Abstract
Molecular markers defining self-renewing pluripotent embryonic stem cells (ESCs) have been identified by relative comparisons between undifferentiated and differentiated cells. Most of analysis has been done under a specific differentiation condition that may present significantly different molecular changes over others. Therefore, it is currently unclear if there are true consensus markers defining undifferentiated hESCs. To identify a set of key genes consistently altered during differentiation of hESCs regardless of differentiation conditions we have performed microarray analysis on undifferentiated hESCs (H1 and H9) and differentiated EB’s and validated our results using publicly available expression array data sets. We constructed consensus modules by Weighted Gene Correlation Analysis (WGCNA) and discovered novel markers that are consistently present in undifferentiated hESCs under various differentiation conditions. We have validated top markers (downregulated: LCK, KLKB1 and SLC7A3; upregulated: RhoJ, Zeb2 and Adam12) upon differentiation. Functional validation analysis of LCK in self-renewal of hESCs by using LCK inhibitor or gene silencing with siLCK resulted in a loss of undifferentiation characteristics- morphological change, reduced alkaline phosphatase activity and pluripotency gene expression, demonstrating a potential functional role of LCK in self-renewal of hESCs. We have designated hESC markers to interactive networks in the genome, identifying possible interacting partners and showing how new markers relate to each other. Furthermore, comparison of these data sets with available datasets from iPSCs revealed that the level of these newly identified markers were correlated to the establishment of iPSCs, which may imply a potential role of these markers in gaining of cellular potency.
Keywords: self-renewal, human embryonic stem cells, WGCNA, interactive molecular network, LCK
Introduction
Embryonic stem cells (ESCs) possess infinite self-renewal capacity and pluripotent capability to differentiate into any cell type in the human body. ESCs hold a great promise for regenerative therapies in a wide range of human diseases 1–3. However, one of the major obstacles in stem cell-based therapies is inducing stem cells to take on a specific form or to differentiate into a particular cell type 4.
Currently, when ESCs are cultured in a defined media, they are not homogenously self-renewing stem cells 5–8. They are composed of heterogeneous cells with gradients in their differentiation status. Moreover, researchers and clinicians have concerns that some of transplanted stem cells will remain in their pluripotent state, which could potentially grow to form teratoma in patients. Thus, in order for ESCs to be useful in a clinical setting, it is essential to identify these guiding signals and understand how they regulate self-renewal and differentiation.
In 2006, Shinya Yamanaka made an important advancement in stem cell research by discovering four key stemness factors essential for self-renewing abilities and developing induced pluripotent stem cells (iPSCs) by transducing mouse fibroblasts with Oct-3/4, Sox2, c-Myc, and Klf4 9. Patient-specific iPSCs have great potential therapeutic merits without the controversial ethical issue of embryonic stem cells. Over the years, extensive efforts have contributed to the development and refinement of iPSC reprogramming, which mostly benefited by advances in our knowledge on the molecular mechanisms underlying reprogramming process. However, we are still lack of detailed knowledge on the reprogramming process and also many reports have demonstrated potential discrepancy between ESCs and iPSCs at molecular levels 10–12.
We understand that there are certain key genes within stemness, yet we still don’t know all the genes and pathways implicated in this process. Several groups have examined stem cell differentiation and performed whole-genome microarray analysis 13–18. We have examined these datasets and assessed if there are certain genes that are consistently altered upon hESC differentiation. We utilized an R package known as Weighted Gene Coexpression Network Analysis (WGCNA) that has the power to analyze all this information and determine clusters of genes known as modules that behave the same across the different datasets 19.
Here we show the list of conserved genes that play a crucial role in the identity of self-renewal from 19 different microarray data 13–18. The top candidates were functionally annotated by DAVID (Database for Annotation, Visualization and Integrated Discovery) analysis 20 and validated by qRT-PCR. Identifying these genes can be helpful to researchers to identify how similar their systems are to hESCs. For example we constructed a consensus list from five microarrays of iPSCs and compared those genes to the conserved gene list in hESCs. We found that 46% of genes were in common between the conserved lists for hESCs and iPSCs including the genes we filtered and considered the “top” genes from this list. Other groups can similarly use WGCNA to develop identifying signatures for their system and compare to the gene lists presented in this work to potentially assess the level of stemness in their system.
Results
Construction of hESC network undergoing differentiation
Using H1 and H9 hESC lines, we performed gene expression microarray analysis (Affymetrix Human Genome U133 Plus 2.0 Array) and analyzed the interaction patterns of 54,614 genes using WGCNA in R programming (Figure 1A). Microarray data are more completely represented by considering the relationships between measured transcripts, which can be assessed by pairwise correlations between gene expression profiles. WGCNA identifies gene modules, and finally uses intramodular connectivity, gene significance, and gene ontology information to identify key genes in the pathways for further validation. Instead of relating thousands of genes, it focuses on the relationship between a few modules and the phenotypic outcome. Modules are constructed without regard to biological status. Because the modules may correspond to biological pathways, focusing the analysis on intramodular hub genes (or the module eigengenes) amounts to a biologically motivated data reduction scheme 19,21.
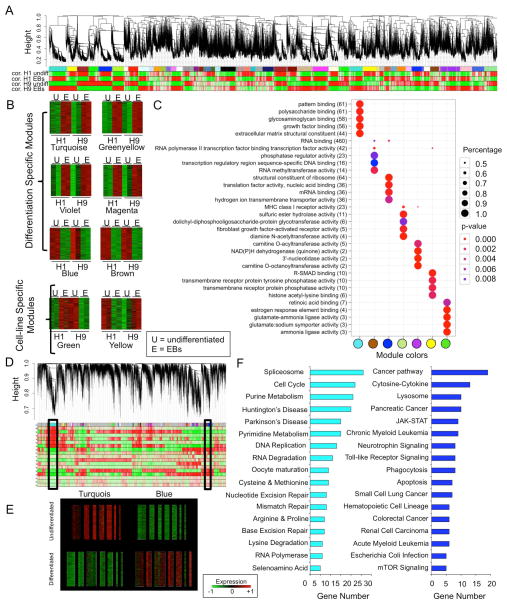
Figure 1. Construction of consensus stem cell network in hESCs.
(A) To identify consensus gene signatures that are coherently associated with self-renewing hESCs, we have performed gene expression microarray analysis with undifferentiated H1 and H9 hESCs and their differentiated EBs. The result was further analyzed by WGCNA analysis to identify coexpression modules. (B) WGCNA identified differentiation specific modules and cell-line specific module and their corresponding heatmaps are shown (the green shows downregulation and the red shows upregulation). (C) Dot plots of seven top modules showed that genes in each module are strongly correlated each other in their potential biological functions. (D) We then further correlated our own dataset to publicly available datasets from Gene Expression Omnibus9–14 as described in the Method section to identify consensus modules defining self-renewal under various differentiation settings. (E) Statistically the turquoise module includes genes significantly downregulated upon differentiation and the blue module includes genes upregulated upon differentiation. (F) DAVID analysis was performed to annotate the genes in the turquois and the blue modules from (D).
We found six modules that showed changes in expression upon differentiation, which were independent of cell lines (H1 vs. H9) (Turquoise, Brown, Blue, Green-yellow, Violet, and Magenta) (Figure 1B). We also found two modules that showed cell line-specific expression changes upon differentiation (bottom panel in Figure 1B). Number of modules and genes within the modules suggest that H1 and H9 hESCs show similar gene signature profiles, however there are small subsets of genes that behave differently upon differentiation, which could be gender-specific genetic differences (H1-male and H9-female). This needs to be further validated with several other gender-specific hESC lines.
To functionally annotate the different modules, seven top statistically significant modules were analyzed using Gene Ontology (GO) terms in R programming. GO result is represented in a dot plot graph (Figure 1C). We found that significant numbers of differentiation-related genes were involved in repeating structure pattern binding, polysaccharide binding, RNA binding, and structural constituent of ribosome. We also found that R-SMAD binding, tyrosine and protein phosphatase activity, and histone acetyl-lysine binding were different between H1 and H9 hESCs upon differentiation (Figure 1C).
Identifying consensus modules in hESC undergoing differentiation
There are certain well-characterized molecular markers that define self-renewing undifferentiated stem cells, however many of genes that are involved in this process are not yet known. We hypothesized that there are molecular markers defining self-renewing stem cells that can be determined by examining hESCs under undifferentiating and differentiating conditions. To determine these molecular markers we compared our own microarray results to 19 different publicly available microarray data in which other labs differentiated hESCs (including H1, H9 and T3ES) to mesodermal, neuronal, hepatic lineages, and other lineages 13–18. Those datasets were all done using the same platform GPL570 (Affymetrix Human Genome U133 Plus 2.0 Array). We examined those datasets with WGCNA and identified consensus modules across the different data sets that represent unique gene signatures that are pertinent to self-renewal and differentiation (Figure 1D) 13–18. These consensus modules as well as the correlation of each trait (differentiated or undifferentiated) are shown in Figure 1D. The top consensus modules based on significance and gene number were found and are represented by the colors of turquoise and blue (Figure 1D, black box). Heatmaps across datasets of turquoise and blue consensus modules are shown in Figure 1E. Consensus turquoise module represents an exclusive set of genes from all 19 microarrays that showed decrease in their expression as different types of stem cells differentiated into various cell types. Consensus blue module contains a list of upregulated genes upon stem cell differentiation (Figure 1E). Furthermore, we performed DAVID analysis on the genes within the turquoise and the blue module (Figure 1F). We observed pathways involved in splicing, cell cycle, repair, and metabolism for the turquoise module (Figure 1F, left), and observed pathways for various cancers and cell death in the blue module (Figure 1F, right). This analysis suggests that some of these pathways are potentially involved in self-renewal and to a certain extent have been understood for sometime 22–24. For example, stem cells have the high proliferation rate compared to other cell types and have unusual cell cycle structure, characterized by a short G1 phase and S-phase preference 22. Cancer cells having stem cell like properties and the existence of cancer stem cells first in leukemia and now in solid tumors are a well known phenomenon 24–26.
Identifying the top genes within the consensus modules based on significance and fold change
Next we wanted to filter and rank the top genes within the turquoise and blue modules to determine what are the best markers for self-renewal and differentiation, respectively. Using fold change and significance we made Volcano plots of the different datasets within each module; representative data sets are shown in Figure 2A. We used two-fold change and a p-value of less than 0.01 as the minimum cutoff. Using multiple data sets from various differentiation conditions (hepatic, hematopoietic, neural and mesenchymal specifications) 13–18, we have found the top genes that are upregulated and downregulated during hESC differentiation (Figure 2B, Table S1 and Table S2). Some of the genes that we found downregulated upon differentiation are already very well known such as OCT4 and NANOG. However, we also found other interesting genes that also met our criteria, such as LCK, KLKB1, and SLC7A3, in the downregulated gene set and RHOJ, ZEB2, and ADAM12 in the upregulated gene set. The criteria we used were quite stringent since all the genes in the consensus modules were significant based on the initial individual WGCNA analysis.
Figure 2. Identification of top genes within consensus modules and validation.
(A) The turquois and blue modules were subjected to Volcano plot to identify gene signatures with significant fold changes. We used the cut off of 2-fold change and the p value of <0.05. (B) The genes with top fold changes across 19 arrays from various differentiation conditions were compared and represented by the Venn-diagrams 9–14. (C) To validate the identified genes we used undifferentiated cells or differentiated EBs from hESCs. Immunofluorescence analysis confirmed a decreased expression of OCT4 as undifferentiated H9 hESC underwent differentiation. (D) We validated three of the top genes upregulated (LCK, KLKB1 and SLC7A3) and downregulated (RHOJ, ZEB2 and ADAM12) in the hESC consensus modules by qRT-PCR in H1 and (E) in H9 hESCs
We then verified some of the top genes from the turquoise and blue consensus modules. We utilized two hESC lines H1, and H9 cells for validation studies 27. To do this we differentiated hESCs as represented in Figure 2C and confirmed differentiation by observing decreased OCT4 expression and changes in morphology. After differentiation of H1 and H9 hESCs we examined expression level of top 6 genes from the consensus modules by qRT-PCR. We validated that all six genes behaved as expected, being downregulated or upregulated upon differentiation (Figure 2D and 2E).
Discovery of intramodular hub genes in the differentiation specific module
Successful generation of induced pluripotent stem cells (iPSCs) has significantly advanced our understanding of stem cell biology, however; the detailed mechanisms behind how these four factors could transform terminally differentiated cells into stem cell like cells still remain elusive. Therefore, we analyzed our microarray data to look for co-regulators of OCT4, NANOG, c-MYC and SOX2. First we calculated interconnectivity values of all genes that are related to the four factors using WGCNA and R-programming (Figure 3A–3D). Then we ranked those genes according to the interconnectivity values and top four genes as shown in Figure 3A–3D.
Figure 3. Interconnectivity analyses on stem cell factors and novel markers.
We have performed interconnectivity analysis to examine molecular regulators that are potentially correlated with stemness markers and our novel markers. (A) TRMT112, RASGEF1, HRNBP3 and SNHG5 are highly connected to OCT4. (B) BLOC1, PCIF1 and ZBTB44 are connected to NANOG. (C) ETV5, FHDC1, HDAC8 and ASMT are connected to c-MYC. (D) TEX10, CDCA5, C16ORF, and RUNX1 are connected to SOX2. (E) A graphic representation of designating six hub genes (OCT4, NANOG, SOX2, LCK1, SLC7A3 and KLKB1) to specific interactive networks in consensus module. (F) HIVE plot on the six factors shows that they are closely related to each other. (G) HIVE plot showing the relationship between the six factors and genes within consensus module. This demonstrates that most of genes in the consensus module are correlatively regulated by the six factors.
After identifying top genes from each module, we identified centrally located genes, which are intramodular hub genes, within the module. The module-centric analysis allows us to focus on the relationship between a few key genes and the self-renewal/differentiation of stem cells instead of relating thousands of unnecessary variables 21. Since the intramodular hub genes are centrally located in the module, they are considered the driving forces of that particular module and have been used to discover therapeutic targets or candidate biomarkers 28–31. Intramodular hub genes also display the highest intramodular connectivity with the rest of genes in the module. Thus the hub genes may function as representative and/or regulatory elements in the module. We measured intramodular connectivity and graphically represented top genes from the differentiation-specific module (turquoise).
We found that there were six intramodular hub genes that are highly connected, SLC7A3, NANOG, SOX2, KLKB1, OCT4, and LCK (Figure 3E). Six intramodular hubs formed mainly four large clusters (represented #1 – #4 in Figure 3E). We analyzed four large clusters by DAVID analysis and found that cluster #1 contained NANOG and OCT4 as the main intramodular hubs and may be involve in the lysine degradation pathway. Cluster #2 contained LCK and SLC7A3 and was possibly responsible for purine metabolism, cell cycle and Huntington’s disease. Cluster #3 contained KLKB1 and may be involved in spliceosome. Cluster #4 included SOX2 as the intramodular hub and may be involved in RNA degradation (Figure 3E).
Genomic analysis is many times difficult to understand and interpret. A HIVE plot offers a number of simple graphing solutions for complex network analyses 32. HIVE plot is to visualize networks by mapping and positioning nodes on radially distributed linear axes based on network structural properties. HIVE plot shows quantitative understanding of important aspects of network structure. First, we used a scalable linear layout HIVE plot to show how top six intramodular hub genes are related to each other (Figure 3F). The HIVE plot confirmed that six intramodular hub genes are highly connected to each other. Moreover, we found that there is a gradient in how close or distant each intramodular hub gene is to one another. For example, KLKB1 is more closely related to OCT4 compared to SLC7A3 based on the genes that particular intramodular gene represents. Second HIVE plot was used to relate those six hub genes to the consensus turquoise module (Figure 3G). We found that some genes from the consensus turquoise module are highly connected to all six hub-genes (x-axis, Figure 3H) and some genes are more hub-specific (y-axis, Figure 3G).
Validating potential involvement of LCK in self-renewal of hESCs
Among three validated upregulated genes, we chose LCK for further functional validation in self-renewal of hESCs since its silencing has been indirectly demonstrated in differentiation of ESC. LCK has been implicated in self-renewal and stem cell maintenance in mESCs 33,34. Differentiation of ES cells to embryoid bodies was associated with rapid transcriptional silencing of LCK and with the loss of the corresponding kinase protein 33,35,36.
To validate the potential function of LCK in maintenance of hESC self-renewal, we have examined the effect of a selective LCK inhibitor (7-cyclopentyl-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3,-d]pyrimidin-4-ylamine) on hESC differentiation 37. It was found that an LCK inhibitor induced spontaneous differentiation of hESCs (Figure 4). As shown in Figure 4A, treatment of undifferentiated hESCs exponentially growing in culture with LCK inhibitor (0.5 and 1 μM) for 72 hours resulted in a morphological change to differentiated cells. Cells lost typical cellular characteristics of embryonic stem cells. The effect was more profound on the edges of colonies with less densely packed cells. Differentiation of hESCs after treatment with LCK inhibitor was further confirmed by reduced alkaline phosphatase staining along with reduced alkaline phosphatase activity that are typically used for marking undifferentiated ESCs (Figure 4B and 4C). We also found that treatment with LCK inhibitor resulted in a significant downregulation of OCT4 expression (Figure 4D). This result demonstrates that activity of LCK is necessary to maintain hESCs in an undifferentiated self-renewing state and downregulation of LCK induces differentiation of hESCs.
Figure 4. Effects of inhibiting LCK on self-renewal and pluripotency of hESCs.
(A) To examine a potential role of LCK in hESC self-renewal, H9 hESCs were treated with 0.1% DMSO (vehicle control), 0.5 mM or 1.0 mM LCK inhibitor (Pyrrolo[2,3-d]pyrimidines) (A-419259, Sigma-Aldrich Corp. St. Louis, MO) for 72 hours. Morphologic assessment showed significant change in the presence of LCK inhibitor. Subsequent images were taken by Leica DMIL inverted light microscope with AxioVision 4.6 software. (B) Alkaline phosphatase (ALP) activity, as indicated by the red staining, was downregulated in LCK inhibitor-treated cells compared with the control DMSO-treated cells in dose dependent manner. ALP staining was followed closely by the recommended manufacturer’s protocol (Sigma-Aldrich Corp. St. Louis, MO). ALP images were taken by using Olympus 1×81 microscope with CellSens 1.8 software. (C) Treatment with LCK inhibitor resulted in reduced ALP activity in hESCs. The lysate and ALP reaction mixture was incubated at 37 degree. OD (405 nm) was measured at 20 min and 40 min. Relative ALP activity was calculated using DMSO (control) as a base value. Bar represents mean ±SD (n=9; * p<0.05 versus control) from triplicate samples. (D) Downregulation of OCT4 expression in hESCs treated with LCK inhibitor. Fold change was calculated using DMSO (control) as a base value. Bar represents mean ±SD (n=9; * p<0.05 versus control). To examine the specific effect of LCK in hESCs we have performed gene knockdown experiment as described in Methods. (E) IF analysis showed that knocking-down of LCK by using 25 nM of siLCK SMARTPool (Thermo Fisher Scientific, Pittsburgh, PA) resulted in the downregulation of OCT4 compared to non-transfected cells or cells transfected with scrambled control siRNA. (F) Quantitative RT-PCR analysis on several hESC pluripotency genes (OCT4, NANOG, FOXD3 and DNMT3B) showed significant reduction of their levels after silencing LCK. Assay was done for triplicate samples to evaluate statistical significance.
To further validate that LCK plays a role in self-renewal of hESCs, we specifically knocked down LCK by siRNA (Figure 4E and 4F). We were able to successfully knock down LCK, which can be seen in its protein level (Figure 4E, left) and in its transcription level (Figure 4F, most left). By performing immunofluorescence, we found that OCT4 level was significantly decreased when LCK siRNA was transfected but not by control or scramble siRNA (Figure 4E, right). Transcriptional analysis of major stem cell markers (OCT4, NANOG, FOXD3, and DNMT3B) showed that knocking down LCK leads to decrease in gene expression of the stem cells markers compared to control and scramble siRNA (Figure 4F). Taken together with the LCK inhibitor data, LCK-specific knockdown experiment proved that LCK is one of key factors in self-renewal of human embryonic stem cells.
Comparing the gene signatures determined in hESCs to iPSCs
Next we assessed how close iPSCs were to hESCs with their molecular signatures. This will help us answer the question as to whether any novel self-renewal factors exist and how significant their molecular roles could be during the acquisition of stemness in iPSCs. To address this we determined consensus modules of five different datasets examining iPSCs and their differentiated cell of origin (Figure 5A) 38–42. This analysis allowed us to determine what were the key molecular signatures that defined an iPSC.
Figure 5. Comparison of consensus modules and molecular hubs correlated with iPSCs and self-renewing hESCs.
(A) Consensus modules were formed using WGCNA by examining iPSCs and their cell of origin across five data sets. These five datasets were obtained from Gene Expression Omnibus from various groups 9–14. (B) Cytoscape interconnectivity analysis (http://www.cytoscape.org/) to visualize molecular interaction networks and pathways revealed SLC7A3, KLKB1 and LCK as top hub genes in the consensus turquoise module of iPSCs. It shows that SLC7A3 and KLKB1 formed closely related molecular hubs, whereas the molecular hub associated with LCK was distal from them. (C) DAVID functional annotation of the best modules from hESCs and iPSCs showed several important biological processes correlated to hESCs, iPSCs, or commonly to both. (D) We have identified and compared interconnectivity genes from hESC hubs and iPSC hubs and annotated the common genes. We found several biological processes that may have critical role in self-renewal and potency of hESCs and iPSCs.
Using WGCNA and R, we found two significant modules- blue and turquois modules. The blue module showed downregulated expression in iPSCs, whereas the turquois module showed upregulated levels in iPSCs. Since we are interested in genes that may have significance in acquiring self-renewal and stem cell characteristics in iPSCs, we focused on the turquois module with upregulated expression. We performed Cytoscape analysis to visualize molecular interaction networks and biological pathways and integrate these networks with annotations, gene expression profiles (http://www.cytoscape.org/). We mapped out interconnectivity of the turquoise module (upregulated in iPSCs vs. parental somatic cell counterparts) to find intramodular hub genes (Figure 5B). We found that SLC7A3, KLKB1, and LCK that we initially identified as novel markers correlated to undifferentiated hESCs were the key genes that might play an important role in self-renewal ability in iPSCs according to our bioinformatics analyses. In addition, it showed that SLC7A3 and KLKB1 could form closely interconnected molecular networks, whereas the molecular hub associated with LCK was somewhat distal from these two factors. This suggests that LCK may be involved in reprogramming process through different molecular function.
To compare hESCs and iPSCs further, we functionally annotated the best representative consensus modules from hESCs and iPSCs (Figure 5C). We found that purine and pyrimidine metabolism was more strongly correlated to hESCs; and alanine, aspartate and glutamate metabolism, endometrial cancer pathway, glycolysis and glucogenesis, and p53 signaling pathway to be more closely associated to iPSCs (Figure 5C). When we identified and compared genes that are associated with hESC hubs and iPSC hubs, we found that the common genes were associated with the spliceosome, cell cycle, RNA degradation and DNA replication (Figure 5D). This suggests that genes involved in these molecular functions have critical roles in the maintenance of self-renewal and potency of hESCs and iPSCs.
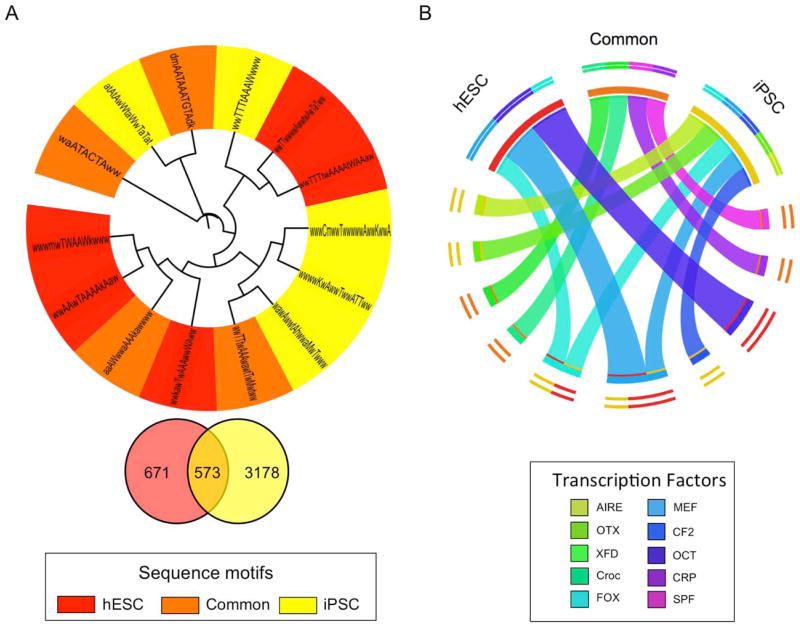
To examine transcriptional hubs that could play critical roles in self-renewal of hESCs and iPSCs, we have analyzed promoter sequence motifs in genes from the best consensus modules correlated to undifferentiated hESCs and iPSCs. The best hESC consensus module contained 671 unique genes and the iPSC module contained 3178. We found that 46% of the genes (573 genes) in the hESC top consensus module matched the iPSC top consensus module (Figure 6A, the gene list is shown in Supplemental Table). By promoter sequence analysis, we found 14 common sequence motifs among promoters of hESC and iPSC genes. Out of 14, hESCs had five unique sequence motifs (red), iPSCs with five unique motifs (yellow) and four in common (orange) (Figure 6A). Based on the promoter sequences, we found transcription factors that may bind differentially to hESCs and iPSCs. The OCT family transcription factor was unique in hESCs and AIRE, OTX and CF2 were unique in iPSCs. XFD, CROC, FOX, MEF, CRP and SPF transcription factors were shared between hESCs and iPSCs (Figure 6B). This data shows that there are indeed common transcription factors that may play critical roles in self-renewal of both hESCs and iPSCs, but it also suggests that molecular regulatory networks in iPSCs may somewhat be different from hESCs that could subsequently have functional effects in the maintenance or downstream manipulation (for example differentiation into a certain lineage) of stem cells from different sources. Knowing these differences will greatly benefit us to design experimental strategy for future utilization of hESCs and iPSCs.
Figure 6. Comparison of sequence motifs and transcription factors associated with consensus genes from iPSCs and self-renewing hESCs.
To examine transcriptional hubs that could play critical roles in self-renewal of hESCs and iPSCs, we have performed promoter sequence analysis and identified sequence motifs of genes from the best consensus modules correlated to undifferentiated hESCs and iPSCs (RSAT:http://rsat.ulb.ac.be/). (A) The best representative modules from hESC and iPSC are shown in Venn diagram. Using the gene lists, we compared common sequence motifs of promoters of hESCs and iPSCs. Five unique sequence motifs were found from consensus modules from hESCs and iPSCs had five unique sequences that are closely correlated to their respective consensus modules (red for consensus modules associated with hESCs, yellow for consensus modules associated with iPSCs and orange for both) (B) Using the common sequence motifs identified in (A), transcription factors related to those sequence motifs were examined. The OCT family transcription factor was strongly correlated to consensus modules from self-renewing hESCs and AIRE, OTX and CF2 were strongly correlated to consensus modules from iPSCs. XFD, CROC, FOX, MEF, CRP and SPF transcription factors were shared between self-renewing hESCs and iPSCs
Discussion
Using gene expression microarray and WGCNA bioinformatics analysis, we have identified new self-renewal factors that are specific to stem cell differentiation from H1 and H9 human embryonic stem cells (hESCs). We constructed consensus modules to compare and validate our results with 19 publicly available microarray data that are specific to stem cell differentiation. These data sets included hESCs guided to differentiate into various somatic cells and lineages such as hemangioblasts, mesoderm, hepatocytes, hematopoietic cells and neural/mesenchymal cells 13–18. The variety of data sets were selected to enhance statistical power for consensus module formation but more importantly to find the set of genes responsible for the identity of a stem cells.
The major consensus module representing a set of genes that are consistently downregulated upon hESC differentiation across datasets includes some of the well-known players in stemness such as OCT4, and NANOG. In addition to these known markers, we have identified a total of 1,244 genes that are consistently downregulated upon hESC differentiation from our own microarray results as well as across the 19 microarrays analyzed. From this list of genes we have identified the top genes based on fold change and significance across datasets. These include genes such as LCK, KLKB1, SLC7A3, as well as others. Furthermore, we validated these genes across three independent hESC lines (H1, H9 and UCLA6) undergoing differentiation. A set of new genes we have identified could potentially be involved in maintenance of self-renewal in undifferentiated stage. Their individual functional role should be experimentally validated.
Next, we asked how OCT4, NANOG, c-MYC and SOX2 connected to other genes to get a glimpse into possible mechanism of these important genes in stem cell and differentiation, which is not yet known. Interestingly, the top genes that are most connected to the four stem cell factors may play a role in epigenetic, transcriptional regulation and male sex characteristics. This in part can be explained by the origin of embryonic stem cells. The human embryo reaches the blastocyst stage after 4–5 days after fertilization. Human embryonic stem cells are derived from the inner cell mass of a blastocyst 43. It is during this blastocyst stage that the embryo is going through major de novo methylation and is getting primed to go through sex determination and other dynamic changes in gene expression 44–46.
When we measured interconnectivities of the turquoise consensus module, we found that SLC7A3, NANOG, SOX2, KLKB1, OCT4 and LCK to be the major foci of downregulated genes upon differentiation, which means these factors are important in maintaining stem cell self-renewal while preventing differentiation. The importance of OCT4, SOX2 and NANOG in these roles has been well documented, but it is not much known about the self-renewing role of SLC7A3, KLKB1 and LCK that we identified from this study. LCK is a lymphocyte-specific protein tyrosine kinase. It is one of eleven members in SRC family kinase 47. Previous work has implicated the SRC family kinase in the self-renewal and differentiation of mouse and human embryonic stem cells 33,34,48,49. Meyn et al. have shown that level of LCK is highly expressed in undifferentiated mouse embryonic stem cells (mESCs) and decreases rapidly upon differentiation into EBs and we were able observed comparable changes in LCK expression during differentiation in hESCs 34. By treating hESCs with physiological concentration of LCK inhibitor (A-419259), we were able to decrease self-renewal capacity of hESCs with morphology of differentiated cells, decrease in ALP activity and OCT4 expression, which in turn suggest LCK plays an important role in SRC family kinase signal transduction that governs stem cell self-renewal. Interestingly, LCK inhibitor (A-419259) of same concentration failed to differentiate mouse embryonic stem cells 34. This result may imply (1) self-renewal capacity affected by LCK inhibitor is minimal in mESCs and significant in hESCs, and (2) mESCs may be able to compensate for decrease in LCK by some other parallel pathways. Although mESCs and hESCs share many basic factors and signaling mechanisms, our LCK inhibitor study and opposite differentiation outcome in presence of leukemia inhibitory factor (LIF) between mESCs and hESCs implicate there are differences in terms of the role LCK plays in self-renewal of these two stem cells. In addition to LCK inhibitor, we specifically knockdown LCK by siRNA and confirmed its role.
We further analyzed the major consensus module to better understand how the stem cell factors relate to each other as well as to the module. The HIVE plot allowed us to detect emerging patterns in our complex human embryonic stem cell’s network. Normalized connectivity of stem cell factors showed how close or far each factor is to one another. The significant overlap between OCT4 (Blue) and KLKB1 (Orange) affirms that KLKB1 could be a major player in differentiation of human embryonic stem cells. Furthermore it may imply a possible shared functionality between OCT4 and KLKB1.
We then examined if our findings using hESC model can be related to iPSCs. Unexpectedly, four Yamanaka factors were not the major hub genes of iPSCs. Although OCT4 and SOX2 did exist in the overall hub gene list, when the stringent cut off we used for hESCs was used for iPSCs, only SLC7A3, KLKB1 and LCK remained. Based on our functional analyses, interconnectivity measurements, transcription factor and sequence motif analyses, we could state that iPSCs may possess certain characteristics of hESCs, however there are distinct molecular differences.
Our analyses may provide an insight into specific pathways and a set of genes critical in hESC self-renewal. The genes discovered could also be used as a tool for other researchers to delineate stemness in a particular system. We examined how close iPSCs were to hESCs and found that 46% (573 genes) behaved the same between the two cell types when examining the top consensus module for hESCs. When the iPSCs top consensus module were compared to the hESCs top consensus module, the iPSCs had 3,178 unique genes suggesting that there may be other pathways in the background dependent upon their somatic cell of origin prior to induction. Detailed knowledge on these novel factors and molecular mechanisms associated with specific cell type or lineage specifications will help us design experimental strategy to improve success rate and efficiency in reprogramming or directed differentiation process.
WGCNA represents a powerful tool to help understand how genes are behaving across datasets and are correlated to each other within specific modules. We have utilized this tool to understand stemness across different datasets. From our analysis we have determined some of the top upregulated and downregulated genes in hESC differentiation that may be useful to researchers in gaining further insight into this field.
Materials and Methods
Cell Culture
Early passage hESC lines H1, H9 and UCLA6 (typically passage 34–36) were obtained from UCLA Stem Cell Core Facility and maintained on mouse embryonic fibroblast feeders in hESC growth medium (hESGM: DMEM/F12 supplemented with 20% Knockout Serum Replacer, 1% non-essential amino acids, 1mM L-glutamine, 0.1mM betamercaptoethanol and 4ng/ml bFGF). Cells were transferred to Matrigel coated dishes (BD Biosciences, San Jose, CA) and grown in mTeSR1 (Stem Cell Technologies, Vancouver, Canada) using standard conditions 50–52. For EB formation, cells were dissociated with Accutase (Stem Cell Technologies, Vancouver, Canada) and plated on ultra low attachment plates (Corning Life Science, Tewksbury, MA) in hESGM (without bFGF) with 10 μM Y-27632 (Chemdea, Ridgewood, NJ) for designated period of time. For adherent differentiation, cells were treated with human differentiation medium (DMEM/F12 supplemented with 20% FBS, 1% glutamate, 1% pyruvate, 1% non essential amino Acids) for 24 hr.
Immunofluorescence analysis
Cells were fixed in 100% methanol for 15 min at room temperature. For staining, samples were permeabilized for 15 min in freshly prepared IF staining solution (PBS containing 0.02% saponin, 1% bovine serum albumin, and 0.05% sodium azide, 0.2% Triton X-100). Samples were incubated in a 37°C water bath for 1h with 1μg/ml of primary antibody diluted in IF solution. Samples were transferred to a 1:500 dilution of Goat anti-mouse IgG Rhodamine (Thermo Scientific, Waltham, MA) in IF solution and incubated for in 37°C water bath for 1h. Processed were mounted on a glass slide with mounting medium with DAPI (Vectashield, Burlingame, CA) and visualized with an inverted light microscope (Olympus IX81 and CellSens Dimension software, Center Valley, PA).
Silencing of LCK in hESCs
Exponentially growing hESCs were collected by using Accutase (Stem Cell Technologies, Vancouver, Canada) and seeded on either cover slips or 6 well plates coated with Matrigel (BD Biosciences, San Jose, CA). Cells were cultured in mTeSR1 medium (Stem Cell Technologies, Vancouver, Canada) supplemented with 10 μM Y-27632 ROCK inhibitor (Chemdea, Ridgewood, NJ) for 24 hrs. On the day of transfection, cells were fed with fresh medium without ROCK inhibitor and transfected with 25 nM ON-TARGETplus human LCK siRNA SMARTpool or non-target scrambled siRNA control (Thermo Fisher Scientific, Pittsburgh, PA) by using DharmaFECT1 (Thermo Fisher Scientific, Pittsburgh, PA). After 48 hr transfection, cells were collected for RNA isolation or fixed with 4% paraformaldehyde/PBS for IF analysis.
Bioinformatics analysis
WGCNA was performed by following the tutorial written by Langfelder and Horvath 19,53. CEL files (Raw data from the UCLA microarray core) were normalized to Affymetrix internal controls by creating MAS5 files using publically available R code from MIT (http://jura.wi.mit.edu/bio/education/bioinfo2007/arrays/array_exercises_1R.html). We pre-processed the MAS5 files to check for excessive missing data values and to identify outliers in the samples according to the WGCNA manual 19,53. All data files have been deposited at the GEO repository (GSE54186). Unsigned co-expression networks were built using the WGCNA package in R software. Clusters of genes that behaved similarly were grouped together into different color modules. These modules were related to specific traits. In heatmaps, red represents genes upregulated within that dataset and green represents genes downregulated within that dataset. The top 1000 connections within a gene network were determined by WGCNA. For the multiple array consensus analysis, we performed WGCNA on individual data set first as suggested by the Langfelder and Horvath’s tutorial. Using “1 step function for network construction and detection of consensus modules”, we chose a default WGCNA soft thresholding power β which co-expression was raised to calculate adjacency of each data set. The soft thresholding power β allowed us to compare each data set by approximate scale-free topology to compensate for scale differences between data sets (Consensus normalization) 19,53.
Cytoscape 2.8.3 was used in making the network and determining the top hub genes within the network 54. For the interconnectivity graphs, we used spring embedded and circular layouts. We installed CytoHubba plugin for more advance network analysis features. The significant modules were annotated by DAVID Functional Annotation Bioinformatics Microarray Tools 55. The differentially expressed gene list was uploaded onto “Enter the gene list” window in DAVID. Official gene symbol was selected as “Identifier”. “Gene list” was selected as List type. After the list was submitted and analyzed, “Pathways” was selected from the Annotation summary results. Under “Pathways” KEGG_Pathway was selected. To generate HIVE Plot, HIVE 0.0.11 was downloaded and ran on Mac OS X 10.7.4. Edges and nodes were normalized to uploaded data.
Supplementary Material
Acknowledgments
We would like to thank Dr. Steve Horvath for his help with his tutorials for WGCNA. We acknowledge the UCLA Clinical Microarray Core for excellent technical service. We also acknowledge the UCLA Broad Stem Cell Institute for providing us with hESC lines. This work was supported by CIRM Basic Biology Award II (RB2-01562), NIH/NIAAA R01 grant award 1R01AA21301) and UCLA SOD SEED Grant to Y.K. and T32 training grant award from NIDCR (T32DE07269 to J.J.K. and O.K.).
Footnotes
Contribution
J.J.K., O.K., and Y.K. participated in designing experiments, execution of described works, statistical analysis, interpretation of data and writing the manuscript.
A.H.N., T.G.T., O.E. and C.L. contributed to the execution of experiments and writing the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Schroeder IS. Potential of pluripotent stem cells for diabetes therapy. Curr Diab Rep. 2012;12:490–498. doi: 10.1007/s11892-012-0292-5. [DOI] [PubMed] [Google Scholar]
- 2.Lindvall O, Kokaia Z, Martinez-Serrano A. Stem cell therapy for human neurodegenerative disorders-how to make it work. Nat Med. 2004;10 (Suppl):S42–50. doi: 10.1038/nm1064. [DOI] [PubMed] [Google Scholar]
- 3.Segers VF, Lee RT. Stem-cell therapy for cardiac disease. Nature. 2008;451:937–942. doi: 10.1038/nature06800. [DOI] [PubMed] [Google Scholar]
- 4.Hu Q, Friedrich AM, Johnson LV, Clegg DO. Memory in induced pluripotent stem cells: reprogrammed human retinal-pigmented epithelial cells show tendency for spontaneous redifferentiation. Stem cells. 2010;28:1981–1991. doi: 10.1002/stem.531. [DOI] [PubMed] [Google Scholar]
- 5.Stewart MH, et al. Clonal isolation of hESCs reveals heterogeneity within the pluripotent stem cell compartment. Nature methods. 2006;3:807–815. doi: 10.1038/nmeth939. [DOI] [PubMed] [Google Scholar]
- 6.Wu J, Tzanakakis ES. Contribution of stochastic partitioning at human embryonic stem cell division to NANOG heterogeneity. PloS one. 2012;7:e50715. doi: 10.1371/journal.pone.0050715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graf T, Stadtfeld M. Heterogeneity of embryonic and adult stem cells. Cell stem cell. 2008;3:480–483. doi: 10.1016/j.stem.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Bhatia S, Pilquil C, Roth-Albin I, Draper JS. Demarcation of stable subpopulations within the pluripotent hESC compartment. PloS one. 2013;8:e57276. doi: 10.1371/journal.pone.0057276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 10.Feng Q, et al. Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem cells. 2010;28:704–712. doi: 10.1002/stem.321. [DOI] [PubMed] [Google Scholar]
- 11.Wang A, et al. Functional modules distinguish human induced pluripotent stem cells from embryonic stem cells. Stem cells and development. 2011;20:1937–1950. doi: 10.1089/scd.2010.0574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowry WE. Does transcription factor induced pluripotency accurately mimic embryo derived pluripotency? Current opinion in genetics & development. 2012;22:429–434. doi: 10.1016/j.gde.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu SJ, et al. GeneChip analysis of human embryonic stem cell differentiation into hemangioblasts: an in silico dissection of mixed phenotypes. Genome Biol. 2007;8:R240. doi: 10.1186/gb-2007-8-11-r240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evseenko D, et al. Mapping the first stages of mesoderm commitment during differentiation of human embryonic stem cells. Proc Natl Acad Sci U S A. 2010;107:13742–13747. doi: 10.1073/pnas.1002077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li SS, et al. Target identification of microRNAs expressed highly in human embryonic stem cells. J Cell Biochem. 2009;106:1020–1030. doi: 10.1002/jcb.22084. [DOI] [PubMed] [Google Scholar]
- 16.Si-Tayeb K, et al. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yung S, et al. Large-scale transcriptional profiling and functional assays reveal important roles for Rho-GTPase signalling and SCL during haematopoietic differentiation of human embryonic stem cells. Hum Mol Genet. 2011;20:4932–4946. doi: 10.1093/hmg/ddr431. [DOI] [PubMed] [Google Scholar]
- 18.Denis JA, Rochon-Beaucourt C, Champon B, Pietu G. Global transcriptional profiling of neural and mesenchymal progenitors derived from human embryonic stem cells reveals alternative developmental signaling pathways. Stem cells and development. 2011;20:1395–1409. doi: 10.1089/scd.2010.0331. [DOI] [PubMed] [Google Scholar]
- 19.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Discher DE, Mooney DJ, Zandstra PW. Growth factors, matrices, and forces combine and control stem cells. Science. 2009;324:1673–1677. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horvath S, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci U S A. 2006;103:17402–17407. doi: 10.1073/pnas.0608396103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White J, Dalton S. Cell cycle control of embryonic stem cells. Stem Cell Rev. 2005;1:131–138. doi: 10.1385/SCR:1:2:131. [DOI] [PubMed] [Google Scholar]
- 23.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 24.Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3:895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- 25.Valent P, et al. Heterogeneity of Neoplastic Stem Cells: Theoretical, Functional, and Clinical Implications. Cancer Res. 2013 doi: 10.1158/0008-5472.CAN-12-3678. [DOI] [PubMed] [Google Scholar]
- 26.Kim CF, Dirks PB. Cancer and stem cell biology: how tightly intertwined? Cell stem cell. 2008;3:147–150. doi: 10.1016/j.stem.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 27.Diaz Perez SV, et al. Derivation of new human embryonic stem cell lines reveals rapid epigenetic progression in vitro that can be prevented by chemical modification of chromatin. Hum Mol Genet. 2012;21:751–764. doi: 10.1093/hmg/ddr506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghazalpour A, et al. Integrating genetic and network analysis to characterize genes related to mouse weight. PLoS Genet. 2006;2:e130. doi: 10.1371/journal.pgen.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gargalovic PS, et al. Identification of inflammatory gene modules based on variations of human endothelial cell responses to oxidized lipids. Proc Natl Acad Sci U S A. 2006;103:12741–12746. doi: 10.1073/pnas.0605457103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oldham MC, Horvath S, Geschwind DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc Natl Acad Sci U S A. 2006;103:17973–17978. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carlson MR, et al. Gene connectivity, function, and sequence conservation: predictions from modular yeast co-expression networks. BMC Genomics. 2006;7:40. doi: 10.1186/1471-2164-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krzywinski M, Birol I, Jones SJ, Marra MA. Hive plots--rational approach to visualizing networks. Briefings in bioinformatics. 2012;13:627–644. doi: 10.1093/bib/bbr069. [DOI] [PubMed] [Google Scholar]
- 33.Meyn MA, 3rd, Schreiner SJ, Dumitrescu TP, Nau GJ, Smithgall TE. SRC family kinase activity is required for murine embryonic stem cell growth differentiation. Molecular pharmacology. 2005;68:1320–1330. doi: 10.1124/mol.104.010231. [DOI] [PubMed] [Google Scholar]
- 34.Meyn MA, 3rd, Smithgall TE. Chemical genetics identifies c-Src as an activator of primitive ectoderm formation in murine embryonic stem cells. Science signaling. 2009;2:ra64. doi: 10.1126/scisignal.2000311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Theus MH, Wei L, Francis K, Yu SP. Critical roles of Src family tyrosine kinases in excitatory neuronal differentiation of cultured embryonic stem cells. Experimental cell research. 2006;312:3096–3107. doi: 10.1016/j.yexcr.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 36.Sun Y, et al. Evolutionarily conserved transcriptional co-expression guiding embryonic stem cell differentiation. PloS one. 2008;3:e3406. doi: 10.1371/journal.pone.0003406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arnold LD, et al. Pyrrolo[2,3-d]pyrimidines containing an extended 5-substituent as potent and selective inhibitors of lck I. Bioorganic & medicinal chemistry letters. 2000;10:2167–2170. doi: 10.1016/s0960-894x(00)00441-8. [DOI] [PubMed] [Google Scholar]
- 38.Marchetto MC, et al. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PloS one. 2009;4:e7076. doi: 10.1371/journal.pone.0007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chin MH, Pellegrini M, Plath K, Lowry WE. Molecular analyses of human induced pluripotent stem cells and embryonic stem cells. Cell stem cell. 2010;7:263–269. doi: 10.1016/j.stem.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim K, et al. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol. 2011;29:1117–1119. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lapasset L, et al. Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes Dev. 2011;25:2248–2253. doi: 10.1101/gad.173922.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma Yu, CL, Gu1 Junjie, Tang Fan, Li Chun, Li Peng, Ping Ping, Yang Shi, Li Zheng, Jin Ying. Aberrant gene expression profiles in pluripotent stem cells induced from fibroblasts of a Klinefelter’s Syndrome patient. The Journal of Biological Chemistry. 2012 doi: 10.1074/jbc.M112.380204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 44.Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- 45.Howlett SK, Reik W. Methylation levels of maternal and paternal genomes during preimplantation development. Development. 1991;113:119–127. doi: 10.1242/dev.113.1.119. [DOI] [PubMed] [Google Scholar]
- 46.Rougier N, et al. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev. 1998;12:2108–2113. doi: 10.1101/gad.12.14.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 48.Anneren C, Cowan CA, Melton DA. The Src family of tyrosine kinases is important for embryonic stem cell self-renewal. J Biol Chem. 2004;279:31590–31598. doi: 10.1074/jbc.M403547200. [DOI] [PubMed] [Google Scholar]
- 49.Betts GN, et al. Prospective technical validation and assessment of intra-tumour heterogeneity of a low density array hypoxia gene profile in head and neck squamous cell carcinoma. Eur J Cancer. 2013;49:156–165. doi: 10.1016/j.ejca.2012.07.028. [DOI] [PubMed] [Google Scholar]
- 50.Xu C, et al. Feeder-free growth of undifferentiated human embryonic stem cells. Nat Biotechnol. 2001;19:971–974. doi: 10.1038/nbt1001-971. [DOI] [PubMed] [Google Scholar]
- 51.Li Y, Powell S, Brunette E, Lebkowski J, Mandalam R. Expansion of human embryonic stem cells in defined serum-free medium devoid of animal-derived products. Biotechnol Bioeng. 2005;91:688–698. doi: 10.1002/bit.20536. [DOI] [PubMed] [Google Scholar]
- 52.Hakala H, et al. Comparison of biomaterials and extracellular matrices as a culture platform for multiple, independently derived human embryonic stem cell lines. Tissue Eng Part A. 2009;15:1775–1785. doi: 10.1089/ten.tea.2008.0316. [DOI] [PubMed] [Google Scholar]
- 53.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- 54.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dennis G, Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.