placentas were 3.5 and 2.6 (day 10) in (B6 × JF1) F1 and (JF1 × B6) F1, respectively, and these values for yolksac samples were 2.2 and 4.9 (day 10) and 2.0 and 4.6 (day 13). Because it is apparent that placenta samples from day 13 contain significant amounts of maternal tissue, we did not analyze these samples. We also cannot exclude the possibility of maternal tissue contamination from placenta samples completely, even when isolated from day 10 conceptus, whereas yolksac samples can be recovered free from maternal contamination. Therefore, we conclude that Neurabin is also a maternally expressed imprinted gene at the embryonic stage, especially in extraembryonic tissues.
The three Pon genes are located adjacent to each other on mouse chromosome 6 and human chromosome 7 (Fig. 1A; Primo-Parmo et al. 1996). Human PON1, PON2, and PON3 share 65% similarity at the amino acid level. PON1 is an enzyme associated with high-density lipoprotein (HDL) that is believed to protect against the early events of atherogenesis via its ability to hydrolyze oxidized phospholipids. It is also involved in the detoxification of organophosphate insecticides, such as parathion and chlorpyrifos. In contrast, PON2 and PON3 lack paraoxonase activity, although they have similar antioxidant properties. PON3 is also found in HDL, whereas PON2 is not associated with HDL (Davies et al. 1996; Shih et al. 1998; Draganov et al. 2000; Ng et al. 2001).
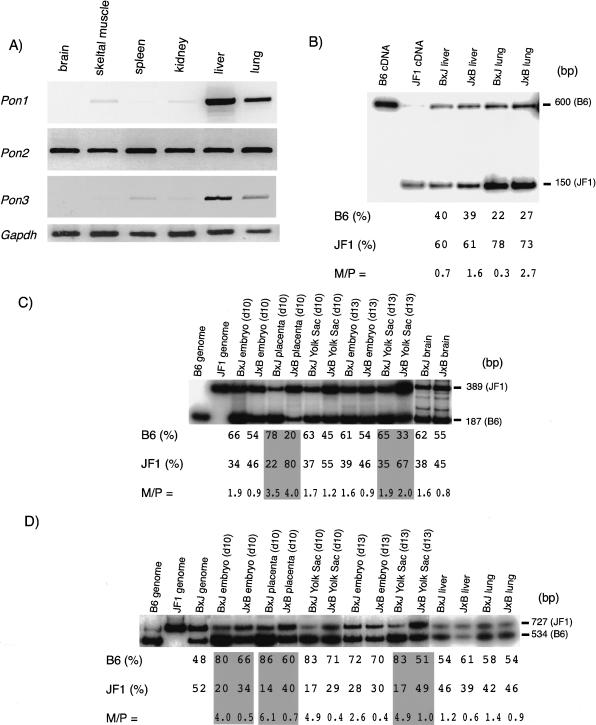
As shown in Figure 4A, Pon1 expression was detected mainly in liver and lung. Therefore, we tested the imprinting status of Pon1 in liver and lung from neonates. As shown in Figure 4B, the same biallelic expression pattern of Pon1 was observed between reciprocal F1 crosses, although it is apparent that the expression of the JF1 allele was constantly higher than that of the B6 allele in both tissues. It has been reported that DNA polymorphisms in human promoter regions affect the levels of IL-4 and TNF-α expression (Song et al. 1996; Wilson et al. 1997). Therefore, it is highly possible that genes showing strain-specific (or strain-biased) expression exist in the mouse. In this experiment, apparent allele-biased expression was also observed in Pon2 and Pon3 (see following). To distinguish imprinted parental allele-specific expression from strain-specific expression, it is very important to analyze the imprinting status in the F1 of both crosses. It was difficult to obtain reliable results from embryos, placentas, and yolksacs because of low expression of Pon1 in these tissues (data not shown). Mouse Pon2 is ubiquitously expressed (Fig. 4A) and biallelic expression was observed in neonatal brain (Fig. 4C, right-hand lanes) and other neonatal tissues (skeletal muscle, spleen, kidney, liver, and lung; data not shown). However, significant maternally biased expression was observed in day 10 placenta (3.5 and 4.0) and to a lesser degree in day 13 yolk sac samples (1.9 and 2.0) in (B6 × JF1) F1 and (JF1 × B6) F1, respectively (Fig. 4C). Therefore, Pon2 is imprinted, at least, in these restricted tissues at the embryonic stages.
Figure 4.
Characterization of Pon1, Pon2, and Pon3. (A) Expression profiles of Pon1, Pon2, and Pon3. RT-PCR products (30 cycles for Pon1, Pon2, and Pon3, and 25 cycles for Gapdh) from the same adult tissues used in Fig. 3A are shown. Pon2 was expressed ubiquitously, whereas Pon1 and Pon3 expression were observed mainly in liver and lung. (B) Biallelic expression of Pon1. In the MspI RFLP experiment with Hot-stop PCR, the B6 and JF1 alleles yielded 600- and 150-bp fragments, respectively. The biallelic expression patterns were unchanged between reciprocal crosses, although JF1 alleles were always expressed more strongly than B6 alleles. (C) Maternal expression of Pon2. In the HaeIII RFLP experiment with Hot-stop PCR, the B6 and JF1 alleles yielded 187-bp and 389-bp fragments, respectively. Preferential maternal expression of Pon2 was observed in placenta and to a lesser degree in day 13 yolk sac. (D) Maternal expression of Pon3. In the PleI RFLP experiment with Hot-stop PCR, the B6 and JF1 alleles yielded 534-bp and 727-bp fragments, respectively. Maternal biases of both the B6 and JF1 alleles were observed in day 10 placenta and day 13 yolk sac and also observed in day 10 embryo to a lesser degree. Note that the B6 allele was expressed more strongly than the JF1 allele in all cases.
As shown in Figure 4A, mouse Pon3 is expressed mainly in liver and lung in the adult. There was no evidence of its imprinting in neonatal liver and lung (Fig. 4D, right-hand lanes). However, Pon3 showed maternally biased expression in day 10 placenta and day 13 yolksac samples, because the B6 (534 bp) and JF1 (727 bp) alleles went up and down reciprocally in these tissues of two different F1s (Fig. 4D). Similar maternal biases were also observed in day 10 embryo to a lesser degree. It was also evident that the expression of B6 alleles in embryo, placenta, and yolksac was much higher than that of JF1 alleles in both reciprocal F1s. Therefore, M/P values are not a good indicator of imprinting in this case.
DISCUSSION
In this study, we demonstrated that at least two paternally expressed genes and four maternally expressed genes are located in a 1-Mb region between Col1a2 and Asb4 in the mouse proximal chromosome 6 and form a large cluster of imprinted genes. Of these, Sgce and Peg10 show paternal expression in an all-or-none fashion, and Asb4 shows mainly maternal expression with a lesser paternal contribution (Piras et al. 2000; Mizuno et al. 2002). One characteristic of this cluster is that three genes (Neurabin, Pon2, and Pon3) show preferential maternal expression in the embryonic stage. They show biallelic expression in neonatal tissues, but their expression in extra-embryonic tissues appears maternally biased. Although most imprinted genes identified so far show clear all-or-none monoallelic expression patterns, some imprinted genes showing weak parental preferences were recently reported, including Tnfrh1 on mouse distal chromosome 7 and Dio3 on mouse distal chromosome 12 (Clark et al. 2002; Tsai et al. 2002). The former shows a weakmaternal bias in several organs and the latter shows preferential paternal expression in embryos and placentas to differing degrees. It is rational to expect that genes showing differential expression of paternal and maternal alleles that are always associated with the same parental biases be classified as imprinted genes.
What is the mechanism of this preferential expression? There are three possibilities: (1) the existence of overlapping transcripts from other gene(s) showing paternal or biallelic expression; (2) the existence of two promoters, one for maternal expression and the other for biallelic expression; and (3) the relaxation of genomic imprinting: the change from an imprinted state to a nonimprinted state occurs in certain situations.
The first possibility, overlapping RNA, such as readthrough products of Peg10, can be excluded, because no other cDNA bands of different sizes or genomic DNA bands of the same size were detected in RT-PCR experiments for Pon1 and Pon2, when the two primers were located in different exons. The amplified bands were directly sequenced to confirm that they were derived from the genes in question, as mentioned. Moreover, as far as the expression profile of Peg10 is concerned, this hypothesis is unlikely, because Peg10 is strongly expressed only in placentas, where the maternal preferential expression of these genes was observed; the genes showed completely biallelic expression in neonatal tissues, where Peg10 expression is very low.
The second possibility is more likely, because several EST clones possessing different transcription start sites for Neurabin have been registered in GenBank(accession no. AW048064, BB617696, and BB639924). There are also several examples in which different promoters show different expression profiles depending on tissue and developmental stage. These include Igf2 and human PEG1/MEST (paternal and biallelic), Gnas, Nesp, and GnasXL (paternal, maternal, and biallelic) and mouse Meg1/Grb10 (maternal and paternal) and human GRB10 (biallelic and paternal; Vu and Hoffman 1994; Peters et al. 1999; Kosaki et al. 2000; Li et al., 2000; Kobayashi et al., 2001; Hikichi et al. 2003). In such cases, intermediate types of expression should be observed where two different transcripts overlap. Further analyses to identify the precise promoters of these genes will be necessary to test this possibility.
It is also worth considering the third possibility, the relaxation of imprinting, because there is evidence for a change in the expression of imprinted genes associated with a change in DNA methylation in DMR regions in some diseases and cancers as a result of mutation or deletions. However, we do not know of a mechanism that relaxes imprinting functions in normal development without changes in DNA sequences.
In conclusion, the maternal bias in expression should be reflected by some imprinting-related mechanism, overlapping transcripts from different promoters showing different parental expression profiles, or the relaxation of imprinting.
There are two conserved imprinted regions in the mouse proximal chromosome 6 and the human syntenic region of the long arm of chromosome 7. In the mouse, the Peg1/Mest region is responsible for perinatal growth retardation, and the region examined in this workis responsible for early embryonic lethality when maternal duplication occurs. However, human maternal disomy of the chromosome 7 containing both imprinted regions is associated with the perinatal growth retardation known as Silver-Russell syndrome, but there is no evidence of early embryonic lethality (Preece et al. 1997). Therefore, two questions arise concerning the latter imprinted region: what gene(s) is responsible for early embryonic lethality in the mouse and why is no severe lethal phenotype observed in humans?
One simple possibility is the existence of a mouse-specific imprinted gene(s) that causes early embryonic lethality. We have confirmed that both human PEG10 and mouse Peg10 are paternally expressed in an all-or-none manner. It has been reported that a mutation in human SGCE on chromosome 7q21 causes myoclonic dystonia (OMIM no.159900), which is characterized by bilateral, alcohol-sensitive myoclonic jerks involving mainly the arms and axial muscles (Zimprich et al. 2001). The family trees of patients show paternal inheritance of this disease, indicating that human SGCE is also a paternally expressed gene. However, it is not known whether it shows completely monoallelic expression, as with mouse Sgce, or paternal expression with a maternal contribution. The imprinting status of the other four human homologs corresponding to the mouse maternally expressed genes, Neurabin, Pon2, Pon3, and Asb4, is not known. Therefore, analyses of these human homologs are required to elucidate phenotype differences between the human and mouse. In this study, we analyzed genes in a 1-Mb region near Sgce and Asb4. It is also probable that there are other imprinted genes responsible for the mouse lethal phenotype downstream from Asb4.
Another explanation for the human–mouse phenotype discrepancy is the difference in the mechanisms producing human uniparental disomies and mouse uniparental duplications. It has been reported that some human chromosomal disomies arise from trisomies, because congenital chromosome mosaicism showing trisomy in placentas containing both paternal and maternal alleles is sometimes observed in human uniparental disomy patients. In contrast, mice with maternal duplication have been constructed by mating mice with Robertsonian translocations or reciprocal translocations at the same locus (see http://www.mgu.har.mrc.ac.uk/imprinting/imprinting.html). Consequently, all the tissues in the embryos and placentas possess two maternal alleles for the duplicated regions. Therefore, it is possible that the mouse early embryonic phenotype results from a placental abnormality, which is not apparent in the human case. In this case, Sgce and Peg10 are most likely involved, because they show complete paternal expression and are strongly expressed in placentas. Gene targeting approaches to these genes are necessary to test this possibility.
In this report, we identified a primary DMR in the region where the first exons of Sgce and Peg10 overlap. Twelve repeat sequences of a 29-bp fragment are located within a CGI just downstream from the Peg10 first exon. It would be very interesting to determine whether this DMR regulates only the two paternally expressed genes or all the genes in the cluster, including the maternally expressed genes, because there are no apparent DMRs in this region. A gene-targeting study of the DMR will be very important for elucidating the mechanism regulating this imprinted region in mouse proximal chromosome 6.
METHODS
Identification of Genes and CGIs From a 1-Mb Region Between Col1a2 and Asb4
The genomic sequence in the 1-Mb region between Mb 1.2 and 2.2 of Mus musculus WGS supercontig Mm6_WIFeb01_97 (GenBankaccession no. NW_000272) containing Sgce and Asb4 was repeat-masked using Repeat Masker, and then used in a BLASTN search of db ESTs to identify transcripts. Five of the identified EST clusters were identical to mouse Col1a2 (GenBankaccession no. NM_007743), Sgce (NM_011360), Pon1 (NM_011134), Pon2 (NM_008896), Pon3 (NM_008897), and Asb4 (NM_023048), and the rest were orthologs of human Cas1 (NM_022900), PEG10 (NM_015068), and rat Neurabin (NM_053473). The mouse gene sequences (registered as Peg10: AB091827, Neurabin: AB091828, and Cas1: AB091829) were confirmed using RT-PCR and nucleotide sequencing. To identify CGIs, we ran the program Cpgplot (http://www.ebi.ac.uk/emboss/cpgplot/) with the following parameters: Observed/Expected ratio > 0.60, Percent C + Percent G > 50.00, Length > 300. Four CGIs were identified: Cas1 CGI (1304815bp-1305668bp; NW_000272), Peg10-Sgce CGI (1453104bp-1453415bp; NW_000272), Neurabin CGI (1609519bp-1610544bp; NW_000272), and Pon2 CGI (2009459bp-2010036bp; NW_000272) corresponding to the promoter regions of these genes.
RT-PCR and 5′-RACE
Genomic DNA and total RNA were prepared from several tissues from the F1 of (B6 × JF1), (JF1 × B6), and (B6 × C3H) mice, using ISOGEN (Nippon Gene), as described previously (Kaneko-Ishino et al. 1995). cDNA was synthesized from 1 μg of total RNA using Superscript II reverse transcriptase (Life Technologies) with an oligodT primer. Gene expression profiles were deduced by agarose gel electrophoresis of RT-PCR products with ethidium bromide (EtBr) staining. The primers used for the expression profiles were the same as those described in the next section. A SMART RACE cDNA Amplification Kit (Clontech) was used for 5′-RACE, following the manufacturer's protocol. RNA was prepared from B6 day 16 embryos, and the gene-specific primer used was GSP: 5′-GTGAGAGGGGCTTCACTCCCCTG-3′. The amplified DNA fragments were purified and sequenced directly.
Allelic Analysis of Gene Expression Combined With Hot-Stop PCR
DNA polymorphisms in six genes between JF1 and C57BL/6 were detected in restriction fragment length polymorphism (RFLP) and single site polymorphism (SSP) analyses. For RT-PCR, 10 ng of cDNA in a 100-μL reaction mixture containing 1× ExTaq buffer (TaKaRa), 2.5 mM dNTP mixture, primers, and 2.5 U of ExTaq (TaKaRa) was subjected to 30–35 PCR cycles. In the case of Pon3, we performed nested PCR and added 10 PCR cycles in the second amplification. PCR was carried out on a Perkin Elmer GeneAmp PCR system 2400 under the following cycle conditions: 96°C for 15 sec, 65°C for 30 sec, and 72°C for 30 sec. The final cycle of PCR was performed in the presence of a primer labeled with [γ-32P] ATP. The PCR product was digested with the appropriate restriction endonuclease and electrophoresed on a 10% polyacrylamide gel. The intensity of the PCR products was measured with a BAS2000 Bioimaging Analyzer (Fuji Film). Experiments were carried out three times and average ratios were shown. Errors in observed values were within 5% in all cases except J × B yolksac (day 13) of Cas1. The following primers were used for DNA amplification: Cas1, 5′-AGCAGAGTGTAACGAACTCCAC-3′ and 5′-CACAGTGGACGGGTGAATGTGC-3′ *; Neurabin, 5′-ACTCTCCTGCCGAGGCTG-3′ and 5′-CAGTTTCAGGGGCTCTCACT-3′ *; Peg10, 5′-GGGTAGATAATCATAAGTATTTTGGGC-3′ and 5′-CAACATTCTAAACTTTATTCCAGCAAC-3′; Pon1, 5′-ACAAGAACCATCGGTCTTCC-3′ * and 5′-CCTTCTGCTACCACCTGGAC-3′; Pon2, 5′-ACGAGCTCCTTCCAAGTGTG-3′ and 5′-ACCTCTGATGCAGGAGGATG-3′ *; Pon3 (the first amplificaton), 5′-TCAGAAGTACTACGCATCCAGG-3′ and5′-CATGGCTGAAGGTAACTGTCC-3′; Pon3 (the second amplification), 5′-GAACAACGGCTCTGTGCTTC-3′ * and 5′-ATGCACCAAGCTAGCTGATG-3′. Asterisks indicate labeled primers with [γ-32P] ATP used in the last PCR cycles.
For RFLP analysis of Cas1, Neurabin, Pon1, Pon2, and Pon3, the PCR products were digested with DdeI, AciI, MspI, HaeIII, and PleI. For SSP analysis of Peg10, the PCR product was sequenced on an ABI 3100 sequencer using Big-Dye terminator chemistry (Applied Biosystems).
Methylation Analyses of Embryos, Placentas, Eggs, and Sperm
Genomic DNA and RNA were isolated from both day 10 embryos and placentas, as well as eggs and sperm, using ISOGEN, as described in the RT-PCR section. Purified genomic DNA was treated with a sodium bisulfite solution, as described previously (Raizis et al. 1995). During this process, cytosine was converted to uracil, except methylated cytosine. The sodium bisulfite-treated DNA was amplified with following the primers: Peg10-Sgce CGI, 5′-GTAAAGTGATTGGTTTTGTATTTTTAAGTG-3′ and 5′-TTAATTACTCTCCTACAACTTTCCAAATT-3′; Cas1 CGI, 5′-GTTTAGGTAGTTGTTAGTTTATTTGGGTATAG-3′ and 5′-CCTCCCTAATAACCTCCTACCTTAATAAC-3′; Neurabin CGI, 5′-GGTGTTTTTTGGTATTAGGTTAGATTG-3′ and 5′-ATAAACACCCTCCCCTCTCC-3′; Pon2 CGI, 5′-AGTGTTTAGGTTTTGGTGGAAGTG-3′ and 5′-CCCAAACTTAACTAAATTAAAAAAACTCC-3′.
The DNA fragments were amplified using ExTaq (TaKaRa) for 35–40 cycles under the following cycle conditions: 96°C for 15 sec, 60°C for 30 sec, and 72°C for 1 min. The amplified fragments were cloned into plasmids and sequenced. DNA polymorphisms in Peg10 promoter region (T/A; B6/JF1;1453020 bp; NW_000272) and DNA polymorphisms in Cas1 promoter region (A/G; B6/JF1;1304769 bp; NW_000272) and in Pon2 promoter region (T/G; B6/JF1; 2009669 bp; NW_000272) were used to determine paternal and maternal alleles of Cas1 CGI and Pon2 CGI, respectively.
Northern Blot Analysis
To analyze Peg10 and Neurabin expression, we used membranes of numerous adult tissues in Northern blot (Seegene, Korea) experiments. The 3′ part of Peg10 was amplified with primers 5′-GGGTAGATAATCATAAGTATTTTGGGC-3′ and 5′-CAACATTCTAAACTTTATTCCAGCAAC-3′ and Neurabin was amplified with primers 5′-ACTCTCCTGCCGAGGCTG-3′ and 5′-CAGTTTCAGGGGCTCTCACT-3′, and used as a DNA probe. Hybridization was performed at 42°C in Ultrasensitive Hybridization Buffer (Ambion) for 18 h. The membrane was then washed with a solution containing SSC and 0.1% SDS at 42°C to a final stringency of 0.1 × SSC.
Acknowledgments
This workwas supported by grants from CREST, the research program of the Japan Science and Technology Cooperation (JST), Asahi Glass Foundation, Uehara Memorial Life Science Foundation and the Ministry of Health, Labour for Child Health and Development (14-C) to F.I.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC Section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.906803.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to DDBJ under accession nos. AB091827–AB091829.]
References
- Beechey, C.V. 2000. Peg1/Mest locates distal to the currently defined imprinting region on mouse proximal chromosome 6 and identifies a new imprinting region affecting growth. Cytogenet. Cell Genet. 90: 309-314. [DOI] [PubMed] [Google Scholar]
- Clark, L., Wei, M., Cattoretti, G., Mendelsohn, C., and Tycko, B. 2002. The Tnfrh1 (Tnfrsf23) gene is weakly imprinted in several organs and expressed at the trophoblast-decidua interface. BMC Genet. 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, H.G., Richter, R.J., Keifer, M., Broomfield, C.A., Sowalla, J., and Furlong, C.E. 1996. The effect of the human serum paraoxonase polymorphism is reversed with diazoxon, soman and sarin. Nat. Genet. 14: 334-336. [DOI] [PubMed] [Google Scholar]
- Draganov, D.I., Stetson, P.L., Watson, C.E., Billecke, S.S., and La Du, B.N. 2000. Rabbit serum paraoxonase 3 (PON3) is a high-density lipoprotein-associated lactonase and protects low-density lipoprotein against oxidation. J. Biol. Chem. 275: 33435-33442. [DOI] [PubMed] [Google Scholar]
- Hikichi, T., Kohda, T., Kaneko-Ishino, T., and Ishino, F. 2003. Imprinting regulation of the murine Meg1/Grb10 and human GRB10 genes; roles of brain-specific promoters and mouse-specific CTCF-binding sites. Nucleic. Acids Res. 31: 1398-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janbon, G., Himmelreich, U., Moyrand, F., Improvisi, L., and Dromer, F. 2001. Cas1p is a membrane protein necessary for the O-acetylation of the Cryptococcus neoformans capsular polysaccharide. Mol. Microbiol. 42: 453-467. [DOI] [PubMed] [Google Scholar]
- Kaneko-Ishino, T., Kuroiwa, Y., Miyoshi, N., Kohda, T., Suzuki, R., Yokoyama, M., Viville, S., Barton, S.C., Ishino, F., and Surani, M.A. 1995. Peg1/Mest imprinted gene on chromosome 6 identified by cDNA subtraction hybridization. Nat. Genet. 11: 52-59. [DOI] [PubMed] [Google Scholar]
- Kobayashi, S., Vemura, T., Kohda, T., Nagai, T., Chinen, Y., Naritomi, K., Kinoshita, E.I., Ohashi, H., Imaizumi, K., Tsukahara, M., et al. 2001. No evidence of PEG1/MEST gene mutations in Silver-Russell syndrome patients. Am. J. Med. Genet. 104: 225-231. [PubMed] [Google Scholar]
- Kosaki, K., Kosaki, R., Craigen, W.J., and Matsuo, N. 2000. Isoform-specific imprinting of the human PEG1/MEST gene. Am. J. Hum. Genet. 66: 309-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre, L., Viville, S., Barton, S.C., Ishino, F., Keverne, E.B., and Surani, M.A. 1998. Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nat. Genet. 20: 163-169. [DOI] [PubMed] [Google Scholar]
- Li, T., Vu, T.H., Zeng, Z.L., Nguyen, B.T., Hayward, B.E., Bonthron, D.T., Hu, J.F., and Hoffman, A.R. 2000. Tissue-specific expression of antisense and sense transcripts at the imprinted Gnas locus. Genomics 69: 295-304. [DOI] [PubMed] [Google Scholar]
- McNally, E.M., Ly, C.T., and Kunkel, L.M. 1998. Human ε-sarcoglycan is highly related to α-sarcoglycan (adhalin), the limb girdle muscular dystrophy 2D gene. FEBS Lett. 422: 27-32. [DOI] [PubMed] [Google Scholar]
- Mizuno, Y., Sotomaru, Y., Katsuzawa, Y., Kono, T., Meguro, M., Oshimura, M., Kawai, J., Tomaru, Y., Kiyosawa, H., Nikaido, I., et al. 2002. Asb4, Ata3, and Dcn are novel imprinted genes identified by high-throughput screening using RIKEN cDNA microarray. Biochem. Biophys. Res. Commun. 290: 1499-1505. [DOI] [PubMed] [Google Scholar]
- Nakanishi, H., Obaishi, H., Satoh, A., Wada, M., Mandai, K., Satoh, K., Nishioka, H., Matsuura, Y., Mizoguchi, A., and Takai, Y. 1997. Neurabin: A novel neural tissue-specific actin filament-binding protein involved in neurite formation. J. Cell Biol. 139: 951-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, C.J., Wadleigh, D.J., Gangopadhyay, A., Hama, S., Grijalva, V.R., Navab, M., Fogelman, A.M., and Reddy, S.T. 2001. Paraoxonase-2 is a ubiquitously expressed protein with antioxidant properties and is capable of preventing cell-mediated oxidative modification of low-density lipoprotein. J. Biol. Chem. 276: 44444-44449. [DOI] [PubMed] [Google Scholar]
- Ono, R., Kobayashi, S., Wagatsuma, H., Aisaka, K., Kohda, T., Kaneko-Ishino, T., and Ishino, F. 2001. A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics 73: 232-237. [DOI] [PubMed] [Google Scholar]
- Peters, J., Wroe, S.F., Wells, C.A., Miller, H.J., Bodle, D., Beechey, C.V., Williamson, C.M., and Kelsey, G. 1999. A cluster of oppositely imprinted transcripts at the Gnas locus in the distal imprinting region of mouse chromosome 2. Proc. Natl. Acad. Sci. 96: 3830-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piras, G., El Kharroubi, A., Kozlov, S., Escalante-Alcalde, D., Hernandez, L., Copeland, N.G., Gilbert, D.J., Jenkins, N.A., and Stewart, C.L. 2000. Zac1 (Lot1), a potential tumor suppressor gene, and the gene for ε-sarcoglycan are maternally imprinted genes: Identification by a subtractive screen of novel uniparental fibroblast lines. Mol. Cell. Biol. 20: 3308-3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulter, R. and Butler, M. 1998. A retrotransposon family from the pufferfish (fugu) Fugu rubripes. Gene 215: 241-249. [DOI] [PubMed] [Google Scholar]
- Preece, M.A., Price, S.M., Davies, V., Clough, L., Stanier, P., Trembath, R.C., and Moore, G.E. 1997. Maternal uniparental disomy 7 in Silver-Russell syndrome. J. Med. Genet. 34: 6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primo-Parmo, S.L., Sorenson, R.C., Teiber, J., and La Du, B.N. 1996. The human serum paraoxonase/arylesterase gene (PON1) is one member of a multigene family. Genomics 33: 498-507. [DOI] [PubMed] [Google Scholar]
- Raizis, A.M., Schmitt, F. and Jost, J.P. 1995. A bisulfite method of 5-methylcytosine mapping that minimizes template degradation. Anal. Biochem. 226: 161-166. [DOI] [PubMed] [Google Scholar]
- Shih, D.M., Gu, L., Xia, Y.R., Navab, M., Li, W.F., Hama, S., Castellani, L.W., Furlong, C.E., Costa, L.G., Fogelman, A.M., et al. 1998. Mice lacking serum paraoxonase are susceptible to organophosphate toxicity and atherosclerosis. Nature 394: 284-287. [DOI] [PubMed] [Google Scholar]
- Song, Z., Casolaro, V., Chen, R., Georas, S.N., Monos, D., and Ono, S.J. 1996. Polymorphic nucleotides within the human IL-4 promoter that mediate overexpression of the gene. J. Immunol. 156: 424-429. [PubMed] [Google Scholar]
- Tsai, C., Lin, S., Ito, M., Takagi, N., Takada, S., and Ferguson-Smith, A. 2002. Genomic imprinting contributes to thyroid hormone metabolism in the mouse embryo. Curr. Biol. 12: 1221. [DOI] [PubMed] [Google Scholar]
- Uejima, H., Lee, M.P., Cui, H., and Feinberg, A.P. 2000. Hot-stop PCR: A simple and general assay for linear quantitation of allele ratios. Nat. Genet. 25: 375-476. [DOI] [PubMed] [Google Scholar]
- Vu, T.H. and Hoffman, A.R. 1994. Promoter-specific imprinting of the human insulin-like growthfactor-II gene. Nature 371: 714-717. [DOI] [PubMed] [Google Scholar]
- Wilson, A.G., Symons, J.A., McDowell, T.L., McDevitt, H.O., and Duff, G.W. 1997. Effects of a polymorphism in the human tumor necrosis factor α promoter on transcriptional activation. Proc. Natl. Acad. Sci. 94: 3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich, A., Grabowski, M., Asmus, F., Naumann, M., Berg, D., Bertram, M., Scheidtmann, K., Kern, P., Winkelmann, J., Muller-Myhsok, B., et al. 2001. Mutations in the gene encoding ε-sarcoglycan cause myoclonus-dystonia syndrome. Nat. Genet. 29: 66-69. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.ebi.ac.uk/emboss/cpgplot/; CpG Island finder and plotting tool.
- http://www.mgu.har.mrc.ac.uk/imprinting/imprinting.html; Mouse imprinting maps and data.