Abstract
Human neocentromeres are fully functional centromeres that arise at previously noncentromeric regions of the genome. We have tested a rapid procedure of genomic array analysis of chromosome scaffold/matrix attachment regions (S/MARs), involving the isolation of S/MAR DNA and hybridization of this DNA to a genomic BAC/PAC array. Using this procedure, we have defined a 2.5-Mb domain of S/MAR-enriched chromatin that fully encompasses a previously mapped centromere protein-A (CENP-A)-associated domain at a human neocentromere. We have independently verified this procedure using a previously established fluorescence in situ hybridization method on salt-treated metaphase chromosomes. In silico sequence analysis of the S/MAR-enriched and surrounding regions has revealed no outstanding sequence-related predisposition. This study defines the S/MAR-enriched domain of a higher eukaryotic centromere and provides a method that has broad application for the mapping of S/MAR attachment sites over large genomic regions or throughout a genome.
The centromere is a specialized structure of the eukaryotic chromosome responsible for the accurate partitioning of sister-chromatids during mitosis and meiosis. The centromeres of most higher eukaryotes are composed of tandemly repetitive DNA. Human centromeres contain tracts of up to 4 Mb of the AT-rich 171-bp α-satellite sequence (Choo 1997). This DNA has been shown to impart centromere forming properties (Harrington et al. 1997).
Centromeric DNA is packaged differently from that of the rest of the genome, with histone H3 being replaced by the centromere-specific histone H3-like protein CENP-A at kinetochores, the structures responsible for attachment of chromosomes to the mitotic spindle (Yoda et al. 2000; Lo et al. 2001a). The higher-order chromatin packaging of centromeres at the level above the 30-nm chromatin fibre also differs from the rest of the genome. Chromatin in the eukaryotic genome has been shown to be organized into loops by attachment to a proteinaceous chromosome scaffold, matrix, or nucleoskeleton structure (Berezney and Coffey 1974; Mirkovitch et al. 1984; Jackson et al. 1988). The most frequently used method to isolate the DNA and protein components of this structure involves the extraction of histones with high salt or mild detergent, followed by restriction enzyme treatment, liberating all except scaffold/matrix-attached DNA and proteins (Paulson and Laemmli 1977; Mirkovitch et al. 1984). The proteinaceous structure left after such treatments has been referred to as a chromosome scaffold or matrix, and the DNA sequences that mediate attachment are termed scaffold/matrix attachment regions or S/MARs. S/MARs are generally AT-rich sequences and include poly(A) tracts, which form a narrow minor groove that is more likely to act as a substrate for S/MAR-binding proteins such as DNA topoisomerase II (Lewis and Laemmli 1982; Earnshaw et al. 1985). S/MARs are localized more often to noncoding regions of DNA, larger than 300 bp, and occur on average every 50–200 kb in the human genome (Pienta et al. 1991; Bode et al. 1995).
Centromeric DNA from Saccharomyces cerevisiae and humans has been shown to contain a significantly increased frequency of S/MAR sites than the rest of the genome (Amati and Gasser 1988; Bickmore and Oghene 1996; Strissel et al. 1996). In humans, the S/MARs in centromeric α-satellite DNA occur every 1.7–1.9 kb (Strissel et al. 1996). Other studies have shown that the presence of tandemly repetitive DNA was not sufficient for scaffold/matrix attachment, as other regions containing large tracts of AT-rich tandem repeats such as satellite III DNA did not behave similarly (Bickmore and Oghene 1996). It is presently unclear whether α-satellite DNA has an intrinsically higher scaffold/matrix attachment predisposition, or if so, why.
In recent years, a new class of centromeres known as neocentromeres are proving to be a useful tool for centromere study hampered previously by the repetitive nature of mammalian centromeric DNA (Choo 2001; Amor and Choo 2002). Human neocentromeres arise at previously noncentromeric chromosomal regions, and although totally devoid of α-satellite repeats, are functionally equivalent to typical centromeres. Detailed sequence analysis of three different human neocentromeres has revealed that, like their normal counter-part, they form at regions of slightly increased AT content (Choo 1997; Lo et al. 2001a,b; Satinover et al. 2001). In addition, current evidence indicates that neocentromere formation is a purely epigenetic event (Karpen and Allshire 1997; Choo 2000).
We have previously delineated the domain for CENP-A association and/or differential DNA replication at two neocentromeres (Lo et al. 2001a,b). Here, we describe the scaffold/matrix attachment properties at one of these neocentromeres, formed on an invdup(20p) marker chromosome derived from inverted duplication of the short-arm of human chromosome 20 (Voullaire et al. 1999). This study involves the use of a newly developed S/MAR analysis technique involving hybridization of isolated chromosomal scaffold/matrix fractions to arrays of contiguous BAC/PACs and validation of the results using an established technique of fluorescence in situ hybridization (FISH) on salt-treated metaphase chromosomes.
RESULTS
Identification of a 2.5-Mb Domain of Enhanced S/MAR Density at a Human Neocentromere
A previous study has identified the CENP-A-binding domain of the neocentromere on an invdup(20p) marker chromosome (Voullaire et al. 1999; Lo et al. 2001b). Here, we have developed a rapid analytical procedure to allow the efficient determination of the underlying higher-order chromatin scaffold/matrix structure at this and other neocentromeres. The S/MAR-array analysis involves the mild detergent fractionation of S/MAR and loop (non-S/MAR) DNA (Mirkovitch et al. 1984) from the invdup(20p) and normal control cell lines, followed by the use of each fraction to probe duplicate dot-blotted genomic arrays of contiguous BACs or PACs spanning 5 Mb around the 20p12 neocentromere region defined in our earlier work (Lo et al. 2001b). The average percentage of scaffold/matrix attachment for each BAC/PAC was determined (see Methods) for each cell line. As a positive control, α-satellite DNA was spotted onto the membranes for comparison. A high scaffold/matrix attachment (>85%) for the α-satellite spots for both cell lines was observed (data not shown) and is comparable with published data (Strissel et al. 1996). As further controls, we have performed experiments in which the isolated S/MAR and loop fractions were run on a gel, Southern blotted, and probed with known centromeric, S/MAR-attached α-satellite DNA sequence Pα3.5 and a previously described loop-DNA PSE16 (Strissel et al 1996), and have demonstrated concurring results (Supplemental Fig. 1, available online at www.genome.org).
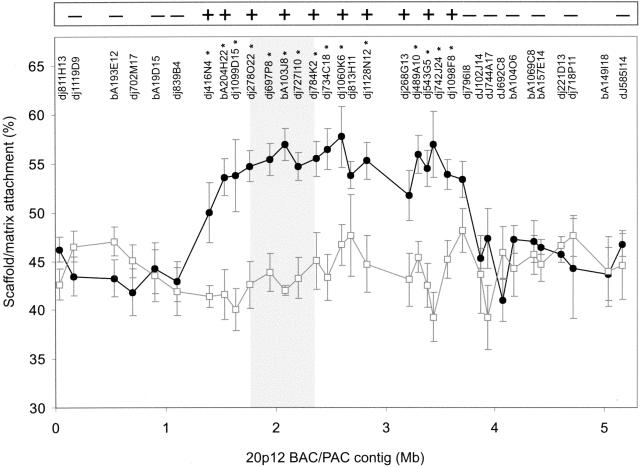
S/MAR-array analysis of the 20p12 BAC/PAC arrays identified a region of significantly enhanced scaffold/matrix attachment in the invdup(20p) cell line compared with the control cell line (P < 0.05) over a region of ∼2.5 Mb, between BAC/PACs dj416N4 and dj109F8 (Fig. 1). This difference was apparent, even though the test cell line contained three copies of the 20p12 region, two residing on the invdup(20p) chromosome (one of which containing the neocentromere), and the third on a normal chromosome 20. As seen in Figure 1, the region of increased scaffold/matrix attachment fully encompassed the previously mapped 460-kb CENP-A-binding domain of the 20p12 neocentromere (Lo et al. 2001b).
Figure 1.
Determination of Scaffold/matrix attachment along the 20p12 BAC/PAC contig using the SIA procedure. Percent matrix/scaffold attachment as determined by array analysis is plotted against BAC/PAC position along the contig for cell lines containing the invdup(20p) chromosome (circles) or normal chromosomes only (squares). Data-points represent the mean ± one standard deviation from at least six independent experiments. The names of the different BAC/PACs are indicated at the top of the graph. BAC/PACs showing a significantly increased scaffold/matrix attachment on the invdup(20p) neocentromere (P <0.05) over the corresponding non-neocentromeric 20p12 region are indicated by asterisks. The shaded area indicates the previously identified CENP-A-binding region. The inset box at the top of the graph represents a summary of the FISH analysis of metaphase chromosomes for a number of the BAC/PACs probes used in the array hybridization analysis, in which - and + denote no difference and an increase in scaffold/matrix attachment, respectively.
Detection of Scaffold/Matrix Attachment by FISH on Metaphase Chromosomes
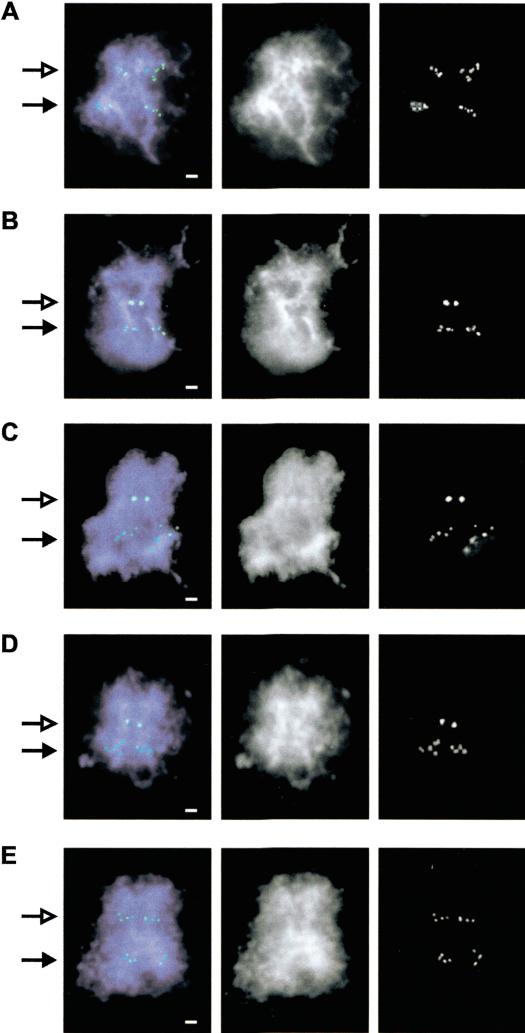
Paulson and Laemmli (1977) first described the visualization of chromosomal scaffold/matrix using electron microscopy after in situ high-salt treatment. In these chromosomes, a halo of liberated loop DNA was seen to emanate from the scaffold/matrix (axial region) of the chromosome. Bickmore and Oghene (1996) later showed, using FISH on human chromosomes treated this way, that specific sequences could be identified at the light microscope level corresponding to the liberated loop DNA or the more tightly scaffold/matrix-attached axial DNA. Figure 2 shows the results obtained by use of this method on a number of pertinent BAC/PACs from our 20p12 contig. The invdup(20p) marker chromosome, being an isochromosome, was readily identifiable by its double set of FISH signals that are present at the primary constriction of the neocentromere and at the nonconstricted 20p12 region. At the nonconstricted normal 20p12 position of the invdup(20p) chromosome, all of the BAC/PACs gave diffuse signals that emanated out into the halo of loops, suggesting that the probed regions were predominantly nonscaffold/matrix attached. In contrast, two types of signals were seen at the constricted neocentromere region. One group of BAC/PACs gave diffuse signals similar to those seen at the normal 20p12 position (Fig. 2A,E), suggesting that the scaffold/matrix packaging of these sequences near the neocentromere was not significantly different from that of the normal chromatin. In the second group, the BAC/PAC signals were seen as a compact pair of spots (Fig. 2B–D), indicating that these sequences were predominantly scaffold/matrix attached. When the FISH results for all of the BAC/PAC probes tested were compared with those obtained using the S/MAR-array analysis, a full concordance of the two data sets was observed (Fig. 1). This provided independent confirmation of the validity of our newly developed S/MAR-array analysis procedure.
Figure 2.
Hybridization of BAC/PAC DNA to salt-extracted invdup(20p) metaphase chromosomes. FISH signals from BAC/PACs were detected using FITC (green) and chromosomes counterstained with DAPI (blue). (A–E) BAC/PAC clones dj839B4, dJ416N4, bA103J8, dj1098F8, and dj79618, respectively. (Left, middle, right) Combined color image, black and white image for DNA staining, and FISH signals, respectively. The neocentromere position is indicated by an open arrow, whereas closed arrow points to the corresponding normal 20p12 region. Scale bar, 1μm.
In Silico Sequence Analysis
S/MARs have been shown to contain higher than average levels of AT-rich DNA and poly(A) tracts (Kas et al. 1989). These and other properties have been used to predict S/MARs from genomic sequences (Singh et al. 1997; Glazko et al. 2001; Frisch et al. 2002). Using in silico sequence analysis, we examined whether the S/MAR-enriched region at the 20p12 neocentromere had a higher predicted S/MAR sequence content. Average values for AT and poly(A) tract contents along with S/MAR content predicted by three different programs were determined for BAC/PACs inside and outside of the observed region of increased S/MARs (Table 1). The average AT content for the domain of increased S/MARs was 60.9% and was not significantly different from the surrounding regions (60.7%). However, this region has an overall slightly higher AT content than the genome average (58.0%) (Smit 1999) and is therefore slightly more akin to that of α-satellite DNA (62.6%) (Choo 1997). When the number of poly(A) tracts (≥[A]4) was determined, no significant difference was seen between the observed S/MAR-enriched domain (1802/100 kb) compared with that of its surrounding domain (1790). Finally, use of three prediction programs has similarly revealed no statistically significant difference in the number of predicted S/MARs between the two domains (Table 1).
Table 1.
Summary of In Silico Analysis of the S/MAR Domain of 20p12 Neocentromere
| S/MAR domain | Non-S/MAR domain | p-value | |
|---|---|---|---|
| AT content | 60.9% | 60.7% | 0.646 |
| Poly(A) tracts | 1802 | 1790 | 0.840 |
| MarWiz | 3.7 | 3.5 | 0.511 |
| Smartest | 10.4 | 9.1 | 0.630 |
| ChrClass | 17.7 | 16.9 | 0.777 |
To facilitate statistical analysis, AT content was calculated for the whole region, whereas the frequency of poly(A) tracts and S/MAR predictions, using MarWiz, Smartest, and ChrClass were calculated per 100 kb. P-values from t-tests comparing the S/MAR-enhanced and non-S/MAR-enhanced domains are given.
DISCUSSION
Development of an Improved Method for S/MAR Analysis
Earlier methods for identifying S/MARs have involved the use of plasmid-sized DNA to probe Southern blots containing fractions of isolated scaffold/matrix-attached and loop DNA (Mirkovitch et al. 1984). A further adaptation of these methods has involved the use of larger probes in FISH analysis of salt-extracted metaphase chromosomes (Bickmore and Oghene 1996). Both methods require the laborious examination of a single probe at a time, thereby limiting their use over large genomic regions. Our present procedure involves the initial isolation of S/MAR and loop DNA, followed by a single-step analysis of the isolated DNA across many BAC/PAC clones displayed on a dot-blot array. This procedure, which we have verified independently using FISH analysis, offers a new, efficient method to allow the determination of S/MAR profiles over large genomic regions.
We have shown that our S/MAR-array analysis is sufficiently sensitive to detect differences in scaffold/matrix attachment that exist only at one of three homologous chromosomal sites within a cell line. This removes any need to separate the chromosome of interest into a somatic cell hybrid background. Furthermore, as the procedure is equally applicable to BACs and PACs ranging in size from 23–177 kb in length used in the present study, it should be possible to increase the resolution of the analysis if significantly smaller DNA sequences are designed and used throughout the array.
In addition to mapping scaffold/matrix attachment domains at neocentromeres, S/MAR-array analysis could be used to map differences in a variety of situations, such as those associated with gene activation, oncogenesis, or possible epigenetic modifications of DNA during development. With the availability of BAC/PAC contigs and data generated by the genome mapping projects, and possibly the future adaptation of dual-color microarray technology to the present method, the scaffold/matrix topography of whole genomes could be determined rapidly at a high resolution.
Definition of a Substantial Domain of Enhanced, Sequence-Independent Scaffold/Matrix Attachment at 20p12 Neocentromere
In earlier work, Bickmore and Oghene (1996) showed that human centromeric α-satellite DNA remains tightly attached to the axial region of a metaphase chromosome, in contrast to the liberated loop DNA seen in other genomic regions of euchromatin or noncentromere-related satellite DNA. However, the method used did not allow the boundaries of the increased centromeric/pericentromeric S/MAR association to be determined. Using a BAC/PAC array of defined sequence and size, we have delineated a 2.5-Mb domain of significantly enhanced S/MAR density at the 20p12 neocentromere that fully encompasses the previously reported kinetichore-specific histone H3-like CENP-A-binding region. This is the first direct measurement of the boundaries of a differential S/MAR domain at any higher eukaryotic centromere.
Our results indicate that neocentromere formation is accompanied by the transformation of a substantial region of normal chromatin into a different type of chromatin that exhibits greatly enhanced scaffold/matrix attachment. It is unclear what the precise roles of such a large region of S/MAR-enriched chromatin domain surrounding CENP-A at the neocentromere or other satellite-DNA-based centromeres. One possibility for enriched S/MARs surrounding this domain relates to the provision of tighter chromatin packaging at the kinetichore and immediately surrounding regions to withstand the high-tensile forces exerted by the spindle microtubules on the kinetochore during cell division.
Sequence analysis suggests that the experimentally identified region of enhanced S/MAR density at the invdup(20p) neocentromere correlates minimally, if at all, with predicted S/MAR-forming potential, suggesting that the formation of such a region is not strictly sequence dependent. This is also borne out by the fact that the S/MAR-enhanced neocentromeric DNA is the same DNA that ordinarily shows scaffold/matrix-binding properties that are indistinguishable from normal chromatin. These observations suggest that, notwithstanding the possibility of some sequence predispositions, such as increased AT content, which is thought to provide innate flexibility and curvature to make a DNA more amenable to scaffold/matrix attachment (Yamamura and Nomura 2001), the overriding determinant in scaffold/matrix attachment appears to be due to some extrinsic epigenetic cue. Several proteins have been shown to be associated with the chromosomal scaffold/matrix and are candidates for such an epigenetic cue. One of the most abundant scaffold/matrix-associated proteins is topoisomerase II (Earnshaw et al. 1985). This protein, which has been shown to occupy a larger region than kinetichores at neocentromeres and the active centromeres of multicentric chromosomes (Saffery et al. 2000; Andersen et al. 2002), plays a key role in disentangling sister chromatids after DNA replication and in centromeric chromatid cohesion (Warburton and Earnshaw 1997; Losada and Hirano 2001). The distribution of topoisomerase II and possibly other scaffold/matrix proteins may explain why such a large domain of S/MAR enrichment is formed substantially beyond the previously described CENP-A binding region. Other proteins enriched at the centromeric and/or pericentromeric regions that are known to be involved with the chromosme scaffold/matrix include CENP-C (Earnshaw et al. 1984), CENP-E (Yen et al. 1991), CENP-G (He et al. 1998), MECP2 (Stratling and Yu 1999), and PARP-1(Galande and Kohwi-Shigematsu 1999; Earle et al. 2000). However, it is unclear how these or other potential proteins (such as cohesin) are distributed on a linear or higher-order centromeric chromatin in relation to the S/MAR domain.
The physical map defined in the present study should provide a useful framework for the localization of these proteins, such as using a previously described, compatible method of chromatin immunoprecipitation and genomic array analysis (Lo et al. 2001a). A direct comparison of the patterns of protein localization with S/MAR distribution may enable us to determine which, if any, of the above proteins are associated with or responsible for the observed chromosome scaffold/matrix attachment behavior at the centromere.
METHODS
Cell Lines
EBV-transformed lymphoblastoid and fibroblastoid cell lines established from a patient with the marker chromosome invdup(20p) (plus one normal chromosome 20) (Voullaire et al. 1999) and a cell line from a normal individual were maintained in RPMI 1640 medium supplemented with 20% FCS, and DMEM 10% FCS, respectively (Thermo Trace Ltd.).
BAC and PAC DNA
BAC prefix “bA” clones were obtained from the human genomic library RPCI-11 (Osoegawa et al. 2001); PAC prefix “dj” DNA clones were obtained from the BACPAC Resources Centre (Oakland).
Isolation of Cell Nuclei and Low-Salt (LIS) Scaffold/Matrix Extraction
This method was essentially as described by Craig et al. (1997). 1 × 106 nuclei were isolated by Dounce homogenization. Chromosome scaffolds/matrices were extracted using 50 mM 3,5-diiodasalicylic acid, lithium salt (Fluka). Restriction enzyme digestion of extracted chromosome scaffolds/matrices was performed using EcoRI, EcoRV, and BamHI in combination, each at 1000 U/mL, and incubated at 37°C for 5 h. The S/MAR fraction of chromatin was pelleted from the digested loop DNA by centrifugation at 2400g for 10 min at 4°C, and both fractions purified using standard DNA extraction techniques.
Scaffold/Matrix Attachment Region (S/MAR)-Array Analysis
Genomic arrays were generated by immobilizing 100 ng of BAC/PAC DNA onto Hybond N+ nylon membranes (AP Biotech) in a dot blot format (Schleicher and Schuell). Identical arrays were preannealed with 5 μg of salmon sperm DNA, and probed with 1μg of S/MAR or loop DNA from the patient and control cell lines, radioactivity labeled by random priming, and pre-annealed with 5 μg of human Cot-1DNA. Standard hybridization was performed and blots washed at high stringency (0.1 × SSC/0.1% SDS, 65°C). All blots were analyzed using a PhosphorImager system and Image QuaNT software (AP Biotech). The signal obtained from each S/MAR DNA spot on the dot-blot BAC/PAC array was compared with that on the duplicate blot hybridized with the loop DNA. Scaffold/matrix attachment for each BAC/PAC for both cell lines was calculated as the proportion of signal on the S/MAR blot compared with the combined signal strengths of the S/MAR and loop signals and expressed as a percentage. Experiments were repeated at least six times for both cell lines and presented graphically by plotting the average percentage scaffold/matrix attachment plus or minus one standard deviation for each BAC/PAC against the midpoint for each BAC/PAC. Student's t-test was performed to determine the significance of the differences in scaffold/matrix attachment between the two cell lines.
Metaphase Chromosome Isolation and In Situ High-Salt Extraction
Human mitotic chromosomes were prepared by modification of the method detailed by Bickmore and Oghene (1996). Exponentially growing fibroblast cell line was treated with 0.1 μg/mL Colcemid (Invitrogen Australia Pty Ltd) for 16 h prior to harvest to obtain a high mitotic index. Cells were harvested by standard mitotic shake off, washed in PBS, and resuspended at a concentration of 2 × 106 cells/mL in 0.075 M KCl for 10 min at 37°C. Cells were then pelleted and resuspended in ice-cold PA buffer (15 mM Tris-HCl, 0.2 mM spermine, 0.5 mM spermidine, 0.5 mM EGTA, 2 mM EDTA, 80 mM KCl, 20 mM NaCl, 0.1mM CuSO4 [pH7.2]) at 8 × 106 cells/mL. Cells were pelleted at 200g for 5 min at 4°C, and resuspended at 1 × 107 cells/mL in cold PA buffer containing 1mg/mL digitonin, and vortexed twice for 15 sec to burst the cell membranes and release mitotic chromosomes. Nuclei were pelleted at 200g for 10 min at 4°C, and the supernatant containing the isolated metaphase chromosomes collected. Isolated metaphase chromosomes were smeared onto precleaned microscope slides and allowed to dry for 16 h. Slides were then lowered horizontally into CIB solution (10 mM Tris, 10 mM EDTA, 0.1% Nonidet P-40, 0.1 mM CuSO4, 20 μg/mL PMSF [pH 8.0]), incubated for 5 min, then extracted in CIB supplemented with 0.5 M NaCl for 5 min. Slides were then fixed in 3:1methanol/acetic acid and air-dried.
FISH Analysis
FISH was carried out using standard techniques (Craig 1999). In brief, 200 ng of BAC/PAC DNA labeled by nick translation with biotin-16-dUTP and preannealed with Cot-1 DNA (Roche) was hybridized to slides at high stringency (50% formamide at 37°C) overnight. Slides were then washed three times in 0.1 × SSC at 60°C. Hybridization was detected by dual-layer detection using avidin-FITC, followed by FITC-conjugated goat anti-avidin. Chromosomes were counterstained using 4,6-diamindo-2-phenylindole (DAPI; 2 μg/mL) in Vectashield antifade mountant (Vector Laboratories). Slides were examined on a Zeiss Axioplan II fluorescence microscope with a 100× oil objective and the appropriate filters. Images were captured with a cooled charge-coupled device video camera (SenSys 2, Photometrics), connected to a PowerMac G4 computer controlled by IPLab software (Scanalytics).
In Silico Sequence Analysis
Sequence data was accessed from the Chromosome 20 Sequencing Group at the Sanger Centre (http://www.sanger.ac.iuk/HGP/Chr20). S/MAR prediction programs were accessed as follows; Mar-Wiz (http://www.futuresoft.org/) SmarTest (http://www.genomatix.de/) and ChrClass (ftp.bionet.nsc.ru/pub/biology/chrclass/chrclas2.zip) and default parameters used.
Acknowledgments
We thank G. Glazko for the ChrClass program and P. Kalitsis for reading the manuscript. H.S. receives an MRS scholarship from the Deptartment of Paediatrics, University of Melbourne. This work was funded by NH&MRC of Australia. K.H.A.C. is a Senior Principal Research Fellow of NH&MRC.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1095903.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Amati, B.B. and Gasser, S.M. 1988. Chromosomal ARS and CEN elements bind specifically to the yeast nuclear scaffold. Cell 54: 967-978. [DOI] [PubMed] [Google Scholar]
- Amor, D.J. and Choo, K.H.A. 2002. Neocentromeres: Role in human disease, evolution, and centromere study. Am. J. Hum. Genet. 71: 695-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, C.L., Wandall, A., Kjeldsen, E., Mielke, C., and Koch, J. 2002. Active, but not inactive, human centromeres display topoisomerase II activity in vivo. Chromosome Res. 10: 305-312. [DOI] [PubMed] [Google Scholar]
- Berezney, R. and Coffey, D.S. 1974. Identification of a nuclear protein matrix. Biochem. Biophys. Res. Commun. 60: 1410-1417. [DOI] [PubMed] [Google Scholar]
- Bickmore, W.A. and Oghene, K. 1996. Visualizing the spatial relationships between defined DNA sequences and the axial region of extracted metaphase chromosomes. Cell 84: 95-104. [DOI] [PubMed] [Google Scholar]
- Bode, J., Schlake, T., Rios-Ramirez, M., Mielke, C., Stengert, M., Kay, V., and Klehr-Wirth, D. 1995. Scaffold/matrix-attached regions: Structural properties creating transcriptionally active loci. Int. Rev. Cytol. 162A: 389-454. [DOI] [PubMed] [Google Scholar]
- Choo, K.H.A. 1997. The centromere. Oxford University Press, Oxford, UK.
- Choo, K.H.A. 2000. Centromerization. Trends Cell. Biol. 10: 182-188. [DOI] [PubMed] [Google Scholar]
- Choo, K.H.A. 2001. Domain organization at the centromere and neocentromere. Dev. Cell 1: 165-177. [DOI] [PubMed] [Google Scholar]
- Craig, J.M. 1999. Isolation of vertebrate metaphase chromosomes and their analysis by FISH. In Chromosome structural analysis: A practical approach (ed. W.A. Bickmore), pp. 59-80. Oxford University Press, Oxford, UK.
- Craig, J.M., Boyle, S., Perry, P., and Bickmore, W.A. 1997. Scaffold attachments within the human genome. J. Cell. Sci. 110: 2673-2682. [DOI] [PubMed] [Google Scholar]
- Earle, E., Saxena, A., MacDonald, A., Hudson, D.F., Shaffer, L.G., Saffery, R., Cancilla, M.R., Cutts, S.M., Howman, E., and Choo, K.H.A. 2000. Poly(ADP-ribose) polymerase at active centromeres and neocentromeres at metaphase. Hum. Mol. Genet. 9: 187-194. [DOI] [PubMed] [Google Scholar]
- Earnshaw, W.C., Halligan, N., Cooke, C., and Rothfield, N. 1984. The kinetochore is part of the metaphase chromosome scaffold. J. Cell. Biol. 98: 352-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw, W.C., Halligan, B., Cooke, C.A., Heck, M.M., and Liu, L.F. 1985. Topoisomerase II is a structural component of mitotic chromosome scaffolds. J. Cell. Biol. 100: 1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch, M., Frech, K., Klingenhoff, A., Cartharius, K., Liebich, I., and Werner, T. 2002. In silico prediction of scaffold/matrix attachment regions in large genomic sequences. Genome Res. 12: 349-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galande, S. and Kohwi-Shigematsu, T. 1999. Poly(ADP-ribose) polymerase and Ku autoantigen form a complex and synergistically bind to matrix attachment sequences. J. Biol. Chem. 274: 20521-20528. [DOI] [PubMed] [Google Scholar]
- Glazko, G.V., Rogozin, I.B., and Glazkov, M.V. 2001. Comparative study and prediction of DNA fragments associated with various elements of the nuclear matrix. Biochim. Biophys. Acta 1517: 351-364. [DOI] [PubMed] [Google Scholar]
- Harrington, J.J., Van Bokkelen, G., Mays, R.W., Gustashaw, K., and Willard, H.F. 1997. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat. Genet. 15: 345-355. [DOI] [PubMed] [Google Scholar]
- He, D., Zeng, C., Woods, K., Zhong, L., Turner, D., Busch, R.K., Brinkley, B.R., and Busch, H. 1998. CENP-G: A new centromeric protein that is associated with the α-1satellite DNA subfamily. Chromosoma 107: 189-197. [DOI] [PubMed] [Google Scholar]
- Jackson, D.A., Yuan, J., and Cook, P.R. 1988. A gentle method for preparing cyto- and nucleo-skeletons and associated chromatin. J. Cell. Sci. 90: 365-378. [DOI] [PubMed] [Google Scholar]
- Karpen, G.H. and Allshire, R.C. 1997. The case for epigenetic effects on centromere identity and function. Trends Genet. 13: 489-496. [DOI] [PubMed] [Google Scholar]
- Kas, E., Izaurralde, E., and Laemmli, U.K. 1989. Specific inhibition of DNA binding to nuclear scaffolds and histone H1 by distamycin. The role of oligo(dA).oligo(dT) tracts. J. Mol. Biol. 210: 587-599. [DOI] [PubMed] [Google Scholar]
- Lewis, C.D. and Laemmli, U.K. 1982. Higher order metaphase chromosome structure: Evidence for metalloprotein interactions. Cell 29: 171-181. [DOI] [PubMed] [Google Scholar]
- Lo, A.W., Craig, J.M., Saffery, R., Kalitsis, P., Irvine, D.V., Earle, E., Magliano, D.J., and Choo, K.H.A. 2001a. A 330 kb CENP-A binding domain and altered replication timing at a human neocentromere. EMBO J. 20: 2087-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, A.W., Magliano, D.J., Sibson, M.C., Kalitsis, P., Craig, J.M., and Choo, K.H.A. 2001b. A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromere DNA. Genome Res. 11: 448-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada, A. and Hirano, T. 2001. Intermolecular DNA interactions stimulated by the cohesin complex in vitro: Implications for sister chromatid cohesion. Curr. Biol. 11: 268-272. [DOI] [PubMed] [Google Scholar]
- Mirkovitch, J., Mirault, M.E., and Laemmli, U.K. 1984. Organization of the higher-order chromatin loop: Specific DNA attachment sites on nuclear scaffold. Cell 39: 223-232. [DOI] [PubMed] [Google Scholar]
- Osoegawa, K., Mammoser, A.G., Wu, C., Frengen, E., Zeng, C., Catanese, J.J., and de Jong, P.J. 2001. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res. 11: 483-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson, J.R. and Laemmli, U.K. 1977. The structure of histone-depleted metaphase chromosomes. Cell 12: 817-828. [DOI] [PubMed] [Google Scholar]
- Pienta, K.J., Getzenberg, R.H., and Coffey, D.S. 1991. Cell structure and DNA organization. Crit. Rev Eukaryot. Gene Expr. 1: 355-385. [PubMed] [Google Scholar]
- Saffery, R., Irvine, D.V., Griffiths, B., Kalitsis, P., Wordeman, L., and Choo, K.H.A. 2000. Human centromeres and neocentromeres show identical distribution patterns of >20 functionally important kinetochore-associated proteins. Hum. Mol. Genet. 9: 175-185. [DOI] [PubMed] [Google Scholar]
- Satinover, D.L., Vance, G.H., Van Dyk, D.L., and Schwartz, S. 2001. Cytogenetic analysis and construction of a BAC contig across a common neocentromeric region from 9p. Chromosoma 110: 275-283. [DOI] [PubMed] [Google Scholar]
- Singh, G.B., Kramer, J.A., and Krawetz, S.A. 1997. Mathematical model to predict regions of chromatin attachment to the nuclear matrix. Nucleic Acids Res. 25: 1419-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, A.F. 1999. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 9: 657-663. [DOI] [PubMed] [Google Scholar]
- Stratling, W.H. and Yu, F. 1999. Origin and roles of nuclear matrix proteins. Specific functions of the MAR-binding protein MeCP2/ARBP. Crit. Rev. Eukaryot. Gene Expr. 9: 311-318. [DOI] [PubMed] [Google Scholar]
- Strissel, P.L., Espinosa III, R., Rowley, J.D., and Swift, H. 1996. Scaffold attachment regions in centromere-associated DNA. Chromosoma 105: 122-133. [DOI] [PubMed] [Google Scholar]
- Voullaire, L., Saffery, R., Davies, J., Earle, E., Kalitsis, P., Slater, H., Irvine, D.V., and Choo, K.H.A. 1999. Trisomy 20p resulting from inverted duplication and neocentromere formation. Am. J. Med. Genet. 85: 403-408. [PubMed] [Google Scholar]
- Warburton, P.E. and Earnshaw, W.C. 1997. Untangling the role of DNA topoisomerase II in mitotic chromosome structure and function. BioEssays 19: 97-99. [DOI] [PubMed] [Google Scholar]
- Yamamura, J. and Nomura, K. 2001. Analysis of sequence-dependent curvature in matrix attachment regions. FEBS Lett. 489: 166-170. [DOI] [PubMed] [Google Scholar]
- Yen, T.J., Compton, D.A., Wise, D., Zinkowski, R.P., Brinkley, B.R., Earnshaw, W.C., and Cleveland, D.W. 1991. CENP-E, a novel human centromere-associated protein required for progression from metaphase to anaphase. EMBO J. 10: 1245-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda, K., Ando, S., Morishita, S., Houmura, K., Hashimoto, K., Takeyasu, K., and Okazaki, T. 2000. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc. Natl. Acad. Sci. 97: 7266-7271. [DOI] [PMC free article] [PubMed] [Google Scholar]