Abstract
Eukaryotic cells are equipped to degrade proteins via the ubiquitin–proteasome system (UPS). Proteins become degraded upon their conjugation to chains of ubiquitin where they are then directed to the 26S proteasome, a macromolecular protease. The transfer of ubiquitin to proteins and their subsequent degradation are highly complex processes, and new research is beginning to uncover the molecular details of how ubiquitination and degradation take place in the cell. We review some of the new data providing insights into how these processes occur. Although distinct mechanisms are often observed, some common themes are emerging for how the UPS guides protein substrates through their final journey.
Keywords: ubiquitin proteasome system, proteolysis, E1-activating, E2-conjugating, E3-ligase
The ubiquitin system: as easy as E1, E2, E3?
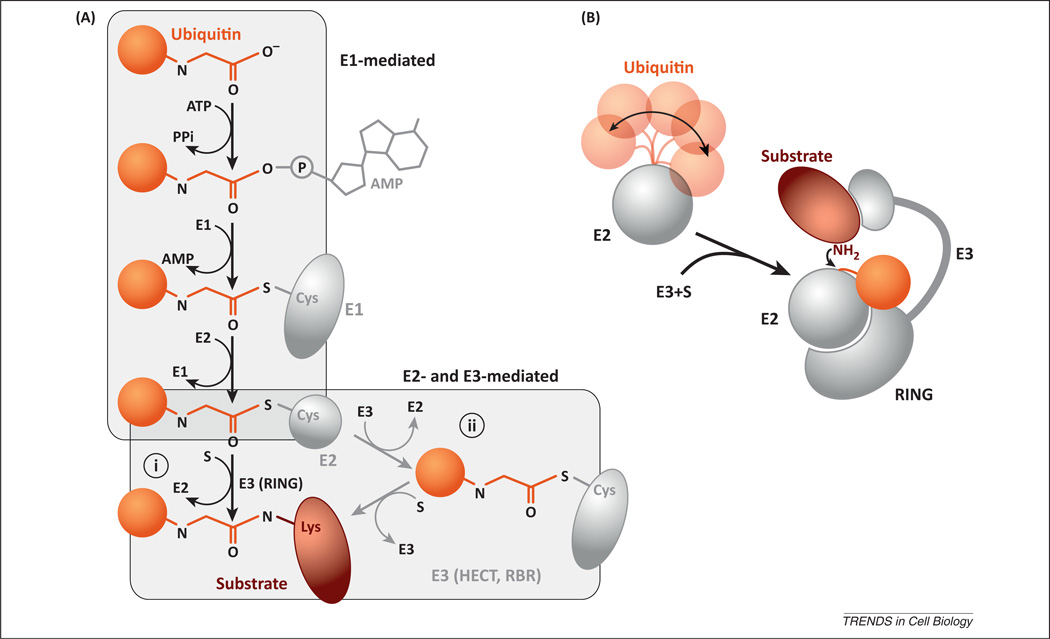
The ubiquitination of proteins involves the hierarchical action of three general families of ubiquitin enzymes (Figure 1A). An E1 enzyme must first activate ubiquitin, a highly conserved, 76 amino acid polypeptide, in an ATP-dependent manner. The E1 forms a covalent bond between the C-terminal end of ubiquitin and a cysteine residue in its active site. The thioesterified ubiquitin passes from the E1 active site to the next member of the cascade, the E2 or ubiquitin-conjugating enzyme. Finally, the E3 ubiquitin ligase binds to both the E2-bound ubiquitin and the protein substrate, promoting the transfer of ubiquitin onto the substrate. Note that in the mammalian ubiquitin E1–E2–E3 cascade, only two members of the E1 family are necessary to tag all E2s with ubiquitin, and the approximately 40 E2s that exist are sufficient to deliver ubiquitin to the more than 600 known E3s [1].
Figure 1.
(A) Schematic representation of the different modifications occurring at the carboxyl end of ubiquitin during ubiquitination. (i) RING (really interesting new gene) and RING-like E3s mediate the direct transfer of ubiquitin from the E2 onto the substrate; (ii) an additional trans-thioesterification step is mediated by HECT (homologous to E6-AP carboxyl terminus) and RBR (ring between ring) E3s before substrate ubiquitination. (B) Model for substrate ubiquitination mediated by RING and RING-like E3s. The conformation of ubiquitin on the E2 can be labile due to its flexible tail. Binding of the E2~ubiquitin (linked via a thioester bond) to the E3 serves to fasten the ubiquitin and its carboxyl tail against the E2, thereby accelerating the rate of ubiquitin transfer to substrate.
In general, proteins are ubiquitinated on lysine residues, where an isopeptide bond forms between the carboxyl end of ubiquitin and the lysine primary amine. Proteins can either be conjugated to one ubiquitin monomer (typically referred to as mono-ubiquitination) or to several ubiquitins to form a poly-ubiquitin chain. Ubiquitin contains seven lysine residues as well as a free amino end, and the primary amines associated with these moieties can all participate as acceptors of additional ubiquitins, thereby enabling the formation of poly-ubiquitin chains with different linkage types. Indeed, all eight of these linkages have been observed in cells [2,3]. To add to the complexity, ubiquitin does not always signal for protein degradation. For instance, ubiquitin can recruit other factors to mediate various cellular responses such as signaling, gene regulation, endocytosis, macro-autophagy, and DNA repair. The partitioning of a ubiquitinated substrate between these cellular responses or degradation is determined by both the protein ubiquitination state (mono- or poly-ubiquitinated) and by the chain linkage type (Box 1) [4].
Box 1.
Ubiquitin chain linkage can influence the fates of modified proteins
The types of linkages for poly-ubiquitin chains can dramatically influence the fate of the protein it is conjugated to [4]. Although it was established early on that chains linked via lysine 48 on ubiquitin target those substrates to the proteasome [41], most linkage types are now known to also mediate proteasomal degradation. Several proteomic analyses were performed to quantify the percentages of ubiquitin chain linkages present in the cell as well as to identify those that accumulate upon inhibition of the proteasome [2,78]. Surprisingly, 30% of the ubiquitin chains in yeast cells were found to be linked through lysine 11 and that this chain type rapidly accumulated upon proteasome inhibition, in both yeast and in human cells. In agreement with these results, previous work showed that the master cell-cycle regulator APC (anaphase-promoting complex), an E3 ligase, promotes the formation of lysine 11 linkages required for the degradation of APC substrates [79]. Similarly, lysine 29 linkages have also been shown to accumulate upon proteasome inhibition, and several E3 ligases have been identified that assemble lysine 29 linked chains that target their substrates to the proteasome for degradation [80,81]. By contrast, lysine 63 linkages make up over one-third of the ubiquitin chains in human cells but are not immediately affected by proteasome inhibition [78]. Indeed, lysine 63 poly-ubiquitin chains are generally not thought to signal for proteasomal degradation in the cell, although a few exceptions have been noted [82]. However, it is not yet clear why this occurs. For example, the proteasome receptors that bind to ubiquitinated substrates show little preference for ubiquitin chain linkage type when isolated from the proteasome; however, purified proteasomes bind preferentially to lysine 48 chains as compared to lysine 63 chains when in the presence of cellular lysate [83]. One possibility is that proteins conjugated to lysine 63 chains rapidly become shielded in the presence of ubiquitin chain binding cofactors. Interestingly, ubiquitin chains are sometimes shortened by deubiquitinases (DUBs) at the proteasome, and lysine 63 chains were shown to be deubiquitinated faster than lysine 48 chains [84]. This may provide additional specificity for the discrimination of which proteins will be degraded in the cell.
As noted above, the size and scope of the repertoire of genes necessary to run the UPS are immense, and there are also 16 ubiquitin-like proteins whose conjugation to protein substrates is controlled by structurally related families of E1, E2, and E3 enzymes [5]. As an example, the ubiquitin-like protein Nedd8 (neural precursor cell expressed, developmentally downregulated gene 8) shares approximately 58% sequence identity with ubiquitin. The conjugation of Nedd8 to the cullin–RING (really interesting new gene) ligases, the largest family of E3s in the human proteome, is responsible for the regulation of their activity [1]. Although substantial structural similarity exists between ubiquitin and the ubiquitin-like proteins, the E1 and E2 enzymes display exquisite selectivity for their cognate substrates, thus avoiding unwanted crosstalk between the ubiquitin machinery and the related ubiquitin-like systems under physiological conditions.
Given the sheer scale of the UPS, it is not surprising that much of the early work on the pathway was centered on the discovery of the protein factors and their roles in ubiquitination and degradation. More recently, many groups in the ubiquitin field have been focusing their efforts on uncovering the molecular details for how proteins in the UPS function. The goal of this review is to summarize some of those findings.
In the beginning: ubiquitin-activating enzymes (E1)
E1s are multidomain enzymes that must activate ubiquitin and efficiently transfer it to the E2 active site. This function is crucial for cellular homeostasis because failure to activate ubiquitin, as seen by the chemical inhibition of E1 activity in the cell, results in the almost immediate shutdown of the entire UPS [6]. Recent work has added to the understanding of the activation and transfer of ubiquitin (or ubiquitin-like proteins) to E2s by E1, thereby making it the best understood process in the UPS [7–13].
The E1-catalyzed reaction involves multiple molecular events, including the ATP-dependent adenylation of the C-terminal carboxyl group of ubiquitin, the formation of a thioester bond between the E1 catalytic cysteine residue and the C-terminus of ubiquitin, and the transfer of ubiquitin onto the E2 catalytic cysteine (Figure 1A). The structural analyses of either E1s for ubiquitin or for the ubiquitin-like proteins Nedd8 and SUMO (small ubiquitin-like modifier) revealed that multiple conformational changes occur during the reaction cycle.
For instance, structural studies of E1s in the presence of adenylated ubiquitin or ubiquitin-like proteins showed conformations where the distances between the E1 cysteine residues that form thioester bonds with either ubiquitin or ubiquitin-like proteins were far too great for this reaction to occur [11–13], suggesting that additional conformational changes would be required. Capturing the short-lived transition state intermediate necessitated the development of chemical methods to trap the enzyme in the proper conformation for thioester formation; however, this challenging work has led to a large increase in the understanding of how E1s function. Indeed, the recent crystallization of the SUMO E1 in complex with a reactive mimic of adenylated SUMO revealed a significant rearrangement of the E1 enzyme domain architecture. These large conformational changes bring the E1 cysteine to the adenylated SUMO moiety and serve to remodel the composition of the active site which reprograms the enzyme to catalyze thioester bond formation rather than SUMO adenylation.
Large conformational changes also occur during the transfer of ubiquitin from the E1 to the E2 active site. For example, it was shown that thioester formation between E1 and Nedd8 causes a large conformational change in the position of the E2-binding domain on the E1 (named the ubiquitin fold domain, UFD) that facilitates E2 binding [7]. Furthermore, a recent ubiquitin-specific E1–E2 co-crystal structure that was stabilized by disulfide crosslinking, as well as the molecular modeling of the transition between the E2 unbound and bound states, were consistent with a mechanism of ubiquitin transfer involving conformational changes [9]. Specifically, upon binding of the E2, the E1 UFD further rotates to juxtapose the E2 active site adjacent to the E1 active site that is conjugated to ubiquitin. The conformational flexibility of the UFD domain may assist in the ability of E1 to accommodate the binding and transfer of ubiquitin or ubiquitin-like proteins to multiple E2s [14]. Indeed, the E1 mechanism must be flexible enough to accommodate the approximately 40 different human E2s, and recent biochemical studies support this contention, demonstrating that the rate of ubiquitin transfer from E1 to E2 is relatively insensitive to the identity of E2 [15]. In conclusion, these structural studies on the mechanism of action of E1 enzymes are uncovering specific conformations that are crucial to E1 function; this information will undoubtedly be useful towards the ongoing development of therapeutics that target E1s [16].
Passing the baton: E2 ubiquitin-conjugating enzymes and E3 ubiquitin ligases
Ubiquitin ligases bind to both a protein substrate and the ubiquitin-conjugating enzyme that is carrying the ubiquitin cargo (commonly written as E2~ubiquitin). In this simple model, E3s act similarly to other protein scaffolds, bringing into proximity the substrate and E2~ubiquitin. However, earlier biochemical investigations on the SCF (Skp, Cullin, F-box containing) ubiquitin ligase complex, the archetypal member of the cullin–RING ligases, indicated that E3s can also positively influence the rate of ubiquitin transfer from the E2 to the protein substrate [17,18]. Indeed, recent studies have provided insight into the mechanism of E3-mediated activation of E2s by trapping and either co-crystallizing or characterizing by nuclear magnetic resonance (NMR) the unstable and transient complexes between E2~ubiquitin and E3. These studies demonstrated that both E2s and E3s can affect the conformation of the ubiquitin on the E2 surface to promote its transfer to the substrate [19–21].
Although the E3s recognize and bind to the protein substrate, E2s bear the responsibility for catalyzing the transfer of ubiquitin to the substrates for almost all ubiquitination reactions in the cell, with a few exceptions noted below. The core of the E2 active site is the thioester bond between the C-terminal end of ubiquitin and the conserved E2 cysteine residue (Box 2). The rate of ubiquitin transfer therefore depends on a mechanism where the thioester bond is positioned in an optimal conformation for the reaction to occur and, in theory, could be accomplished by the E2, the E3, or a combination.
Box 2.
How E2s catalyze ubiquitin transfer and promote chain linkage specificity
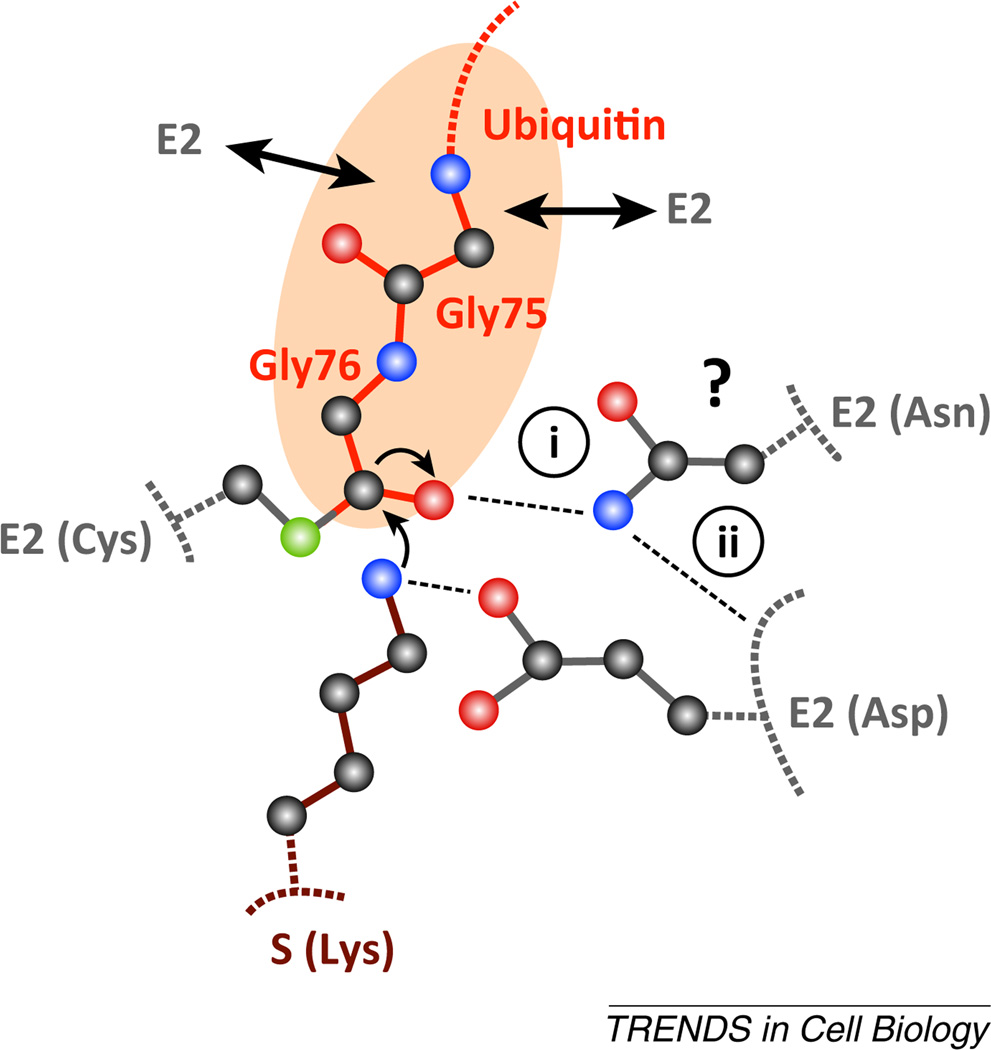
Isopeptide bond formation between lysine (on either protein substrate or on ubiquitin; denoted S in Figure I) and the E2~ubiquitin is catalyzed by several conserved residues in the E2 active site. The reaction proceeds through a tetrahedral intermediate where the carbonyl oxygen (shown as a red sphere) of the thioesterified terminal glycine of ubiquitin forms an oxyanion. For a long time, it was assumed that the E2 mechanism of catalysis would involve a stabilizing interaction between the oxyanion and a residue on the E2 [(i) in Figure I]. Cecile Pickart and colleagues were the first to demonstrate the functional importance of an invariably conserved asparagine residue in the active site, and its mutation caused severe defects in the ability of E2 to promote catalysis [85]. More recently, several structural studies have elucidated the molecular role for this residue [19,86,87]. Rather than having a direct role in stabilizing the oxyanion, the asparagine forms hydrogen bonds with an adjacent loop on the E2 that contains several residues that are involved in catalysis (ii) [86]. In particular, a conserved aspartic acid residue located in this loop in Ubc9 (the E2 responsible for SUMO conjugation to proteins) was identified [87]. Together with additional conserved residues, this aspartate forms a microenvironment that promotes the deprotonation of the substrate lysine, a prerequisite step before isopeptide bond formation can occur. Thus, E2 catalysis is promoted in at least two ways: by restricting the conformation of the thioester bond (see section on E2s and E3s; double arrows in the Figure I represent the presence of multiple stabilizing interactions between the ubiquitin C-terminal tail and E2) and by aiding in the deprotonation of substrate lysine.
Furthermore, it is of interest to understand how E2s select lysine residues on ubiquitin to form specific chain linkage types. For instance, Ubc13 is an E2 that generates ubiquitin chains with lysine 63 specificity. The mechanism of lysine selectivity involves an additional protein subunit, Mms2 (methyl methanesulfonate sensitive 2 transactivation; also known as ubiquitin-conjugating enzyme E2 variant 2, Ube2v2), which binds to both ubiquitin and Ubc13 and orients the ubiquitin such that lysine 63 is within close proximity of the Ubc13 active site [88]. Another example is Ube2S, an E2 that generates lysine 11 chains. Here, a TEK box motif on ubiquitin forms primarily electrostatic interactions with a complementary surface on Ube2S such that lysine 11 is presented to the active site [24].
It was originally shown that the E2 Ubc1 (ubiquitin-conjugating enzyme E2 1) can form a non-covalent interface with a ubiquitin that is thioesterified to the active site [22]. Recent work has shown that the E2s Cdc34 (cell division cycle 34) and Ube2S (ubiquitin-conjugating enzyme E2S) also form noncovalent interfaces with ubiquitin in addition to the covalent thioester bond [23,24], suggesting that E2s catalyze ubiquitin transfer at least in part by holding ubiquitin against an interface on the E2 surface that optimizes the position of the thioester bond in the active site. Even when considering these findings, the tether between the E2~ubiquitin conjugate was shown to be fairly flexible, in at least two cases (Figure 1B) [25], such that optimal placement of the thioester bond in the E2 active site may necessitate the interaction of additional factors with the ubiquitin.
Some E3s can also stabilize the conformation between ubiquitin and the E2, thereby accelerating further the rate of conjugation of ubiquitin to proteins. Note that most E3s possess a small subunit named RING, which recruits the E2 enzyme. Indeed, three recent structural studies observed the E2~ubiquitin conformation in the presence of either RING domains or the structurally related U-box domain [19–21]. In all the structures, ubiquitin was found to contact part of the E3, consequently stabilizing its position on the E2 (Figure 1B). Additional elements on the E3 may also aid ubiquitin binding in a regulated manner. For example, a phosphorylated residue located in a domain adjacent to the RING domain of the CBL-B (Casitas B lineage lymphoma) ubiquitin ligase was also found to interact with ubiquitin [26]. In summary, mechanisms of E2 activation involve the binding of ubiquitin to surfaces on the E2 and/or E3, thereby allowing an optimal conformation of ubiquitin on the E2 surface.
Despite recent work uncovering the role of the RING domain in ubiquitin binding, not all E3 families contain RING domains, and recent structural work on the HECT (homologous to E6-AP carboxyl terminus) and RBR (ring between ring) E3s have begun to elucidate the mechanisms of action of non-RING-dependent ubiquitin binding to E3. Both HECT and RBR E3s are unique in comparison to the RING ligases because they catalyze an additional trans-thioesterification step, where ubiquitin is transferred from the E2 to the E3 before its conjugation to the protein substrate (Figure 1A). Several HECT ligase structures were found to display different conjugation steps [27–32], indicating that these events are accompanied by striking domain rearrangements. The HECT domain is divided into two structural elements, the N- and C-lobes, connected by a flexible linker. The E2~ubiquitin first binds to the N-lobe to transfer ubiquitin onto the catalytic cysteine residue located on the C-lobe, which causes the C-lobe~ubiquitin to rotate approximately 130 degrees towards the substrate lysine residue [27]. In this new conformation, a large binding site for ubiquitin forms, locking its flexible carboxyl tail into place, and catalyzing the reaction through a mechanism similar to those described above. Interestingly, recent structural studies on the RBR E3s also revealed that large conformational changes are likely to occur during catalysis, suggesting a common theme for how these related enzymes work [33–37]. One can speculate a biological rationale for why maximal E2 activity only occurs in the presence of E3s: it may reduce the chance that free E2~ubiquitin (whose concentrations may be as high as 10 µM in the cell for some E2s) [38,39] will ubiquitinate proteins that are not true substrates of the UPS, and thereby minimize non-productive hydrolysis of the E2~ubiquitin conjugate.
In addition to using structural biology to probe the mechanism of action of E2s and E3s, classical enzymological methods have recently been employed to understand better the ubiquitination reaction. Determining the kinetic parameters of the ubiquitination reaction is also biologically relevant because the outcome of ubiquitination is partially controlled by the number of ubiquitins added onto the substrate (note that poly-ubiquitin chains must contain a minimum of four ubiquitins to be efficiently degraded by the proteasome) [40,41]. Determining the mechanism of ubiquitin chain formation would ideally involve the detection of each transfer event; however, because of the fast nature of ubiquitination reactions, the quantitative examination of these events may be difficult. To overcome this difficulty, researchers used a quench flow apparatus that could detect product formation at the millisecond time-scale [42] and determined that, once the first ubiquitin is transferred to the substrate, the rate of substrate dissociation from E3 is nearly always sufficiently slow to allow the building of ubiquitin chains competent for proteasomal degradation. One key take-home message is that many E3 ligases likely evolved to be processive, meaning that they can conjugate multiple ubiquitins onto a substrate before it is released from enzyme. This understanding is important because the ubiquitinated substrate will be exposed to deubiquitinases (see below) upon its dissociation from the E3, and an inadequately ubiquitinated substrate may risk never achieving the four-ubiquitin threshold needed for its targeting to the proteasome.
Born under a bad sign: potential targets of the UPS
Since the discovery of cyclin-B as a substrate of the UPS, and the role of its degradation in controlling cell cycle progression [43], the ubiquitin field has been traditionally focused on proteasome substrates that are targeted for proteolysis for regulatory purposes. However, a growing body of evidence suggests that scores of ubiquitinated substrates destined for proteasome-mediated degradation are presumably misfolded proteins (typically corresponding to abundant proteins) [44,45]. Misfolded proteins can aggregate with time, and protein quality-control pathways are tasked with their ubiquitination and subsequent degradation [46,47]. In the cytosol, ribosomes coexist with protein chaperones (that attempt to assist in protein folding) as well as proteasomes, and as such all newly synthesized proteins are potential targets for proteolysis. Indeed, earlier reports suggest that as many as 30% of newly synthesized proteins are degraded (presumably due to misfolding) in dividing cells grown in tissue culture [48]. More recent studies are also in alignment with past findings suggesting that 2–15% of ribosome-bound polypeptides become ubiquitinated [49,50]. Interestingly, the ubiquitination of nascent proteins is more prominent for longer polypeptides, and it was recently proposed that ribosome-associated chaperones may at least partially shield misfolded segments from the ubiquitination machinery when exiting the ribosome [50].
It is not yet clear how the folding and degradation systems cooperate; however, a balance must be struck between the rapid removal of misfolded proteins versus those that may achieve the correct fold if given enough time. It was proposed that the kinetic partitioning of proteins between the folding and degradation systems may be key for the triaging of misfolded proteins [51]. It can be speculated then that the rate of poly-ubiquitin chain formation may be crucial in determining the fate of a misfolding protein. For example, the formation of poly-ubiquitin chains on regulatory proteins typically occurs at a fast rate [42], and these effectively serve as an irreversible targeting signal for degradation. By contrast, the formation of poly-ubiquitin chains on some misfolded proteins may occur at much slower rates (presumably due to the absence of a consensus E3-binding sequence motif), and this could then provide additional time for folding and/or ubiquitin chain editing to take place.
On the path to destruction: a rocky road may lie ahead
The assembly of a ubiquitin chain onto a protein substrate is necessary but not always sufficient for its degradation. Some ubiquitinated substrates require additional factors that can assist either in the disassembly of proteins in complex with the substrate or in the unfolding of substrate during degradation. For example, yeast Cdc48 (p97 in mammalian cells) is a conserved multisubunit enzyme that plays a major role in dissociating ubiquitinated proteins from their binding partners to promote their degradation by the proteasome [52]. Indeed, much effort has been put into discovering substrates of the UPS that depend on Cdc48/p97 activity, and this growing list includes star molecules such as RNA polymerase and stalled nascent polypeptides attached to ribosomes [53–55]. Cdc48/p97 generally acts downstream of ubiquitin ligases, although its activity may also promote ubiquitination in some cases; a major challenge is to delineate further the mechanism of action of this multipurpose enzyme.
Some substrates of the proteasome also depend on various shuttling factors that ensure that the substrates will safely arrive at the proteasome instead of unwanted outcomes such as aggregation with other proteins. For example, proteins with exposed hydrophobic domains, that are at risk of either aggregation or binding nonspecifically to other proteins, bind to the mammalian Bag6/Bat3 (BCL2-associated athanogene 6/HLA-B-associated transcript 3) ‘holdase’ chaperone. Specifically, Bag6 recognizes and binds to long hydrophobic stretches of the polypeptide chain of misfolded proteins targeted by the ERAD (endoplasmic reticulum associated protein degradation) pathway [56–58]. In addition, Bag6 also mediates the degradation of newly translated proteins that normally reside in the plasma membrane but have become mislocalized [59]. Because Bag6 interacts with both E3 ligases as well as the proteasome [57,58,60], it may promote both the ubiquitination and delivery of misfolded proteins to the proteasome while preventing their aggregation. Many other proteins are known to associate with the proteasome [61], and we suspect that some of these may also assist the proteasome with substrate processing before proteolysis in a manner similar to Bag6.
Although the formation of a ubiquitin chain on a protein substrate may target its degradation by the proteasome, cells contain many isopeptidases that can quickly ruin the hard work of an E3. These enzymes, referred to as deubiquitinases (DUBs), cleave ubiquitin from conjugated proteins or edit ubiquitin chains by trimming their lengths [62]. The seemingly subtle distinction of the length of a poly-ubiquitin chain can have profound consequences when considering the ability of E3 to assemble the ubiquitin chain within a particular time-frame. A recently developed complex, reconstituted in vitro system that recapitulates the degradation of transmembrane proteins showed that a small difference in the binding affinity between a substrate and its E3 ligase can have a large effect on chain length when in the presence of DUBs. Therefore, only higher-affinity substrates acquire the necessary ubiquitin chain length for targeting to the proteasome [63].
DUBs can also associate with the proteasome and may participate in the ‘molecular triage’ of ubiquitinated proteins. It was initially believed that the presence of DUBs at the proteasome (such as Uch37/UchL5 and Usp14) would promote the degradation of proteins through facilitating substrate processing (see also the next section). Indeed, the chemical inhibition of these DUBs was recently shown to impair the degradation of protein substrates by the proteasome [64]. However, recent studies have shown that, by contrast, upon chemical inhibition of the proteasome-bound DUB Usp14, the degradation of aggregation-prone substrates in mammalian tissue culture cells significantly increased [65]. Similarly, the inactivation of the evolutionarily related DUB in yeast, Ubp6, increases the cellular fitness of aneuploid cells, which have been shown to incur increased proteotoxic stresses due to chromosomal imbalance [66]. These results indicate that although the trimming of ubiquitin chains at the proteasome may ensure that only proteins with long ubiquitin chains become degraded, it may also limit the degradation of proteins conjugated to shorter chains, potentially including some of the misfolded proteins targeted by quality-control pathways.
The final frontier: the 26S proteasome
The 26S proteasome is the final destination for ubiquitinated proteins targeted for degradation. Its structure is best described as two multisubunit components: the 20S core and the 19S regulatory particle. The core adopts a highly symmetrical, hollow barrel-like structure containing proteolytically active subunits, which face the interior of the chamber and are responsible for the digestion of protein substrates. The two narrow tunnels, located at either end of the barrel, serve as entrances for substrates, and access is gated by the regulatory particle that assembles onto both ends of the 20S barrel.
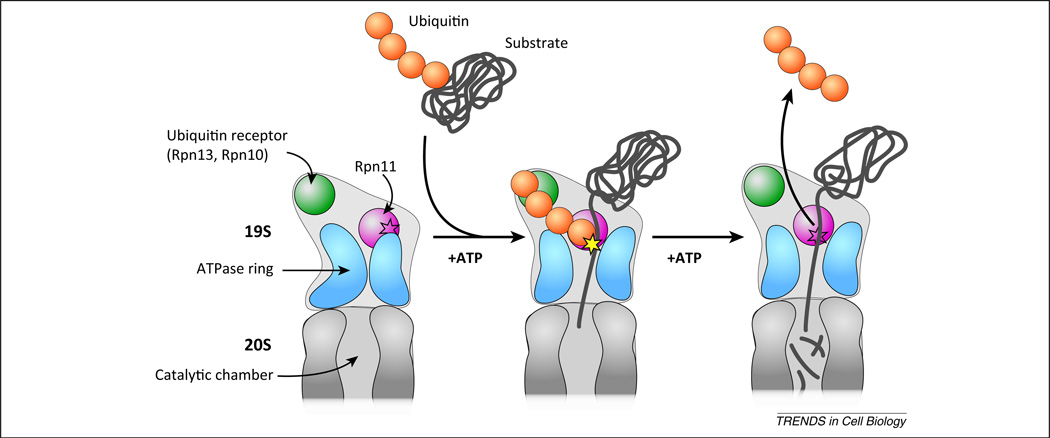
Biochemical investigations on the mechanism of 26S-mediated degradation demonstrated its vast complexity, and a lack of detailed structural information on the 19S had until recently precluded the molecular description of those findings. Recent advances in high-resolution cryo-electron microscopy (EM) have now provided close to atomic structures of the 26S proteasome and have precipitated compelling explanations for two key biochemical observations: why proteasome substrates are required to contain poly-ubiquitin chains with four or more ubiquitins [40], and why proteasome-mediated cleavage of the poly-ubiquitin chain is coupled to translocation of the substrate into the proteasome core. Before discussing how the 26S structure may explain these two observations, two relevant molecular events must be described. First, ubiquitinated proteins are docked to the proteasome through receptors associated to the 19S [67,68], including the proteasome proteins Rpn10 (proteasome regulatory particle base subunit; also known as Psmd4 – proteasome 26S subunit, non-ATPase, 4) and Rpn13 (also known as Adrm1, adhesion regulating molecule 1), that bind to the poly-ubiquitin chains [69,70]. Second, Rpn11/Poh1 (also known as Psmd14) is a subunit in the 19S regulatory particle that cleaves the entire, intact ubiquitin chain from the protein substrate [71,72]. This activity promotes both the recycling of chains back into the free cellular pool of ubiquitin and creates space for the protein substrate to enter the 20S core. In theory, if the distal end of poly-ubiquitin chains can remain bound to the 19S receptors, while the proximal end simultaneously binds to the Rpn11 active site, this bidentate interaction could increase the affinity between the proteasome and ubiquitinated substrate, providing additional time for the initial threading of the ubiquitinated protein into the 20S core (Figure 2).
Figure 2.
Proposed model for substrate recognition and processing by the proteasome. The schematic represents a cutaway view through the center of the 26S proteasome. When the proteasome is not bound to a ubiquitinated substrate (left), the 19S conformation places the Rpn11 (proteasome regulatory particle base subunit 11) active site (represented by the star outline) in a position that is inaccessible to the substrate translocation pore. Furthermore, the channel formed by a ring of six ATPases that promote substrate translocation (blue) is not properly aligned with the 20S aperture. When the proteasome binds to a ubiquitinated substrate, the conformation of the 19S is rearranged such that the unfolded protein can enter the 20S chamber and the Rpn11 active site (yellow star) can cleave the poly-ubiquitin chain. In this model, only substrates conjugated to poly-ubiquitin chains containing four or more protomers can be efficiently processed because only these chains have the minimal length that is necessary to span the distance between one of the two ubiquitin receptors (Rpn10 or Rpn13) and Rpn11. Upon removal of the ubiquitin chain, the substrate is translocated by the ATP-dependent action of the ATPases into the 20S subunit where it is hydrolyzed into short peptides.
If this model is correct, it necessitates that the length of the poly-ubiquitin chain must minimally span the distance between Rpn11 and the ubiquitin receptors. The recently determined cryo-EM structures of the 26S proteasome demonstrated that the span between Rpn11 and the ubiquitin chain receptors closely matched the length of an extended lysine 48-linked poly-ubiquitin chain with four protomers [73,74]. This observation suggests that chain binding and cleavage from the protein substrate are tightly coordinated by the proteasome, and may help to prevent situations such as premature chain cleavage by Rpn11, which could result in the release of the substrate before it becomes actively engaged to the proteasome.
Another key early finding was that the cleavage of the ubiquitin chain from the substrate was ATP-dependent and was coupled to the translocation of the protein substrate into the 20S core. Recently, the structure of the 26S proteasome in complex with protein substrates containing a poly-ubiquitin chain was solved [75]. It revealed a dramatic rearrangement of the 19S subunits, similar to findings from an independent study where 26S proteasomes were incubated in the presence of a non-hydrolyzable ATP analog [76]. The positions of nearly every subunit in the 19S shifted substantially, including Rpn11, where the active site was brought within close proximity of the substrate translocation pore into the 20S (in the structure of the 26S holoenzyme the Rpn11 active site was occluded from the 20S entrance channel; Figure 2). Thus, the binding of substrate to the 26S and its concomitant translocation into the 20S results in the activation of Rpn11 through a conformational change.
There is a strong biological rationale for regulating Rpn11 activity deep within the 19S structure. Because eukaryotic cells typically contain high concentrations of proteasomes, constitutive Rpn11 activity could result in the unwanted editing of ubiquitin chains on proteins that are not destined for the proteasome, assuming that these chains were granted access to an active Rpn11 enzyme.
Concluding remarks
There are still many unresolved questions in the ubiquitin field. For instance, the vast majority of E3s have yet to be characterized, and an even greater number of protein substrates await discovery. One highly promising area is the targeting of the UPS for human therapy. Inhibition of the proteasome blocks several essential cellular processes, such as cell cycle progression and cell survival mediated by the NF-κB pathway, and leads to an increase in proteotoxicity due to the accumulation of misfolded proteins [77]. Indeed, there are now two FDA-approved drugs, Bortezomib and Carfilzomib, that target the proteasome to treat cancer (because rapid cell growth is thought to place a greater demand on the proteasome). As we gain greater insight into how the UPS functions in the cell, it will undoubtedly open up new pipelines for drug discovery, including the search for molecules that are both more specific and selective for smaller portions of the UPS. It is clear that the rate of new discoveries in the UPS continues to rise, bringing forth optimism that new links between important biological processes and the UPS will be uncovered soon. Furthermore, we hope that their underlying mechanisms will be dissected faster based on the unifying themes described here.
Figure I.
Catalysis of ubiquitin transfer.
Acknowledgments
We thank Drs Rachel Klevit and Rati Verma for comments on the manuscript. T.M. is supported by grants from the Natural Sciences and Engineering Research Council of Canada and the Canada Institutes of Health Research (CIHR), and by New Investigator Career Awards from the CIHR and the Michael Smith Foundation for Health Research. G.K. is supported by grants from the National Center for Research Resources (5P20RR016464-11) and from the National Institute of General Medical Sciences (8 P20 GM103440-11) of the National Institutes of Health.
References
- 1.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 2.Xu P, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tokunaga F, et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat. Cell Biol. 2009;11:123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 4.Komander D, Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 5.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Y, et al. Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res. 2007;67:9472–9481. doi: 10.1158/0008-5472.CAN-07-0568. [DOI] [PubMed] [Google Scholar]
- 7.Huang DT, et al. Basis for a ubiquitin-like protein thioester switch toggling E1–E2 affinity. Nature. 2007;445:394–398. doi: 10.1038/nature05490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olsen SK, et al. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature. 2010;463:906–912. doi: 10.1038/nature08765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olsen SK, Lima CD. Structure of a ubiquitin E1–E2 complex: insights to E1–E2 thioester transfer. Mol. Cell. 2013;49:884–896. doi: 10.1016/j.molcel.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lake MW, et al. Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB–MoaD complex. Nature. 2001;414:325–329. doi: 10.1038/35104586. [DOI] [PubMed] [Google Scholar]
- 11.Lee I, Schindelin H. Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell. 2008;134:268–278. doi: 10.1016/j.cell.2008.05.046. [DOI] [PubMed] [Google Scholar]
- 12.Lois LM, Lima CD. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 2005;24:439–451. doi: 10.1038/sj.emboj.7600552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walden H, et al. The structure of the APPBP1–UBA3–NEDD8–ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol. Cell. 2003;12:1427–1437. doi: 10.1016/s1097-2765(03)00452-0. [DOI] [PubMed] [Google Scholar]
- 14.Huang DT, et al. E2–RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol. Cell. 2009;33:483–495. doi: 10.1016/j.molcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tokgoz Z, et al. E1–E2 interactions in ubiquitin and Nedd8 ligation pathways. J. Biol. Chem. 2012;287:311–321. doi: 10.1074/jbc.M111.294975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soucy TA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 17.Ohta T, et al. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 18.Tan P, et al. Recruitment of a ROC1–CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of I kappa B alpha. Mol. Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- 19.Dou H, et al. BIRC7–E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat. Struct. Mol. Biol. 2012;19:876–883. doi: 10.1038/nsmb.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pruneda JN, et al. Structure of an E3:E2~Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol. Cell. 2012;47:933–942. doi: 10.1016/j.molcel.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plechanovova A, et al. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamilton KS, et al. Structure of a conjugating enzyme–ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure. 2001;9:897–904. doi: 10.1016/s0969-2126(01)00657-8. [DOI] [PubMed] [Google Scholar]
- 23.Saha A, et al. Essential role for ubiquitin–ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol. Cell. 2011;42:75–83. doi: 10.1016/j.molcel.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wickliffe KE, et al. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell. 2011;144:769–781. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pruneda JN, et al. Ubiquitin in motion: structural studies of the ubiquitin-conjugating enzyme approximately ubiquitin conjugate. Biochemistry. 2011;50:1624–1633. doi: 10.1021/bi101913m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dou H, et al. Essentiality of a non-RING element in priming donor ubiquitin for catalysis by a monomeric E3. Nat. Struct. Mol. Biol. 2013;20:982–986. doi: 10.1038/nsmb.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamadurai HB, et al. Mechanism of ubiquitin ligation and lysine prioritization by a HECT E3. eLife. 2013;2:e00828. doi: 10.7554/eLife.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamadurai HB, et al. Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin–HECT(NEDD4L) complex. Mol. Cell. 2009;36:1095–1102. doi: 10.1016/j.molcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verdecia MA, et al. Conformational flexibility underlies ubiquitin ligation mediated by the WWP1 HECT domain E3 ligase. Mol. Cell. 2003;11:249–259. doi: 10.1016/s1097-2765(02)00774-8. [DOI] [PubMed] [Google Scholar]
- 30.Huang L, et al. Structure of an E6AP–UbcH7 complex: insights into ubiquitination by the E2–E3 enzyme ascade. Science. 1999;286:1321–1326. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- 31.Maspero E, et al. Structure of a ubiquitin-loaded HECT ligase reveals the molecular basis for catalytic priming. Nat. Struct. Mol. Biol. 2013;20:696–701. doi: 10.1038/nsmb.2566. [DOI] [PubMed] [Google Scholar]
- 32.Ogunjimi AA, et al. Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol. Cell. 2005;19:297–308. doi: 10.1016/j.molcel.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 33.Riley BE, et al. Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat. Commun. 2013;4:1982. doi: 10.1038/ncomms2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trempe JF, et al. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 2013;340:1451–1455. doi: 10.1126/science.1237908. [DOI] [PubMed] [Google Scholar]
- 35.Wenzel DM, et al. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474:105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spratt DE, et al. A molecular explanation for the recessive nature of parkin-linked Parkinson’s disease. Nat. Commun. 2013;4:1983. doi: 10.1038/ncomms2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duda DM, et al. Structure of HHARI, a RING–IBR–RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism. Structure. 2013;21:1030–1041. doi: 10.1016/j.str.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleiger G, et al. The acidic tail of the Cdc34 ubiquitin-conjugating enzyme functions in both binding to and catalysis with ubiquitin ligase SCFCdc4. J. Biol. Chem. 2009;284:36012–36023. doi: 10.1074/jbc.M109.058529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yew PR, Kirschner MW. Proteolysis and DNA replication: the CDC34 requirement in the Xenopus egg cell cycle. Science. 1997;277:1672–1676. doi: 10.1126/science.277.5332.1672. [DOI] [PubMed] [Google Scholar]
- 40.Thrower JS, et al. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chau V, et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 42.Pierce NW, et al. Detection of sequential polyubiquitylation on a millisecond timescale. Nature. 2009;462:615–619. doi: 10.1038/nature08595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evans T, et al. Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell. 1983;33:389–396. doi: 10.1016/0092-8674(83)90420-8. [DOI] [PubMed] [Google Scholar]
- 44.Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mayor T, et al. Quantitative profiling of ubiquitylated proteins reveals proteasome substrates and the substrate repertoire influenced by the Rpn10 receptor pathway. Mol. Cell. Proteomics. 2007;6:1885–1895. doi: 10.1074/mcp.M700264-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Comyn SA, et al. False start: cotranslational protein ubiquitination and cytosolic protein quality control. J. Proteomics. 2013 doi: 10.1016/j.jprot.2013.08.005. http://dx.doi.org/10.1016/j.jprot.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 47.Brodsky JL. Cleaning up: ER-associated degradation to the rescue. Cell. 2013;151:1163–1167. doi: 10.1016/j.cell.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schubert U, et al. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 49.Wang F, et al. A cotranslational ubiquitination pathway for quality control of misfolded proteins. Mol. Cell. 2013;50:368–378. doi: 10.1016/j.molcel.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duttler S, et al. Principles of cotranslational ubiquitination and quality control at the ribosome. Mol. Cell. 2013;50:379–393. doi: 10.1016/j.molcel.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turner GC, Varshavsky A. Detecting and measuring cotranslational protein degradation in vivo. Science. 2000;289:2117–2120. doi: 10.1126/science.289.5487.2117. [DOI] [PubMed] [Google Scholar]
- 52.Dantuma NP, Hoppe T. Growing sphere of influence: Cdc48/p97 orchestrates ubiquitin-dependent extraction from chromatin. Trends Cell Biol. 2012;22:483–491. doi: 10.1016/j.tcb.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 53.Defenouillere Q, et al. Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proc. Natl. Acad. Sci. U.SA. 2013;110:5046–5051. doi: 10.1073/pnas.1221724110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verma R, et al. Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. eLife. 2013;2:e00308. doi: 10.7554/eLife.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brandman O, et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–1054. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu Y, et al. SGTA recognizes a noncanonical ubiquitin-like domain in the Bag6–Ubl4A–Trc35 complex to promote endoplasmic reticulum-associated degradation. Cell Rep. 2012;2:1633–1644. doi: 10.1016/j.celrep.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Q, et al. A ubiquitin ligase-associated chaperone holdase maintains polypeptides in soluble states for proteasome degradation. Mol. Cell. 2011;42:758–770. doi: 10.1016/j.molcel.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Minami R, et al. BAG-6 is essential for selective elimination of defective proteasomal substrates. J. Cell Biol. 2010;190:637–650. doi: 10.1083/jcb.200908092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hessa T, et al. Protein targeting and degradation are coupled for elimination of mislocalized proteins. Nature. 2011;475:394–397. doi: 10.1038/nature10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lehner B, et al. Analysis of a high-throughput yeast two-hybrid system and its use to predict the function of intracellular proteins encoded within the human MHC class III region. Genomics. 2004;83:153–167. doi: 10.1016/s0888-7543(03)00235-0. [DOI] [PubMed] [Google Scholar]
- 61.Guerrero C, et al. Characterization of the proteasome interaction network using a QTAX-based tag-team strategy and protein interaction network analysis. Proc. Natl. Acad. Sci. U.SA. 2008;105:13333–13338. doi: 10.1073/pnas.0801870105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Komander D, et al. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 63.Zhang ZR, et al. Deubiquitinases sharpen substrate discrimination during membrane protein degradation from the ER. Cell. 2013;154:609–622. doi: 10.1016/j.cell.2013.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.D’Arcy P, et al. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat. Med. 2011;17:1636–1640. doi: 10.1038/nm.2536. [DOI] [PubMed] [Google Scholar]
- 65.Lee BH, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Torres EM, et al. Identification of aneuploidy-tolerating mutations. Cell. 2010;143:71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Verma R, et al. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin–proteasome system. Cell. 2010;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 68.Elsasser S, et al. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J. Biol. Chem. 2004;279:26817–26822. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- 69.Husnjak K, et al. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Nocker S, et al. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol. Cell. Biol. 1996;16:6020–6028. doi: 10.1128/mcb.16.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 72.Verma R, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 73.Lander GC, et al. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beck F, et al. Near-atomic resolution structural model of the yeast 26S proteasome. Proc. Natl. Acad. Sci. U.SA. 2012;109:14870–14875. doi: 10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matyskiela ME, et al. Conformational switching of the 26S proteasome enables substrate degradation. Nat. Struct. Mol. Biol. 2013;20:781–788. doi: 10.1038/nsmb.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sledz P, et al. Structure of the 26S proteasome with ATP-gammaS bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proc. Natl. Acad. Sci. U.SA. 2013;110:7264–7269. doi: 10.1073/pnas.1305782110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hideshima T, et al. Mechanism of action of proteasome inhibitors and deacetylase inhibitors and the biological basis of synergy in multiple myeloma. Mol. Cancer Ther. 2011;10:2034–2042. doi: 10.1158/1535-7163.MCT-11-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dammer EB, et al. Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. J. Biol. Chem. 2011;286:10457–10465. doi: 10.1074/jbc.M110.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jin L, et al. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133:653–665. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chastagner P, et al. Itch/AIP4 mediates Deltex degradation through the formation of K29-linked polyubiquitin chains. EMBO Rep. 2006;7:1147–1153. doi: 10.1038/sj.embor.7400822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Johnson ES, et al. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- 82.Saeki Y, et al. Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J. 2009;28:359–371. doi: 10.1038/emboj.2008.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nathan JA, et al. Why do cellular proteins linked to K63-polyubiquitin chains not associate with proteasomes? EMBO J. 2013;32:552–565. doi: 10.1038/emboj.2012.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jacobson AD, et al. The lysine 48 and lysine 63 ubiquitin conjugates are processed differently by the 26 s proteasome. J. Biol. Chem. 2009;284:35485–35494. doi: 10.1074/jbc.M109.052928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu PY, et al. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J. 2003;22:5241–5250. doi: 10.1093/emboj/cdg501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Berndsen CE, et al. A conserved asparagine has a structural role in ubiquitin-conjugating enzymes. Nat. Chem. Biol. 2013;9:154–156. doi: 10.1038/nchembio.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yunus AA, Lima CD. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat. Struct. Mol. Biol. 2006;13:491–499. doi: 10.1038/nsmb1104. [DOI] [PubMed] [Google Scholar]
- 88.Eddins MJ, et al. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat. Struct. Mol. Biol. 2006;13:915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]