Abstract
Description of the molecular phenotypes of pathobiological processes in vivo is a pressing need in genomic biology. We have implemented a high-throughput real-time PCR strategy to establish quantitative expression profiles of a customized set of target genes. It enables rapid, reproducible data acquisition from limited quantities of RNA, permitting serial sampling of mouse blood during disease progression. We developed an easy to use statistical algorithm—Global Pattern Recognition—to readily identify genes whose expression has changed significantly from healthy baseline profiles. This approach provides unique molecular signatures for rheumatoid arthritis, systemic lupus erythematosus, and graft versus host disease, and can also be applied to defining the molecular phenotype of a variety of other normal and pathological processes.
Expression profiling promises to provide insight into normal biological and pathological processes (Alizadeh et al. 2000; Shaffer et al. 2001; van't Veer et al. 2002). The hope is that knowledge obtained from gene expression patterns will predict disease outcome or indicate individualized courses of therapy. The two technologies that have emerged as the most promising gene expression tools are hybridization-based microarrays and quantitative real-time RT-PCR (QPCR)analysis (Duggan et al. 1999; Lockhart and Winzeler 2000; Giulietti et al. 2001; Green et al. 2001). Microarrays permit the simultaneous analysis of a large number of genes, but extensive replicate sampling can be labor-intensive. Additionally, samples with limiting RNA (such as mouse peripheral blood or laser-capture microdissection samples)can only be used following cDNA amplification (Wang et al. 2000), which adds another processing step that could introduce bias. This makes longitudinal microarray analysis of peripheral blood samples from an experimental cohort technically challenging.
QPCR platforms using gene-specific primers provide highly sensitive and reproducible expression quantification from small amounts of starting material (Gibson et al. 1996; Heid et al. 1996; Schmittgen et al. 2000), but have been limited in the number of genes analyzed practically. Therefore, we combined the multiple gene approach of microarrays with the sensitivity of QPCR to produce a high-throughput customized “ImmunoQuantArray” (IQA), the first generation of which consists of 96 gene-specific QPCRs designed to monitor genes associated with immune processes.
QPCR instruments monitor gene-specific amplicons with fluorescent dye chemistry. The amplification curves typically have a sigmoidal shape in which the exponential amplification phase reveals the number of PCR cycles required to achieve a certain fluorescence intensity. A cycle threshold or Ct value for each reaction is the number of cycles at which the reaction crosses the fluorescence threshold. The fewer cycles required to reach a certain fluorescence intensity, the lower the Ct value and the greater the initial amount of input target cDNA. Genes that do not amplify during the 40-cycle PCR are considered “off” and are given a Ct value of 40 (Heid et al. 1996).
QPCR data are usually interpreted as fold changes in gene expression. Changes in gene expression are derived by normalizing the expression of a gene to that of an appropriate “housekeeping” gene (assumed to be invariant; Livak and Schmittgen 2001). This relative normalization procedure is presently regarded to be the only practical option available for interpreting QPCR data. An alternative, accurate quantification of input RNA/cDNA is challenging when input RNA is of limiting quantities and impractical for scale-up (Morrison et al. 1998).
To more reliably evaluate expression changes in QPCR data, we developed a novel statistical algorithm—Global Pattern Recognition (GPR)—to reveal significant changes in gene expression patterns. Inspired by triangulation techniques to determine positional information in cartography and astronomy, GPR goes through several iterations to compare the change of expression of a gene normalized to every other gene in the IQA. By comparing the expression of each gene to every other gene in the array, a global pattern is established, and significant changes are identified and ranked. Importantly, GPR takes advantage of biological replicates to extract significant changes in gene expression, thus providing a novel alternative to the use of relative normalization in QPCR experiments.
We show here that the IQA/GPR approach is a reliable analytical tool that can be used to establish immunological gene expression profiles of normal mice and in mice developing rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), and graft versus host disease (GVHD). Moreover, we show that one can obtain temporal expression profiles from unamplified blood cDNA samples from individual mice, thus making it possible to establish the relationship between gene expression pattern and individual disease severity.
RESULTS AND DISCUSSION
Design and Validation of the ImmunoQuantArray
We generated a first-generation QPCR-based IQA consisting of 96 PCR amplicons that survey, at the transcriptional level, genes associated with a broad spectrum of immunological processes. We therefore selected sentinel genes, whose altered expression correlates with innate or adaptive immune responses, T-cell-mediated (T helper 1 and T helper 2)responses, humorally mediated responses, and/or general inflammatory responses (Table 1; primer sequences are available online at www.genome.org). The SYBR Green detection system was used because it obviates the need for expensive gene-specific TaqMan probes.
Table 1.
Genes Included in the ImmunoQuantArray
| Gene | GenBank accession # | Name | ||
|---|---|---|---|---|
| Aopoptosis/antiapoptosis | ||||
| Bad | NM_007522 | Bcl-associated death promoter | ||
| Bax | NM_007527 | Bcl2-associated X protein | ||
| Bcl2 | NM_009741 | B-cell leukemia/lymphoma 2 | ||
| Bcl21 | NM_009743 | Bcl2-like [Bcl-x] | ||
| Bid | U75506 | BH3-interacting domain death agonist | ||
| Fadd | NM_010175 | Fas-associated via death domain | ||
| Cflar | U97076 | CASP8 AND FADD-like apoptosis regulator [FLIP(L)] domain death agonist | ||
| Tnfrsf6 | NM_007987 | Tumor necrosis factor receptor superfamily, member 6 [Fas] | ||
| Tnfsf6 | NM_010177 | Tumor necrosis factor (ligand) superfamily, member 6 [FasL] | ||
| Pfp | M23182 | Pore-forming protein [perforin] | ||
| Costimulatory/activation cell curface ligands | ||||
| Cd28 | NM_007642 | CD28 antigen | ||
| Cd44 | M27130 | CD44 antigen | ||
| Cd80 | AF065894 | CD80 antigen | ||
| Cd86 | NM_019388 | CD86 antigen | ||
| Icosl | AF216747 | ICOS-ligand | ||
| Mox2 | AF231126 | Antigen identified by monoclonal antibody MRC OX-2 | ||
| Leukocyte cell surface differentiation markers | ||||
| Cd4 | NM_013488 | CD4 antigen | ||
| Cd5 | NM_007650 | CD5 antigen | ||
| Cd34 | S69293 | CD34 antigen | ||
| Cd3e | M23376 | CD3 antigen, epsilon polypeptide | ||
| Cd8a | AJ131778 | CD8 antigen, α-chain | ||
| Cd8b | NM_009858 | CD8 antigen, β-chain | ||
| Ptprc | NM_011210 | Protein tyrosine phosphatase, receptor type, C | ||
| Art2b | AF016465 | ADP-ribosyltransferase 2b | ||
| Fc receptors | ||||
| Fcer1a | NM_010184 | Fc receptor, IgE, high affinity I, α polypeptide | ||
| Fcer1g | NM_010185 | Fc receptor, IgE, high affinity I, γ polypeptide | ||
| Fcgrt | NM_010189 | Fc receptor, IgG, α chain transporter | ||
| Cytokines | ||||
| Il1b | NM_008361 | Interleukin 1 β | ||
| Il2 | NM_008366 | Interleukin 2 | ||
| Il4 | NM_021283 | Interleukin 4 | ||
| Il5 | NM_010558 | Interleukin 5 | ||
| Il6 | M20572 | Interleukin 6 | ||
| Il7 | NM_008371 | Interleukin 7 | ||
| Il10 | NM_010548 | Interleukin 10 | ||
| Il12a | NM_008351 | Interleukin-12 [p35 subunit] | ||
| Il12b | NM_008352 | Interleukin-12 [p40 subunit] | ||
| Il15 | NM_008357 | Interleukin 15 | ||
| Il18 | NM_008360 | Interleukin 18 | ||
| Csf1 | NM_007778 | Colony-stimulating factor 1 (macrophage) | ||
| Csf3 | NM_009971 | Colony-stimulating factor 3 (granulocyte) | ||
| Tgfb1 | AJ009862 | Transforming growth factor, β 1 | ||
| Lta | NM_010735 | Lymphotoxin A | ||
| Ltb | NM_008518 | Lymphotoxin B | ||
| Ifna1 | NM_010502 | Interferon α family, gene 1 | ||
| Ifnb | NM_010510 | Interferon β, fibroblast | ||
| Ifng | K00083 | Interferon γ | ||
| Tnf | X02611 | Tumor necrosis factor | ||
| Tnfsf11 | AF013170 | Tumor necrosis factor (ligand) superfamily, member 11 | ||
| Tnfrsf1a | NM_011609 | Tumor necrosis factor receptor superfamily, member 1a | ||
| Tnfrsf1b | NM_011610 | Tumor necrosis factor receptor superfamily, member 1b | ||
| Tnfrsf11b | NM_008764 | Tumor necrosis factor receptor superfamily, member 11b [osteoprotegerin] | ||
| Cytokine receptors | ||||
| Il1r1 | NM_008362 | Interleukin 1 receptor, type I | ||
| Il2ra | NM_008367 | Interleukin 2 receptor, α chain | ||
| Il2rg | NM_013563 | Interleukin 2 receptor, γ chain | ||
| Il4ra | NM_010557 | Interleukin 4 receptor, α | ||
| Il7r | NM_008372 | Interleukin 7 receptor | ||
| Il10ra | NM_008348 | Interleukin 10 receptor, α | ||
| Il12rb2 | NM_008354 | Interleukin 12 receptor, β 2 | ||
| Ltbr | NM_010736 | Lymphotoxin B receptor | ||
| Ifngr2 | NM_008338 | Interferon γ receptor 2 | ||
| Leukocyte adhesion | ||||
| Sell | NM_011346 | Selectin, lymphocyte | ||
| Itgal | AF065901 | Integrin α L | ||
| Itgam | NM_008401 | Integrin α M | ||
| Itgax | NM_021334 | Integrin α X | ||
| Innate immune cell surface receptor/ligands | ||||
| Tlr2 | AF165189 | Toll-like receptor 2 | ||
| Tlr4 | AF185285 | Toll-like receptor 4 | ||
| Klrb1d | AF338322 | Killer cell lectin-like receptor subfamily B member 1D | ||
| Cd1d1 | M63695 | CD1d1 antigen | ||
| Immune activation/signal transduction | ||||
| Zap70 | NM_009539 | ζ-chain (TCR) associated protein kinase (70 kD) | ||
| Cd3z | U17267 | T cell receptor-ζ chain | ||
| Il1rak | NM_008363 | Interleukin 1 receptor-associated kinase | ||
| C2ta | NM_007575 | Class II transactivator | ||
| Btk | NM_013482 | Bruton agammaglobulinemia tyrosine kinase | ||
| Jun | NM_010591 | Jun oncogene | ||
| Fyn | NM_008054 | Fyn proto-oncogene | ||
| Lck | M12056 | Lymphocyte protein tyrosine kinase | ||
| Map2k1 | NM_008927 | Mitogen-activated protein kinase 1 [MEK1] | ||
| Map2k2 | NM_023138 | Mitogen-activated protein kinase 2 [MEK2] | ||
| Nfkb1 | NM_008689 | Nuclear factor of κ light chain enhancer in B-cells 1, p105 | ||
| Hcph | NM_013545 | Hemopoietic cell phosphatase | ||
| Myd88 | NM_010851 | Myeloid differentiation primary response gene 88 | ||
| Csk | NM_007783 | C-src tyrosine kinase | ||
| Chemokines and chemokine receptors | ||||
| Scyc1 | U15607 | Small inducible cytokine subfamily C, member [lymphotactin] | ||
| Lcp2 | NM_010696 | Lymphocyte cytosolic protein 2 | ||
| Scya19 | Af307988 | Small inducible cytokine A19 | ||
| Scya20 | NM 016960 | Small inducible cytokine subfamily A20 | ||
| Scyd1 | NM_009142 | Small inducible cytokine subfamily D, 1 | ||
| Ccxcr1 | NM_011798 | Chemokine (C motif) XC receptor 1 | ||
| Stress response | ||||
| Tra1 | NM_011631 | Tumor rejection antigen gp96 | ||
| Hsp70-2 | NM_008301 | Heat shock protein, 70 kDa 2 | ||
| Hsp70-1 | M35021 | Heat shock protein, 70 kDa 1 | ||
| Other | ||||
| Rn18s | X00686 | 18S RNA [ribosomal] | ||
| Hprt | NM_013556 | Hypoxanthine guanine phosphoribosyl transferase | ||
| Gapd | NM_008084 | Glyceraldehyde-3-phosphate dehydrogenase | ||
| Tbp | NM_013684 | TATA-box-binding protein | ||
| Zfp106 | AF060246 | Zinc finger protein 106 | ||
| Rag1 | NM_009019 | Recombination activating gene 1 | ||
| Terc | AF047387 | Telomerase RNA component | ||
| Tert | NM_009354 | Telomerase reverse transcriptase | ||
| Nos2 | NM_010927 | Nitric oxide synthase 2, inducible, macrophage | ||
Primer sequences available in Supplemental Material; available online at http://www.genome.org
Reproducibility and Sensitivity of the IQA
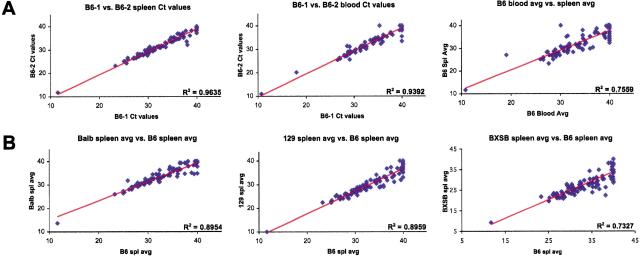
We tested the ability of this system to generate reproducible data. cDNAs derived from the samples being compared were analyzed using the IQA, and the raw cycle threshold (Ct)values of each amplicon represented as a scatterplot. The farther a gene deviates from the linear regression best-fit line, the greater the difference in its level of expression between the two samples being compared. Figure 1A is a representative experiment comparing “biological replicate” cDNAs from two 8-week-old C57BL/6J males from splenocytes (first panel)and whole blood (second panel). This strong correlation was maintained whenever two similar samples (e.g., age-, sex-, and strain-matched spleen cDNAs from two animals)were compared (data not shown), indicating that variability between genetically identical biological replicates is low. However, the correlation breaks down when comparing splenocyte with blood Ct values, emphasizing the contribution of tissue source to the gene expression pattern (Fig. 1A, third panel). RNA degradation and inconsistency of the reverse transcription reaction were not significant issues when samples were collected and processed quickly and uniformly (see Methods). These results demonstrated that the IQA system is capable of generating reproducible data between biological replicates not only from mouse spleen but also from microsamples (75 μL)of blood.
Figure 1.
Reproducibility and sensitivity of the ImmunoQuantArray. Raw Ct values for each of the 96 genes in the IQA are plotted. The linear regression best-fit line is shown and its correlation coefficient indicated. (A) Spleen cDNAs (left panel) or blood cDNAs (middle panel) from two C57BL/6J males. The average of the two spleen cDNA Cts is compared with the average two blood cDNA Cts (right panel). (B) The average spleen cDNA Cts of five BALB/cJ (left), three 129X1/SvJ (middle), or five BXSB/MpJ-Yaa+ (right) males was compared with the average spleen cDNA Cts of five C57BL/6J males. The point lying closest to the ordinate is 18S rRNA.
The influence of background genetics on the molecular phenotypes was also tested. Using splenocyte cDNA from 8-week-old male animals, the average expression level (average Ct values)of cDNAs prepared from five C57BL/6J animals was compared with the average from five BALB/cJ, three 129X1/SvJ, and five BXSB/MpJ animals (Fig. 1B). The departure of the correlation coefficient from unity indicated that genetic background altered the gene expression patterns.
Rationale for Implementing the GPR Algorithm
Although scatterplot analysis (Fig. 1)provided some insight into gene expression patterns, it was restricted to one-by-one gene comparisons, and would only be expected to provide reliable data after accurate quantification of input RNA/cDNA. Biological replicate comparison of two cohorts of animals could be performed by plotting the average Ct values for each gene (as shown in Fig. 1B), but it was a suboptimal method for identifying significant changes in gene expression between two experimental groups.
The common mode of comparative analysis for QPCR data is the use of a single normalizer gene with which the expression of all genes is compared. This mode of analysis is greatly complicated by the fact that housekeeping genes commonly used as normalizers (e.g., GAPDH, β-actin, and HPRT) themselves can change in expression when comparing tissues or cells in different states of activation (Bustin 2000; Schmittgen et al. 2000; Goidin et al. 2001; Hamalainen et al. 2001). 18S rRNA is another normalizer that intuitively and experimentally seems more stable, but for unknown reasons, even 18S can vary in comparison to other genes when analyzed by sensitive QPCR techniques (e.g., Fig. 1B, left panel; Bustin 2000). Any small variation in the normalizer amplification would therefore compromise the analysis of the complete QPCR data set.
Ideally, we wanted to compare the expression profiles of experimental groups of animals with those of the control groups such that the comparison was not contingent on the expression stability of a single normalizer gene. Using the expression data from the 96 genes in the IQA as a foundation, we developed the GPR algorithm to discern statistically significant changes in gene expression. After filtering the data (see Methods), GPR normalizes each eligible gene against every other gene that is eligible as a normalizer, thus eliminating the reliance on single-gene normalization. Conceptually, GPR resembles standard ANOVA techniques but differs in its implementation (Kerr and Churchill 2001). We initially applied ANOVA techniques to our QPCR data sets (data not shown). In ANOVA, to normalize sample-to-sample variability, the average Ct value of the 96 genes for each sample is subtracted from each gene's Ct value. However, PCR dropouts or genes that are “off” (with a Ct of 40)are necessarily included in this average, adversely skewing the entire data set. Because GPR considers each gene individually and filters out such null data, it is not adversely affected by PCR dropouts as is ANOVA. In a typical experiment, a 96-well IQA QPCR is run for each of up to 10 samples—five control and five experimental biological replicates. GPR then uses a T-test to evaluate gene expression between control and experimental group biological replicates on a gene-by-gene basis. Because GPR ranks genes based on biological replicate consistency, those genes whose expression differs significantly when comparing control and experimental cohorts will be identified regardless of whether the changes are large or small.
Validation of the GPR Algorithm by Bootstrap Analysis
We used bootstrap analysis (Efron and Tibshirani 1998)to evaluate the reliability of GPR to detect nonrandom changes in gene expression. After using GPR to analyze a set of IQA results, we shuffled the data on a gene-by-gene basis for 250 iterations and analyzed the randomized data set with GPR after each shuffling. This random resampling generated a bootstrap probability distribution of GPR scores. The GPR scores obtained by analyzing the experimental data (observed scores)were tested to see if they could have arisen simply by chance in a randomized data set. If the observed GPR score did not appear once in 250 shufflings of the data set, the probability of that particular gene having significantly changed by chance alone is less than 1/250, or p < 0.004.
The KRN T-cell receptor transgenic strain, when bred to NOD/Lt, produces transgene-bearing F1 mice that develop a severe autoimmune disorder with distinct similarities to RA (Kouskoff et al. 1996; Korganow et al. 1999). Table 2A lists the 12 top-ranked genes identified by GPR and associated bootstrap analysis when comparing blood cDNAs from transgenic (KRNxNOD) F1 and nontransgenic control littermate cohorts. The highest GPR scores were also highly significant (p ≤ 0.004)when compared with the bootstrap scores generated by randomly resampling the data set.
Table 2.
GPR Analysis and Validation by the Bootstrap Method
| A. (KRNxNOD)F1 transgenic animals compared with healthy nontransgenic littermatesa | ||||||||
|---|---|---|---|---|---|---|---|---|
| Gene | GPR score | 99.5th %tile | p-value | Fold change | ||||
| Cd4 | 0.841 | 0.827 | 0.004 | 0.12 | ||||
| Il7r | 0.773 | 0.447 | <0.004 | 0.29 | ||||
| Zap70 | 0.750 | 0.408 | <0.004 | 0.25 | ||||
| Fcer1g | 0.636 | 0.371 | <0.004 | 2.90 | ||||
| Art2b | 0.636 | 0.363 | <0.004 | 0.36 | ||||
| Cd3e | 0.614 | 0.442 | <0.004 | 0.25 | ||||
| Cd5 | 0.614 | 0.409 | <0.004 | 0.25 | ||||
| Bid | 0.523 | 0.385 | <0.004 | 2.46 | ||||
| Ltb | 0.523 | 0.251 | <0.004 | 0.57 | ||||
| Tnfsf6 | 0.432 | 0.244 | <0.004 | 0.48 | ||||
| Myd88 | 0.409 | 0.341 | <0.004 | 1.98 | ||||
| CD8a | 0.386 | 0.203 | <0.004 | 0.62 | ||||
| B. C57BL/6J malesb | ||||||||
| Tnf | 0.489 | 0.135 | <0.004 | 0.33 | ||||
| Csk | 0.149 | 0.246 | 0.064 | 1.96 | ||||
| Fcgrt | 0.149 | 0.403 | 0.100 | 3.11 | ||||
| Fcer1g | 0.128 | 0.185 | 0.044 | 0.82 | ||||
| Tnfrsf1a | 0.128 | 0.093 | <0.004 | 0.63 | ||||
| Tnfrsf1b | 0.128 | 0.101 | <0.004 | 0.86 | ||||
| Ly55d | 0.128 | 0.236 | 0.084 | 2.13 | ||||
| Il4ra | 0.106 | 0.136 | 0.028 | 0.78 | ||||
| Itgam | 0.106 | 0.110 | 0.012 | 0.99 | ||||
| Ltb | 0.106 | 0.226 | 0.096 | 1.89 | ||||
| Il10ra | 0.085 | 0.290 | 0.084 | 2.06 | ||||
| Ptprc | 0.085 | 0.203 | 0.096 | 2.98 | ||||
The top 12 genes from the GPR output are shown. GPR parameters were Cycle cutoff = 37.5 and threshold p-value = 0.05. The 99.5th percentile score from the bootstrap distribution (250 random resamplings) is shown in the third column. The p-value (fourth column) was compared by dividing the number of scores in the bootstrap probability distribution that were higher than the observed GPR score by the number of random resamplings in the bootstrap analysis (250). The expression fold change (fifth change) was derived after normalization to 18S rRNA. Although GPR ranks genes according to the statistical significance of their change in expression, it does not provide a fold change in gene expression. Thus, following GPR analysis, we computed the fold change normalized to 18S rRNA to gain a sense for the direction and magnitude of change of the highest ranking genes
Blood cDNAs from five 8-week-old (KRNxNOD)F1 transgenic animals were compared with five healthy nontransgenic littermates. The IQA gene set was identical to that shown in Table 1, except Fcer1a, Il1r1, Btk, and Scyd1 were replaced by Tbp, Art2b, Hprt, and Gapd, respectively
Triplicate blood cDNAs from two 8-week-old C57BL/6J males were compared on the IQA (genes tested as in Table 1)
As a negative control, we then subjected an IQA data set with minimal expected expression differences to similar bootstrap analysis. Table 2B compares GPR results derived from three consecutive bleeds of one C57BL/6J mouse compared with consecutive bleeds of another C57BL/6J mouse. In contrast to the (KRNxNOD) F1 blood data, 96-gene GPR analysis of the C57BL/6J blood cDNAs yielded only a single difference (Tnf, GPR score 0.489, indicating that this gene was significantly different from 49% of the eligible normalizer genes). In analyzing more than 50 IQA data sets, we observed that genes with GPR scores falling below 0.4 lose reliability regarding their change in expression because the values are based on too few normalizers. Because the bootstrap distribution was generated by randomizing of the GPR scores, genes falling well below 0.4, such as Tnfrsf1a and Tnfrsf1b in Table 2B, occasionally appear as significant. Typically, these genes have a very low level of expression (i.e., Ct values close to 40)and/or are statistical noise. However, as shown in Table 2B, genes with GPR scores ≥0.4 are always highly significant by bootstrap analysis. Taken together, the results indicate that the GPR can reliably identify genes with expression changes between biological replicates in control and experimental cohorts.
Molecular Phenotype of the (KRNxNOD) F1 Model for RA
Blood samples from (KRNxNOD) F1 transgenic animals showed reduced levels of the T-cell-specific genes Cd4, Cd3e, Cd5, and Zap70 expression compared with nontransgenic littermates (Table 2A). This result is consistent with the fact that adult transgenic animals have lower numbers of CD4 T-cells compared with nontransgenic littermates (Kouskoff et al. 1996). Up-regulation of the antibody Fc receptor common γ-chain (Fcer1g)used by the inflammatory Fc receptors FcγRI and FcγRIII also correlates with the presence of the transgene and disease. Notably, these proinflammatory Fc receptors are required to precipitate disease in the (KRNxNOD) F1 serum transfer model (Ji et al. 2002). Other genes reported as significantly changed in Table 2A are interesting candidates for further study. Analysis of other lymphoid tissues and longitudinal peripheral blood analysis of these mice may identify other genes transcriptionally activated/repressed at specific stages of disease progression. The results show that IQA/GPR analysis from blood samples can reveal expression alterations in genes consistent with the progression of autoimmune arthritis in the (KRNxNOD) F1 model.
Serial Molecular Phenotyping of BXSB-Yaa SLE
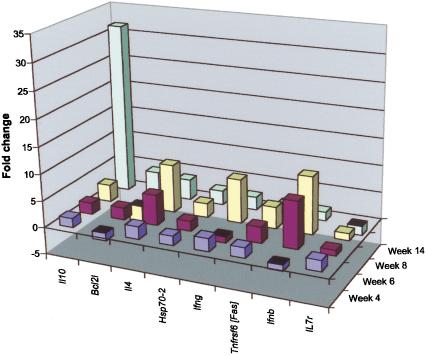
SLE is a heterogeneous disease syndrome with common features of B- and T-cell activation leading to the elaboration of pathogenic autoantibodies. The BXSB/MpJ strain develops a chronic form of SLE that is severely aggravated in males carrying the SB allele at the Y-linked autoimmune accelerator (Yaa) locus (Murphy and Roths 1979). Using BXSB male mice carrying a wild-type Y-chromosome as controls, we examined splenocyte cDNA samples from cohorts of BXSB-Yaa males over a 14-wk time course (Fig. 2). Most notably, Il10 expression increases substantially at week 14, a time at which the disease first becomes evident. Increased IL10 production is strongly associated with SLE in both humans and mouse SLE models (Grondal et al. 2000; Moore et al. 2001). Expression of Il4, Ifnb, Ifng, and Tnfrsf6 (Fas), all of which have been associated with SLE (Nousari et al. 1998; Wong et al. 2000; Bijl et al. 2001; Theofilopoulos et al. 2001), are up-regulated prior to 14 wk, but with some oscillation. Because the animals in the two groups were genetically matched except for the Yaa locus, these data indicate a molecular phenotype of the Yaa-driven acceleration of the BXSB SLE disease model.
Figure 2.
Serial molecular phenotyping of BXSB-Yaa SLE. Spleen cDNAs from cohorts of three to five BXSB/MpJ-Yaa males and age-matched BXSB.B6-Yaa+ controls were subjected to IMQ/GPR analysis at weeks 4, 6, 8, and 14. The fold changes (normalized to 18S rRNA) of genes that received a GPR score ≥0.4 and were significant after normalization to 18S rRNA are plotted.
Molecular Phenotype of Mice Undergoing GVHD
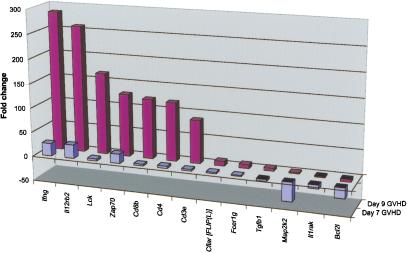
GVHD is a prototypic T-cell-mediated disease in which donor CD4 and CD8 T-cells respond to host alloantigens, proliferate, attack, and destroy multiple host organs, and undergo apoptotic cell death. C57BL/6J-derived bone marrow and spleen cells cause acute GVHD when transferred into lethally irradiated allogenic male 129P3/J recipients (Korngold and Sprent 1983). To understand the transcriptional changes associated with GVHD, we sampled and analyzed peripheral blood leukocytes from mice undergoing acute GVHD and compared them with the same source of leukocytes transferred into irradiated syngeneic C57BL/6J mice. Figure 3 depicts the fold changes of genes identified as significant by GPR analysis. Of the 96 genes analyzed, markers of T-cell activation Lck, Zap70, Cd4, Cd8, and Cd3 were strongly up-regulated, as was the key acute GVHD cytokine Ifng (Ferrara 2000)and the receptor IL-12Rb1, which regulates IFN-γ production (Losana et al. 2002). The cell cycle control gene Map2k2 and the apoptotic/antiapoptotic gene Bcl2l were down-regulated as a consequence of allogenic cell transfer (Fig. 3). These results highlight the amount of coherent information that can be obtained from serial blood analysis.
Figure 3.
Molecular phenotype of GVHD progression. Bone marrow and splenocytes from C57BL/6J females were used to induce GVHD in five allogenic 129P3/J males (C57BL6/J → 129P3/J). Five syngeneic transplant recipients (C57BL6/J → C57BL6/J) were used as controls. cDNAs were prepared from samples collected on days 7 and 9 after the transplant and were subjected to IMQ/GPR analysis. The fold changes of genes that received a GPR score ≥0.4 and were significant after normalization to 18S rRNA are shown.
Although the approach outlined here is a logical method for confirmation and accurate quantification of genes whose expression appears to have changed based on microarray analyses, a more general application is as a routine analytical tool to perform high-throughput quantitative expression analysis of a customized gene set. Major strengths of this approach include the sensitivity of QPCR techniques to accurately assess the expression of a customized gene set from limited RNA sources (e.g., mouse peripheral blood), the exploitation of multiple biological replicates to extract significant expression changes, and the obviation of the need for single-gene-based normalization. Significant expression changes were evident even though the blood and spleen samples analyzed were comprised of heterogeneous cell types. Despite the fact that the expression changes observed could thus be a consequence of variation of cell types and/or changes in expression level on a per cell basis, the varied gene expression patterns observed were consistent with the pathological processes analyzed. Longitudinal analysis from limiting biological samples is not yet practical with microarrays without amplification of the cDNA (Wang et al. 2000). However, the limited sample needed for QPCR allows serial sampling of individual mice to arrive at molecular profiles that predict disease onset and severity. Importantly, the IQA requires no sophisticated equipment other than quantitative PCR equipment and a Microsoft Excel-capable computer. The platform is flexible such that genes can be added or subtracted from the set according to the needs of the investigator, and can readily be expanded to a 384-well format. Finally, although we have applied the system to probing the molecular signatures of immunological diseases, the same approach can be used to establish accurate molecular phenotypes of a wide variety of nonimmunological normal and disease processes.
METHODS
Mice
Age- and sex-matched C57BL/6J, BALB/cJ, 129X1/SvJ, 129P3/J, BXSB/MpJ-Yaa+, and BXSB/MpJ-Yaa mice were obtained from our research colony or from the Jackson Research System's production facility at the Jackson Laboratory, Bar Harbor, Maine. KRN T-cell receptor transgenic mice were a kind gift from D. Mathis and C. Benoist. Hemizygous KRN transgenic males (on a C57BL/6J background)were bred to NOD/LtJ females, and progeny were typed by PCR. Nontransgenic (arthritis-free)animals were compared with transgenic (arthritic)littermates for IQA experiments. All animal experiments were approved by the Jackson Laboratory's Animal Care and Use Committee (ACUC).
Induction of GVHD
Eight-week-old recipient male 129P3/J (experimental group) and female C57BL/6J mice (control group)were irradiated with split doses of 450 cGy from a 137Cs source within a 4-h interval, and injected with a mixture of bone marrow and spleen cells from female C57BL/6J mice as described (Choi et al. 2002).
cDNA Synthesis
To minimize sample preparation variation, all samples for a given experiment (from both control and experimental groups)were processed in parallel. Solid tissues were collected into RNALater (Ambion), used immediately, or stored at -20°C for not more than 3 d. Total RNA was purified from solid tissue using the RNAqueous 4-PCR kit (Ambion)and DNase-treated following the manufacturer's recommendations. Total RNA was purified from 75 μL of blood, collected by a retro-orbital bleed into heparinized 100-μL capillary tubes, using the 6100 RNA Prep Station (ABI), and DNase-treated following the manufacturer's recommendations. Synthesis of cDNA from 5–10 μL of total RNA was carried out using the Retroscript kit (Ambion)following the manufacturer's recommendations. To minimize variation in sample preparation, the cDNA was stored at -20°C and was used for QPCR within 3 d of preparation.
PCR Amplicon Development
Primer sets (MWG Biotech)were designed using Primer Express v1.5 (Applied Biosystems, ABI)following recommendations appropriate for use in the ABI Prism 7700 Sequence Detection System. Selected primers were searched against GenBank via the NCBI BLAST algorithm to ensure specificity to the desired gene target. Each PCR product was subjected to bidirectional sequencing using each end-specific primer on the ABI Prism 3700 Sequencer. SYBR Green dissociation curves were generated via the 7700 to further ensure the generation of a single PCR product under experimental reaction conditions. Primer sequences are available online at www.genome.org.
Real-Time Quantitative PCR
ImmunoQuantArray 96-well plates were prepared by the addition of 0.7 μL of 1 μM Primers per well. To each well was then added 9.3 μL of PCR master mix, which contained 525 μL of 2× SYBR Green Master Mix (ABI), 384 μL of dH2O, and 70.4 μL of cDNA (typically a 1:10 for blood or 1:20 for spleen dilution of stock cDNA). The plate was sealed using an Optical Adhesive Cover (ABI), and the fluid was spun down in a swinging bucket centrifuge. Real-time PCR data were collected on the ABI Prism 7700 Sequence Detection System v1.7 using the default reaction conditions (1 cycle at 50°C for 2 min, 1 cycle at 95°C for 10 min, 40 cycles at 95°C for 15 sec and at 60°C for 1 min). The baseline and threshold were set to experimentally determined values and the Experimental Report data (a table of Ct values for each of the 96 reactions)were exported for GPR analysis.
Global Pattern Recognition Algorithm
The GPR algorithm is implemented as a Microsoft Excel macro to identify significant changes in gene expression profiles within a 96-well real-time PCR data set using the Cycle Threshold (Ct)values generated by the ABI Prism 7700. GPR compares the Ct of each candidate gene individually with the Ct of every other gene in the 96-gene IQA data set that is eligible as a normalizer. Doing so allows stratification of genes both as a function of the magnitude of the change and the reproducibility of the Ct values within each of the two experimental groups.
GPR first filters data into overlapping gene and normalizer bins. This filtering process is controlled by a user-defined Cycle Cutoff (CC)value (set at 37.5 for all experiments shown). The CC is the PCR cycle number above which data are disregarded. A number of 37 cycles approaches single-copy detection, and thus leads to variable data. Consequently, using the CC eliminates these noisy data. Using the CC, GPR culls the data with the Normalizer Filter and the Gene Filter. A gene passes through the Normalizer Filter if all observations in both control and experimental groups fall below the cycle cutoff value (e.g., an eligible normalizer—Group 1 Ct values: 33.4, 31.1, 31.5; Group 2 Ct values: 33.9, 34.2, 33.6). A gene passes through the Gene Filter if all observations in either control or experimental groups fall below the cycle cutoff value (e.g., an eligible gene, but not an eligible normalizer— Group 1 Ct values: 32.4, 33.1, 31.8; Group 2 Ct values: 37.9, 39.1, 40). Each eligible gene is then normalized in turn to each eligible normalizer by computing a ΔCt value (ΔCtgene = Ctgene - Ctnormalizer). For each gene-normalizer combination, the individual ΔCt values generated for the control and experimental groups are compared by a two-tailed heteroscedastic (unpaired)Student's t-test, and a “hit” is recorded if the p-value from the t-test falls below a user-defined p-value (e.g., 0.05). Thus data from biological replicates are compared directly at the ΔCt level at each round of normalization. Each candidate gene, when processed through GPR, is significantly different when compared with certain normalizers and insignificant when compared with others. The total number of normalizer “hits” for each gene is tallied and used to sort the genes in the 96-well array with the genes changed in comparison to the largest number of normalizer genes ranking highest. The GPR score indicates the fraction of normalizer genes against which the candidate gene was found to be significantly different. Analysis of more than 50 data sets indicates that a GPR score of 0.4 or higher (statistically different when compared with 40% or more of the eligible normalizers)reliably identifies the genes having undergone significant change (see Results and Discussion). After ranking genes by GPR score, the direction and magnitude of change of a particular gene with respect to the control group can then be approximated by comparing the average ΔCt values of the two groups after normalization to 18S rRNA by the 2-ΔΔCt method (Livak and Schmittgen 2001). The GPR algorithm implemented in Excel (and documentation)is available for download at http://www.jax.org/research/roop/gene_expression.html.
Bootstrap Probability Distribution
Ct values from IQA data sets for the two comparison groups were randomly resampled on a gene-by-gene basis and then processed by the GPR algorithm. The resultant GPR scores for each gene were recorded for each of the 250 random resamplings to generate a bootstrap probability distribution (Efron and Tibshirani 1998). An observed GPR score above the 99.5th percentile of the gene-specific bootstrap probability distribution was considered significant (bootstrap p < 0.005, corresponding to ≤1 value in the bootstrap distribution being higher than the observed GPR score). The p-value for each gene was computed as the number of scores in the bootstrap distribution higher than the observed GPR score divided by the number of random resamplings.
Acknowledgments
The authors thank Gary Churchill and Jason Stockwell for expert advice and ANOVA analysis; Thomas Sproule for expert mouse colony management; and Carol Bult, Wayne Frankel, and Jason Stockwell for manuscript review. This work was supported by grants from the National Institute for Diabetes, Digestive and Kidney Diseases (NIDDK) and the Alliance for Lupus Research (ALR). S.A. was supported by a fellowship from the Shelby Cullom Davis Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.533003.
Footnotes
[Supplemental material—The primer sequences for genes included in the ImmunoQuant Array (and listed in Table 1) are available online at www.genome.org. The GPR algorithm, documentation, and sample data sets are available at http://www.jax.org/staff/roopenian/labsite/index.html.The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: D. Mathis and C. Benoist.]
References
- Alizadeh, A.A., Eisen, M.B., Davis, R.E., Ma, C., Lossos, I.S., Rosenwald, A., Boldrick, J.C., Sabet, H., Tran, T., Yu, X., et al. 2000. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403: 503-511. [DOI] [PubMed] [Google Scholar]
- Bijl, M., Horst, G., Limburg, P.C., and Kallenberg, C.G. 2001. Fas expression on peripheral blood lymphocytes in systemic lupus erythematosus (SLE): Relation to lymphocyte activation and disease activity. Lupus 10: 866-872. [DOI] [PubMed] [Google Scholar]
- Bustin, S.A. 2000. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25: 169-193. [DOI] [PubMed] [Google Scholar]
- Choi, E.Y., Christianson, G.J., Yoshimura, Y., Jung, N., Sproule, T.J., Malarkannan, S., Joyce, S., and Roopenian, D.C. 2002. Real-time T cell profiling identifies H60 as a major minor histocompatibility antigen in murine graft-versus-host disease. Blood 100: 4259-4265. [DOI] [PubMed] [Google Scholar]
- Duggan, D.J., Bittner, M., Chen, Y., Meltzer, P., and Trent, J.M. 1999. Expression profiling using cDNA microarrays. Nat. Genet. 21: 10-14. [DOI] [PubMed] [Google Scholar]
- Efron, B. and Tibshirani, R. 1998. An introduction to the bootstrap. CRC Press, Boca Raton, FL.
- Ferrara, J.L. 2000. Pathogenesis of acute graft-versus-host disease: Cytokines and cellular effectors. J. Hematother. Stem Cell Res. 9: 299-306. [DOI] [PubMed] [Google Scholar]
- Gibson, U.E., Heid, C.A., and Williams, P.M. 1996. A novel method for real time quantitative RT-PCR. Genome Res. 6: 995-1001. [DOI] [PubMed] [Google Scholar]
- Giulietti, A., Overbergh, L., Valckx, D., Decallonne, B., Bouillon, R., and Mathieu, C. 2001. An overview of real-time quantitative PCR: Applications to quantify cytokine gene expression. Methods 25: 386-401. [DOI] [PubMed] [Google Scholar]
- Goidin, D., Mamessier, A., Staquet, M.J., Schmitt, D., and Berthier-Vergnes, O. 2001. Ribosomal 18S RNA prevails over glyceraldehyde-3-phosphate dehydrogenase and β-actin genes as internal standard for quantitative comparison of mRNA levels in invasive and noninvasive human melanoma cell subpopulations. Anal. Biochem. 295: 17-21. [DOI] [PubMed] [Google Scholar]
- Green, C.D., Simons, J.F., Taillon, B.E., and Lewin, D.A. 2001. Open systems: Panoramic views of gene expression. J. Immunol. Methods 250: 67-79. [DOI] [PubMed] [Google Scholar]
- Grondal, G., Gunnarsson, I., Ronnelid, J., Rogberg, S., Klareskog, L., and Lundberg, I. 2000. Cytokine production, serum levels and disease activity in systemic lupus erythematosus. Clin. Exp. Rheumatol. 18: 565-570. [PubMed] [Google Scholar]
- Hamalainen, H.K., Tubman, J.C., Vikman, S., Kyrola, T., Ylikoski, E., Warrington, J.A., and Lahesmaa, R. 2001. Identification and validation of endogenous reference genes for expression profiling of T helper cell differentiation by quantitative real-time RT-PCR. Anal. Biochem. 299: 63-70. [DOI] [PubMed] [Google Scholar]
- Heid, C.A., Stevens, J., Livak, K.J., and Williams, P.M. 1996. Real time quantitative PCR. Genome Res. 6: 986-994. [DOI] [PubMed] [Google Scholar]
- Ji, H., Ohmura, K., Mahmood, U., Lee, D.M., Hofhuis, F.M., Boackle, S.A., Takahashi, K., Holers, V.M., Walport, M., Gerard, C., et al. 2002. Arthritis critically dependent on innate immune system players. Immunity 16: 157-168. [DOI] [PubMed] [Google Scholar]
- Kerr, M.K. and Churchill, G.A. 2001. Statistical design and the analysis of gene expression microarray data. Genet. Res. 77: 123-128. [DOI] [PubMed] [Google Scholar]
- Korganow, A.S., Ji, H., Mangialaio, S., Duchatelle, V., Pelanda, R., Martin, T., Degott, C., Kikutani, H., Rajewsky, K., Pasquali, J.L., et al. 1999. From systemic T cell self-reactivity to organ-specific autoimmune disease via immunoglobulins. Immunity 10: 451-461. [DOI] [PubMed] [Google Scholar]
- Korngold, R. and Sprent, J. 1983. Lethal graft-versus-host disease across minor histocompatibility barriers in mice. Clin. Haematol. 12: 681-693. [DOI] [PubMed] [Google Scholar]
- Kouskoff, V., Korganow, A.S., Duchatelle, V., Degott, C., Benoist, C., and Mathis, D. 1996. Organ-specific disease provoked by systemic autoimmunity. Cell 87: 811-822. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔC(T) method. Methods 25: 402-408. [DOI] [PubMed] [Google Scholar]
- Lockhart, D.J. and Winzeler, E.A. 2000. Genomics, gene expression and DNA arrays. Nature 405: 827-836. [DOI] [PubMed] [Google Scholar]
- Losana, G., Rigamonti, L., Borghi, I., Assenzio, B., Ariotti, S., Jouanguy, E., Altare, F., Forni, G., Casanova, J.L., and Novelli, F. 2002. Requirement for both IL-12 and IFN-γ signaling pathways in optimal IFN-γ production by human T cells. Eur. J. Immunol. 32: 693-700. [DOI] [PubMed] [Google Scholar]
- Moore, K.W., de Waal Malefyt, R., Coffman, R.L., and O'Garra, A. 2001. Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 19: 683-765. [DOI] [PubMed] [Google Scholar]
- Morrison, T.B., Weis, J.J., and Wittwer, C.T. 1998. Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. Biotechniques 24: 954-962. [PubMed] [Google Scholar]
- Murphy, E.D. and Roths, J.B. 1979. A Y chromosome associated factor in strain BXSB producing accelerated autoimmunity and lymphoproliferation. Arthritis Rheum. 22: 1188-1194. [DOI] [PubMed] [Google Scholar]
- Nousari, H.C., Kimyai-Asadi, A., and Tausk, F.A. 1998. Subacute cutaneous lupus erythematosus associated with interferon β-1a. Lancet 352: 1825-1826. [DOI] [PubMed] [Google Scholar]
- Schmittgen, T.D., Zakrajsek, B.A., Mills, A.G., Gorn, V., Singer, M.J., and Reed, M.W. 2000. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: Comparison of endpoint and real-time methods. Anal. Biochem. 285: 194-204. [DOI] [PubMed] [Google Scholar]
- Shaffer, A.L., Rosenwald, A., Hurt, E.M., Giltnane, J.M., Lam, L.T., Pickeral, O.K., and Staudt, L.M. 2001. Signatures of the immune response. Immunity 15: 375-385. [DOI] [PubMed] [Google Scholar]
- Theofilopoulos, A.N., Koundouris, S., Kono, D.H., and Lawson, B.R. 2001. The role of IFN-γ in systemic lupus erythematosus: A challenge to the Th1/Th2 paradigm in autoimmunity. Arthritis Res. 3: 136-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van't Veer, L.J., Dai, H., van de Vijver, M.J., He, Y.D., Hart, A.A., Mao, M., Peterse, H.L., van der Kooy, K., Marton, M.J., Witteveen, A.T., et al. 2002. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415: 530-536. [DOI] [PubMed] [Google Scholar]
- Wang, E., Miller, L.D., Ohnmacht, G.A., Liu, E.T., and Marincola, F.M. 2000. High-fidelity mRNA amplification for gene profiling. Nat. Biotechnol. 18: 457-459. [DOI] [PubMed] [Google Scholar]
- Wong, C.K., Ho, C.Y., Li, E.K., and Lam, C.W. 2000. Elevation of proinflammatory cytokine (IL-18, IL-17, IL-12)and Th2 cytokine (IL-4)concentrations in patients with systemic lupus erythematosus. Lupus 9: 589-593. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.jax.org/staff/roopenian/labsite/index.html; access to the GPR algorithm and documentation.