Abstract
Analysis of membrane protein interactions is difficult because of the hydrophobic nature of these proteins, which often renders conventional biochemical and genetic assays fruitless. This is a substantial problem because proteins that are integral or associated with membranes represent approximately one-third of all proteins in a typical eukaryotic cell. We have shown previously that the modified split-ubiquitin system can be used as a genetic assay for the in vivo detection of interactions between the two characterized yeast transmembrane proteins, Ost1p and Wbp1p. This so-called split-ubiquitin membrane yeast two-hybrid (YTH) system uses the split-ubiquitin approach in which reconstitution of two ubiquitin halves is mediated by a protein–protein interaction. Here we converted the split-ubiquitin membrane YTH system into a generally applicable in vivo screening approach to identify interacting partners of a particular mammalian transmembrane protein. We have demonstrated the effectiveness of this approach by using the mammalian ErbB3 receptor as bait and have identified three previously unknown ErbB3-interacting proteins. In addition, we have confirmed one of the newly found interactions between ErbB3 and the membrane-associated RGS4 protein by coimmunoprecipitating the two proteins from human cells. We expect the split-ubiquitin membrane YTH technology to be valuable for the identification of potential interacting partners of integral membrane proteins from many model organisms.
The recent advances in analyses of completely sequenced genomes of numerous model organisms, and also the human genome, have revealed that approximately one-third of all predicted gene products of a given organism are likely to be associated with membranes (Auerbach et al. 2002b). These proteins execute a variety of essential cellular tasks, which include cell signaling, transport of membrane-impermeable molecules, and cell adhesion.
The central position of membrane proteins in mediating signaling across the membrane makes them one of the most important pharmacological targets today. Receptor tyrosine kinases, such as the epidermal growth factor (EGF) family (also known as the ErbB receptor family; Riese II and Stern 1998), mediate signals influencing essential cellular responses such as differentiation, proliferation, and survival. Currently, there are four members of the ErbB family: ErbB1 (also termed EGFR or HER1), ErbB2 (also termed HER2 or Neu), ErbB3 (also termed HER3), and ErbB4 (also termed HER4). The importance of ErbB receptors, in particular ErbB1, ErbB2, and ErbB3, in the development and malignancy of human cancer has been amply demonstrated (Olayioye et al. 2000).
Interactions among proteins are essential for proper cellular functioning. By associating an uncharacterized protein with other proteins of known function, deductions about its potential role in the cell can often be made (von Mering et al. 2002). Traditionally, biochemical methods such as coimmunoprecipitation, cross-linking, and copurification by chromatography have been used to investigate the composition of protein complexes. However, these biochemical methods require harsh treatments for cell disruption and therefore may not preserve weak and/or transient interactions.
To address technical difficulties associated with the biochemical characterization of physical protein–protein interactions, alternative genetic methods have been developed. The most powerful genetic method for the study of protein–protein interactions is the yeast two-hybrid (YTH) system, which is based on reconstitution of a functional transcription factor through a defined protein–protein interaction (Fields and Song 1989). Since its description, various modifications of the YTH system have been described, which include the SOS-recruitment system (Aronheim et al. 1994), Ras-recruitment system (RRS; Broder et al. 1998), reverse RRS (Hubsman et al. 2001), split-ubiquitin assay (Johnsson and Varshavsky 1994), and a G protein fusion method (Ehrhard et al. 2000).
Despite significant progress in development of the YTH system, the analysis of interactions between membrane proteins remained a significant challenge because of the hydrophobic nature of these proteins (Auerbach et al. 2002a; Stagljar and Fields 2002). In addition, integral and membrane-associated proteins often undergo posttranslational modifications or oligomerize via interactions between their transmembrane domains, all of which is unfavorable for a traditional YTH assay. Consistent with these problems, two independently performed genome-wide YTH screens have shown that the coverage of membrane protein interactions is poor (Uetz et al. 2000; Ito et al. 2001).
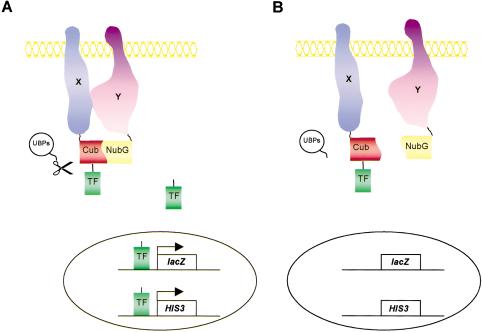
We have shown previously that the modified split-ubiquitin system can be used as a genetic assay for the in vivo detection of interactions between two essential subunits of the yeast oligosaccharyl transferase membrane protein complex, Ost1p and Wbp1p (Stagljar et al. 1998; Stagljar and te Heesen 2000). This so-called split-ubiquitin membrane YTH system takes advantage of the split-ubiquitin approach first described by Johnsson and Varshavsky (1994). It is based on the ability of the N- and C-terminal halves of ubiquitin, Nub and Cub, to reassemble into a quasi-native ubiquitin (split-ubiquitin). Ubiquitin-specific proteases (UBPs), present in the cytosol and nucleus of all eukaryotic cells, recognize such reconstituted ubiquitin but not its halves and cleave off a reporter protein that is linked to the C terminus of Cub. The assay is designed in such a way that the association of Nub and Cub is only efficient if the ubiquitin halves are linked to two proteins that interact in vivo. In the split-ubiquitin membrane YTH system, one protein of interest, the bait X, is fused to the Cub domain, followed by an artificial transcription factor (TF), and the other, the prey Y, is fused to NubG (Fig. 1). To detect a potential interaction between these two proteins X and Y, the two plasmids are introduced into the yeast reporter strain. Interaction of both proteins results in the assembly of split-ubiquitin and the proteolytic release of TF by UBPs, allowing TF to enter the nucleus, leading to the activation of the yeast reporter genes (Stagljar et al. 1998; Thaminy and Stagljar 2002). The split-ubiquitin membrane YTH technology has also been used to study the interaction between the yeast α 1,2-mannosidase Mns1p and Rer1p in the ER (Massaad and Herscovics 2001), to investigate the influence of mutations on the assembly of fragments of presenilin (Cervantes et al. 2001), and with plant proteins to study intra- and intermolecular interactions between sucrose transporters (Reinders et al. 2002) as well as between TOM2A and TOM1 transmembrane proteins (Tsujimoto et al. 2003).
Figure 1.
Outline of the split-ubiquitin membrane yeast two-hybrid system. (A) A membrane bait protein of interest X is fused to Cub followed by an artificial transcription factor (TF), while another membrane (or cytoplasmic) protein Y is fused to the NubG domain (Y-NubG). On interaction of the X and Y proteins, ubiquitin reconstitution occurs, leading to proteolytic cleavage by UBPs, and the subsequent release of the transcription factor. This factor activates reporter genes to result in HIS3+/lacZ+ yeast cells. (B) If X and Y do not interact, there is no ubiquitin reconstitution and thus no UBP-mediated cleavage, resulting in HIS-/lacZ- yeast cells.
In this study, we have generated a new set of reagents for the expression of heterologous proteins in the split-ubiquitin membrane YTH system and converted the system into a generally applicable in vivo screening approach for identification of interacting partners of a particular mammalian transmembrane protein. We have demonstrated the effectiveness of this approach using the mammalian ErbB3 receptor as bait, and have identified three previously unknown ErbB3-interacting proteins. We expect the split-ubiquitin membrane YTH technology to be valuable for the identification of interacting partners of many integral membrane proteins from any model organism, and it may also prove useful for drug discovery purposes.
RESULTS
New Bait and Prey Vectors for the Split-Ubiquitin Membrane Yeast Two-Hybrid System
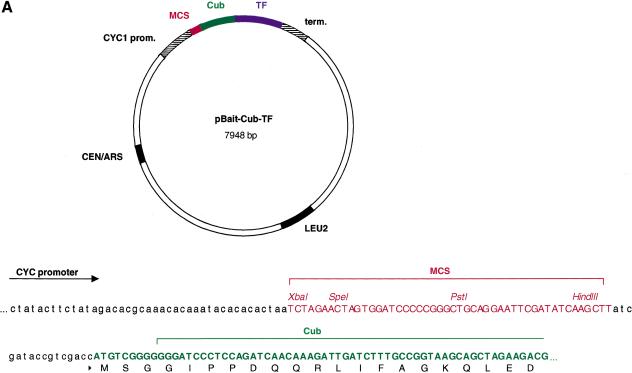
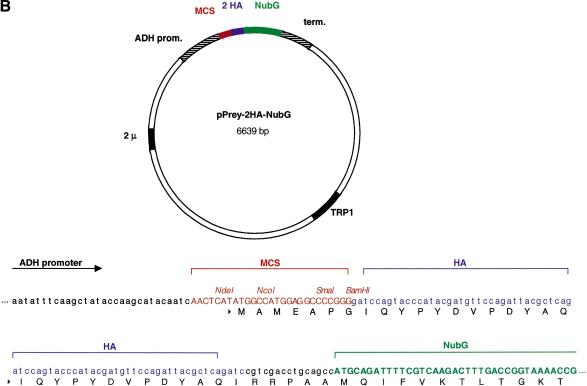
Our central goal was to use the split-ubiquitin membrane YTH technology to identify proteins that interact with a given mammalian membrane target protein. To this end, we have constructed a new set of bait and prey vectors that allow the constitutive expression of heterologous membrane proteins in yeast (Fig. 2). The bait vector pCYC-BAIT-Cub-TF is a centromeric vector containing a CEN/ARS origin of replication, a Cytochrome-C oxidase (CYC1) promoter, which provides low-level expression of the bait fusion protein, a multicloning site (MCS), the C-terminal (Cub) domain of yeast ubiquitin fused in frame to an artificial transcription factor consisting of Staphylococcus aureus Protein A, Escherichia coli LexA, and the Herpes simplex virus transactivator VP16 (collectively called TF), as well as the LEU2 gene for selection in yeast (Fig. 2A). The prey vector pADH-PREY-2HA-NubG carries a 2 μm origin of replication, the ADH1 promoter, a MCS followed by two hemagglutinin (HA) epitope tags fused in frame to the mutated N-terminal (NubG) domain of yeast ubiquitin, the ADH terminator, and the TRP1 gene for selection in yeast (Fig. 2B). Both pCYC-BAIT-Cub-TF and pADH-PREY-2HA-NubG vectors are suitable for the expression of Type I transmembrane protein in the split-ubiquitin membrane YTH system. We have also generated novel bait (pCYC-TF-Cub-BAIT) and prey (pADH-NubG-HA-PREY) vectors for the analysis of Type II transmembrane proteins. The construction and application of these vectors in the split-ubiquitin membrane YTH system will be described elsewhere (S. Thaminy and I. Stagljar, unpubl.).
Figure 2.
Maps of novel vectors for the expression of Type I transmembrane bait and prey proteins in the split-ubiquitin membrane yeast two-hybrid system. (A) The bait vector pCYC-BAIT-Cub-TF is a LEU2-based low copy number (CEN/ARS) vector bearing a weak yeast CYC1 promoter, the MCS, and the Cub domain followed by the TF. The foreign cDNA sequence encoding a transmembrane bait protein of interest is introduced into the MCS in frame to Cub-TF portion. Also shown is the MCS sequence upstream of the Cub-TF fusion containing the unique XbaI, SpeI, PstI, and HindIII restriction sites. (B) The prey vector pADH-PREY-2HA-NubG is a TRP1-based multicopy (2μ) vector bearing a strong yeast ADH1 promoter, the MCS, and two HA tags followed by the NubG domain. The cDNA or a library of genomic or cDNA fragments is fused in frame to the NubG cassette. Also shown is the MCS sequence upstream of the two HA-NubG cassettes containing the unique restriction sites NdeI, NcoI, SmaI, and BamHI. Both bait and prey vectors were constructed as described in the Methods section.
Expression of ErbB3 Bait Within the Yeast Membrane
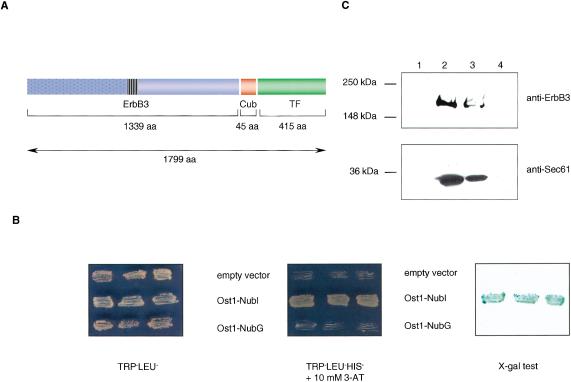
Mammalian ErbB3 protein was the first heterologous bait to be tested in the split-ubiquitin membrane YTH system because it has an important function in cell signaling and is an interesting drug target (Olayioye et al. 2000; de Bono and Rowinsky 2002). To generate an ErbB3 bait, we fused the full-length rat ErbB3 protein (aa 1–1339) N terminally to Cub-TF, thus generating ErbB3-Cub-TF (Fig. 3A). We then examined whether coexpression of ErbB3-Cub-TF with either an empty prey vector or a noninteracting yeast membrane protein Ost1p (Stagljar et al. 1998) fused to the NubI or NubG domains would activate the yeast gene reporter system. Expression of ErbB3-Cub-TF with a noninteracting Ost1-NubG and empty prey vector resulted in HIS-/lacZ- cells, indicating that the ErbB3-Cub-TF bait is not self-activating (Fig. 3B). In contrast, coexpression of ErbB3-Cub-TF and a control plasmid pADH-Ost1–2HA-NubI, containing the wild-type Nub sequence (NubI), resulted in the split-ubiquitin formation and activation of the yeast reporter system. This occurs because any NubI protein fusion associates with Cub independent of additional protein–protein interactions (Johnsson and Varshavsky, 1994; Stagljar et al. 1998).
Figure 3.
(A) The structure of the ErbB3-Cub-TF bait protein used in this study. Like other members of the ErbB family, ErbB3 is a type I transmembrane protein consisting of an extracellular ligand binding domain (blue dotted box), a single membrane-spanning region (striped box), and a cytoplasmic protein tyrosine kinase domain (blue open box). The ErbB3 bait was fused to Cub (red box), followed by an artificial transcription factor (TF; green box). The number of amino acids of ErbB3, Cub, and TF portions are indicated. (B) Growth of yeast cells expressing ErbB3-Cub-TF bait with various Nub-fusions on agar plates lacking tryptophan and leucine (left), and tryptophan, leucine, and histidine containing 10 mM 3-aminotriazole (3-AT; middle). The L40 yeast reporter strain was cotransformed with the ErbB3-Cub-TF bait and indicated prey plasmids, and three independent colonies were grown on Leu-Trp- and Leu-Trp-His-selective plates prior to assessment of β-galactosidase activity using X-gal filter test (right). (C) ErbB3 is localized within the yeast membrane. Cytosolic (lanes 1 and 3) and membrane (lanes 2 and 4) fractions of yeast cells expressing the ErbB3-Cub-TF bait were subjected to SDS-PAGE. The insoluble fraction (lane 2) was treated with 1% Triton X-100 to solubilize the proteins, and centrifuged to separate the soluble (lane 3) from insoluble proteins (lane 4). The ErbB3-Cub-TF bait and a control endogenous yeast membrane protein Sec61p were detected by immunoblot analysis using a mouse monoclonal anti-ErbB3 antibody (upper panel), and an anti-Sec61 polyclonal antibody (lower panel). The positions of molecular markers are indicated.
To demonstrate that the bait is targeted to the yeast membrane, ErbB3-Cub-TF was expressed in the L40 yeast reporter strain. Subsequently, yeast cells were lysed, then centrifuged to resolve the membrane and cytosolic fractions, and separated by SDS-PAGE. A protein of approximately 200 kD corresponding to ErbB3-Cub-TF fusion was detected within the membrane fraction (Fig. 3C, lane 2, upper panel) but not in the cytosolic fraction (Fig. 3C, lane 1, upper panel). To control our fractionation experiment, we examined the distribution of an endogenous yeast ER membrane protein Sec61p. Similar to the ErbB3-Cub-TF bait, Sec61p was found exclusively within the membrane fraction (Fig. 3C, lane 2, lower panel) and not in the cytosolic fraction (Fig. 3C, lane 1, lower panel). We then treated the membrane fraction (containing the insoluble proteins) with the detergent TritonX-100, and centrifuged the extract to separate soluble and insoluble fractions. After detergent treatment, both the ErbB3-Cub-TF bait and Sec61p were only detected within the soluble fraction (Fig. 3C, lane 3, upper and lower panels) and not within the insoluble fraction (Fig. 3C, lanes 4, upper and lower panels). In conclusion, these biochemical experiments indicate that the ErbB3-Cub-TF bait is expressed and correctly localized within the yeast membrane.
Screening of a Human Brain cDNA Library Fused to NubG
To identify novel ErbB3 interacting partners, we constructed a human brain cDNA library. The cDNAs were inserted N terminally to two HA tags followed by the NubG sequence, thus generating the library in Y-NubG orientation (where Y is an insert cDNA). This human brain cDNA library was transformed into the L40 yeast reporter strain (Vojtek et al. 1993) that expressed ErbB3-Cub-TF as bait. From 1.5 × 107 transformants, 170 clones displayed histidine prototrophy and β-galactosidase activity. The library plasmids from these HIS3+/lacZ+ colonies were recovered and transformed into E. coli for amplification. They were then reintroduced into L40, expressing either the ErbB3-Cub-TF bait or a control bait consisting of the yeast oligosaccharyl transferase subunit Wbp1 fused to Cub-TF (Stagljar et al. 1998). One hundred and forty-five independent clones failed to display a HIS3+/lacZ+ phenotype with either the ErbB3-Cub-TF bait or the Wbp1-Cub-TF control bait. These clones represent false positives that most probably arise from several distinct mechanisms during the initial growth selection process. Such false positives have also been observed in other genetic systems such as the traditional YTH system (Serebriiskii et al. 2000). Possible reasons for the occurrence of this class of false positives in our screen are given in the Discussion section. Twenty of the 170 independent clones interacted with both the ErbB3-Cub-TF and Wbp1-Cub-TF control bait. These plasmids encode a class of false positives that apparently bind to the TF portion of the bait proteins. Importantly, five of the 170 independent clones interacted specifically with the ErbB3-Cub-TF but not with the Wbp1-Cub-TF control bait (Table 1). Two cDNAs encoded RGS4, a member of a protein family termed Regulators of G-protein Signaling (RGS) that act as negative regulators within G protein pathways (Hollinger and Hepler 2002). RGS4 was shown to exist as a membrane-bound protein (Srinivasa et al. 1998), but it was also found in the cytosol of human neuronal NG108 cells (Druey et al. 1998). Recently, it was reported that RGS16, another member of the RGS family, interacts with the epidermal growth factor (EGFR) receptor (Derrien and Druey 2001), a member of the ErbB family. Two additional clones encoded the hypothetical zinc finger protein ZNF207. So far, the function and localization of ZNF207 in human cells remain elusive (Pahl et al. 1998). The last clone identified encoded Egr-1 (Early Growth Response protein-1). Egr-1 is a zinc-finger transcription factor that can be localized both to the nucleus and cytosol of human cells (Matheny et al. 1994). Many biological functions have been attributed to Egr-1, which include neurite outgrowth, wound repair, growth control, and apoptosis (Beckmann and Wilce 1997). Furthermore, recent studies have shown that Egr-1 is a regulator of the platelet-derived growth factor receptor (Khachigian et al. 1995) and EGF receptor signaling pathways (Amorino et al. 2002).
Table 1.
ErbB3-Interacting Proteins Identified in the Split-Ubiquitin Membrane Yeast Two-Hybrid Screen
| Name of the encoded gene | No. of clones identified | Acc. no.a | Protein sequence fused to NubGb | Localization in human cells | References |
|---|---|---|---|---|---|
| RGS4 (Regulator of G protein Signaling) | 2 | XM_034023 | aa 1-178 (205) | Membrane | Srinivasa et al. 1998 |
| Cytosol | Druey et al. 1998 | ||||
| Egr-1 (Early Growth Response 1) | 1 | NM_001964.1 | aa 169-443 (543) | Nucleus, cytosol | Matheny et al. 1994 |
| Zinc finger protein ZNF207 | 2 | XM_008462 | aa 143-334 (493) | Unknown | Pahl et al. 1998 |
For each identified clone, the GenBank database accession number is given
The number of amino acids corresponding to the full-length protein is indicated in parentheses
ErbB3 and RGS4 Form a Complex in Human Cells
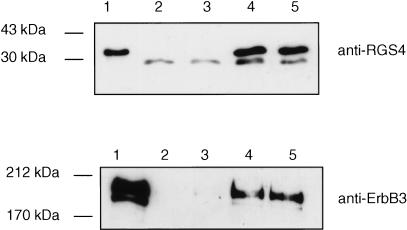
Given the fact that ErbB3 interacts with the membrane-associated RGS4 protein in the split-ubiquitin membrane YTH system, we wanted to test whether ErbB3 forms a complex with RGS4 in vivo. To this end, ErbB3 and RGS4 were transiently overexpressed in HEK293T cells and coimmunoprecipitation experiments were performed (Fig. 4). ErbB3 was immunoprecipitated with RGS4 using the anti-RGS4 antibody (lower panel, lanes 4 and 5), but not with the control IgG antibody (lower panel, lanes 2 and 3). Moreover, the interaction between ErbB3 and RGS4 was not affected by increasing the salt concentration from 100 mM to 150 mM (lower panel, lanes 4 and 5). We conclude from the split-ubiquitin membrane YTH system and coimmunoprecipitation assays that ErbB3 interacts with RGS4 both in yeast and human cells.
Figure 4.
ErbB3 and RGS4 form a complex in human cells. Cell lysates of HEK293T cells coexpressing ErbB3 and RGS4 were immunoprecipitated with either IgG antibody (lanes 2 and 3) or goat polyclonal anti-RGS4 antibody (lanes 4 and 5) using either 100 mM (lanes 2 and 4) or 150 mM NaCl (lanes 3 and 5), and analyzed by immunoblotting with either rabbit polyclonal anti-RGS4 antibody (upper panel) or mouse monoclonal anti-ErbB3 antibody (lower panel). One-tenth of the same extract was used as the input control (lane 1).
Mapping of the ErbB3 Interaction Region Within RGS4
To investigate which region of RGS4 is required for binding to ErbB3, several RGS4 truncations were generated and tested for their ability to interact with ErbB3-Cub-TF in the split-ubiquitin membrane YTH assay (Fig. 5A). Binding to the ErbB3-Cub-TF bait was assessed using a quantitative β-galactosidase assay (Fig. 5B), and the correct expression and localization of all RGS4 deletion mutants were confirmed by immunoblotting analysis using an anti-HA antibody (Fig. 5C; Fig. 1 Supplementary Material, available online at www.genome.org).
Figure 5.
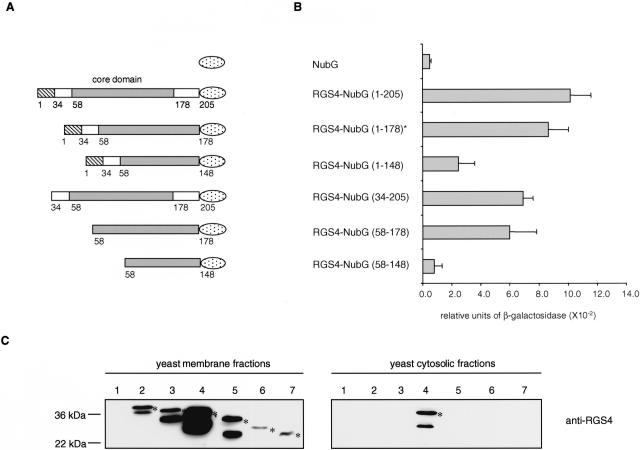
Mapping of the ErbB3 interaction domain within RGS4. (A) RGS4-NubG deletion constructs. RGS4 contains an N-terminal amphipathic region important for membrane localization (amino acid 1–33, striped box) followed by the RGS box (amino acid 58–178, grey box) and the C-terminal tail (amino acid 179–205). The NubG domain of ubiquitin is shown as a dotted ellipse. (B) Quantitative β-galactosidase assay showing the binding of different RGS4-NubG deletion mutants to the ErbB3-Cub-TF bait. Values given are the mean of four transformants, each assayed at least twice. The asterisk indicates the original RGS4 clone isolated in the split-ubiquitin membrane yeast two-hybrid screen. (C) Expression and localization of the RGS4–2HA-NubG deletion mutants. The RGS4–2HA-NubG deletion mutants were generated by PCR followed by in vivo recombination into the prey plasmid pADH-PREY-2HA-NubG previously digested with NdeI. Western blot analyses were performed using membrane (left panels) and cytosolic (right panels) fractions of the yeast L40 reporter strain expressing ErbB3-Cub-TF and the prey NubG (lane 1), RGS4-NubG (1–178; lane 2), RGS4-NubG (1–205; lane 3), RGS4-NubG (34–205; lane 4), RGS4-NubG (1–148; lane 5), RGS4-NubG (58–178; lane 6), and RGS4-NubG (58–148; lane 7). The blots were incubated with anti-RGS4 antibody. The positions of molecular markers are indicated. The bands that are not marked with an asterisk are considered as degradation products of RGS4. The RGS4-NubG (1–178) protein found in the split-ubiquitin membrane yeast two-hybrid screen (lane 2) migrates more slowly than the RGS4-NubG (1–205; lane 3) because it contains an additional 45 amino acids upstream of the initial RGS4 start codon.
The results of our interaction region mapping experiment indicate that the RGS4-NubG clone (aa 1–178) identified in the split-ubiquitin membrane YTH assay, as well as the full-length RGS4-NubG (aa 1–205), strongly interacts with ErbB3-Cub-TF. Deletion of the N-terminal 33 amino acids of RGS4 (aa 34–205), known to be necessary for membrane localization of the protein in yeast (Srinivasa et al. 1998), slightly decreased the binding of RGS4 to ErbB3. Immunoblot analysis of membrane and cytosolic fractions isolated from yeast cells expressing RGS4-NubG (aa 34–205) showed that this deletion mutant was localized both in the cytosol and membrane fractions, whereas the full-length RGS4-NubG was exclusively expressed in the membrane fraction (Fig. 1 Supplementary material). The RGS4 core domain alone (aa 58–178), which is shared by all RGS family members (Srinivasa et al. 1998), also showed reduced binding to ErbB3 compared with full-length RGS4. However, a deletion affecting the C-terminal 30 amino acids of the core domain (amino acids 1–148 and amino acids 58–148) strongly decreased the interaction with ErbB3. These results indicate that residues important for the interaction of RGS4-NubG with ErbB3-Cub-TF are located in the C-terminal region of the RGS box.
DISCUSSION
In this study, we describe a novel yeast-based screening technology, the split-ubiquitin membrane YTH system, designed for the study of membrane protein interactions. We have demonstrated the utility of this technology to detect three novel proteins associated with the mammalian ErbB3 tyrosine kinase receptor. In addition, we have confirmed one of the newly found interactions between ErbB3 and membrane-associated RGS4 protein by coimmunoprecipitating the two proteins from human cells. We have also shown that the split-ubiquitin membrane YTH technology can be used to map the region of RGS4 that mediates the interaction with the ErbB3 receptor. However, the functional importance of the ErbB3/RGS4 and other two interactions identified in our ErbB3 screen still remains to be determined.
The split-ubiquitin membrane YTH technology has several advantages over the conventional YTH (Fields and Song 1989). Unlike the YTH, the split-ubiquitin membrane YTH is not limited to the analysis of soluble proteins or subdomains of membrane proteins; thus, screening with full-length integral membrane proteins or membrane-associated proteins offers the opportunity to identify protein–protein interactions as they take place in their natural setting. In addition, the split-ubiquitin membrane YTH system can be applied to any transmembrane bait protein as long as the Cub-TF and NubG modules that are fused to the protein of interest are located in the cytoplasm. Moreover, in the split-ubiquitin membrane YTH system genomic or cDNA libraries can be screened in both orientations (Y-NubG or NubG-Y). In this way, it is possible to identify both Type I (Y-NubG orientation) and Type II (NubG-Y orientation) transmembrane proteins that interact with a particular membrane bait protein. To test the feasibility of this approach, we have recently generated a NubG-Y human brain cDNA library and screened it for proteins interacting with the human β-2 adrenergic receptor, a G-protein coupled receptor (D. Auerbach and I. Stagljar, unpubl.). We have also extended the split-ubiquitin membrane YTH approach to detect novel protein interactors of the yeast oligosaccharyl transferase subunit Wbp1 by screening the yeast random genomic NubG-fused libraries (D. Auerbach and I. Stagljar, unpubl.).
Like other genetic screening systems, a major drawback of the split-ubiquitin membrane YTH is a high number of false positives that arise during the selection process. Several independent factors may account for such a high rate of false positives in the split-ubiquitin membrane YTH system. Promoter mutations may activate either the HIS3 or lacZ reporter genes, leading to growth on selective medium in the absence of a protein–protein interaction. In addition, certain proteins, when overexpressed in yeast, may unspecifically activate the HIS3 and lacZ genes (Serebriiskii et al. 2000). Furthermore, interactions between integral membrane proteins in our system are especially sensitive to protein levels and transiently increased expression levels, which, for example, may occur because of copy number variations of the library plasmids, thus producing unspecific interactions (D. Auerbach. and I. Stagljar, unpubl.). However, these false positives can be eliminated using a set of genetic criteria that can be rapidly tested: plasmids encoding putative interactors are rapidly recovered from yeast and transformed back into the original yeast reporter strain, together with a plasmid encoding the bait protein-Cub-TF fusion, or with a plasmid encoding a noncognate bait protein-Cub-TF fusion. Putative interactors that are positive with the original bait, but negative with the noncognate bait, are considered as true positives and are selected for further study. Thus, when using automated procedures that facilitate the recovery of library plasmids from yeast, false positives from a split-ubiquitin membrane YTH screen can be eliminated quickly.
Despite the fact that we expressed the full-length ErbB3 receptor in its natural setting, we did not identify any of the components associated with the ErbB signaling cascade. ErbB homo- or heterodimerization is induced by binding of extracellular ligands to their respective receptors, which then leads to phosphorylation of the cytosolic kinase domains and recruitment of adaptor proteins (Yarden 2001). However, ErbB3 is an exception because it possesses no intrinsic kinase activity and, consequently, signaling via ErbB3 requires heterodimerization with another ErbB family member. As all adaptor proteins that have been identified as components of ErbB signaling cascades so far bind exclusively to activated (e.g., phosphorylated) receptor tails (Olayioye et al. 2000), this may explain why we did not find those proteins in our screen.
Nrdp1 (also called FLRF) is a ubiquitin ligase involved in regulation of steady-state ErbB receptor levels (Diamonti et al. 2002; Qiu and Goldberg 2002). Nrdp1 has been shown to interact with the nonactivated ErbB3 receptor cytoplasmic tail in a YTH assay (Diamonti et al. 2002). For this reason, we would expect Nrdp1 to bind to the ErbB3 receptor when expressed in yeast. However, we failed to identify Nrdp1 in our library screen. The reason for this is presently unclear but may be connected to an underrepresentation or absence of Nrdp1 in the cDNA library we used.
Like the YTH system, the split-ubiquitin membrane YTH system is prone to false positives and thus, interactions that have been identified in a screen should be confirmed in an independent system. Coimmunoprecipitation or pull-down assays using recombinantly expressed proteins or protein fragments have traditionally been used most often to confirm YTH results. However, both assays are problematic when used with integral membrane proteins. In a first step, it may therefore be advantageous to directly assess newly found interactions in the split-ubiquitin membrane YTH system by means of a competition experiment, in which untagged bait is overexpressed together with the original bait-Cub-TF/prey-NubG combination. The activation of reporter genes is then expected to be abolished by overexpression of the competing partner. Alternatively, methods that are more suitable for use with membrane proteins, such as colocalization experiments or fluorescence resonance energy transfer, may also be used to confirm an interaction.
Among alternative yeast-based screening approaches developed for membrane proteins, the reverse RRS is also capable of detecting interactions involving a membrane protein as bait (Hubsman et al. 2001). However, interactions involving two integral membrane proteins cannot be detected in the RRS system, because the use of a prey membrane protein would activate the system in the absence of any protein–protein interaction. Recently, another split-ubiquitin-based genetic approach has been described (Wittke et al. 1999), which uses a destabilized version of the yeast Ura3 protein (termed rUra3) as a reporter moiety. The interaction of two proteins fused to Cub-rUra3 and NubG, respectively, leads to the cleavage of rUra3, followed by its degradation by enzymes of the N-end rule pathway (Varshavsky 1996). Use of the counterselectable compound 5-fluororotic acid allows the selection of clones where a protein–protein interaction has led to the complete degradation of the rUra3 protein. In the past, the rUra3 based split-ubiquitin assay has been used to analyze changes in protein conformation and stability of the Saccharomyces cerevisiae ER membrane protein Sec62 (Dunnwald et al. 1999) and to map the interactions between several S. cerevisiae integral membrane proteins (Wittke et al. 1999). Furthermore, this system has recently been used in a screening format to identify proteins interacting with Gal4p and Tup1p, two yeast transcription factors (Laser et al. 2000). However, to our knowledge, the rUra3-based split-ubiquitin assay has not yet been reported to work as a screening system for membrane proteins.
The successful application of the split-ubiquitin membrane YTH system as a screening system for membrane proteins, a class of proteins that to date has been difficult to analyze using conventional biochemical and genetic assays, represents a step forward in the analysis of physical protein–protein interactions. The next challenge lies in the adaptation of the split-ubiquitin membrane YTH technology for pharmacological purposes. Using the reagents described in this study, it may be possible to design selection systems that identify peptides, single-chain antibodies, or small molecules that specifically inhibit the interaction between two particular transmembrane proteins. Last but not least, the further development of a split-ubiquitin membrane YTH system, combined with more sophisticated vectors, libraries, and reporter genes, will also enable its adaptation to a high-throughput format to elucidate interactions between membrane and cytosolic proteins on a genome-wide scale. Taken together, these studies will undoubtedly broaden our knowledge of how membrane proteins interact in a cell.
METHODS
Construction of Plasmids
Plasmid sequences and detailed construction schemes of all constructs used in this paper are available on request. All plasmids were verified by sequencing, and the expression of all constructs was checked by Western blot analysis using suitable antibodies.
pCYC-BAIT-Cub-TF
A DNA fragment encoding the Cub-TF portion was amplified by PCR from pY-Cub-PLV (Reinders et al. 2002) and cloned in the multiple cloning site of the HindIII- and NaeI-digested p415CYC1 vector (Mumberg et al. 1995).
pCYC-ErbB3-Cub-TF
The cDNA encoding the rat ErbB3 was amplified by PCR from pBS-ErbB3 (kindly provided by John Koland) and cloned in frame to Cub-TF portion of the SpeI- and HindIII-digested pCYC-BAIT-Cub-TF.
pADH-PREY-2HA-NubG
Two oligos, X-HA-NubG-up (5′-GATCCAGTACCCATACGATGTTCCAGATTACGCTCA-3′) and X-HA-NubG-low (5′-GATCTGAGCGTAATCTGGAACATCGTATGGGTACTG-3′), were ligated to each other and inserted into the BamHI site of the pX-NubG vector. To construct the pX-NubG vector, the DNA sequence encoding the NubG portion was amplified by PCR from pOST1-NubG (Stagljar et al. 1998) and inserted into the PstI site of the Gal4-DNA binding-domain-less pAS2–1 vector (BD Biosciences).
pADH-Ost1–2HA-NubG and pADH-Ost1–2HA-NubI
The entire open reading frame encoding the Ost1p was amplified directly from yeast genomic DNA and cloned into the NcoI and BamHI sites of pADH-PREY-2HA-NubG and pADH-PREY-2HA-NubI, respectively, to yield pADH-Ost1–2HA-NubG and pADH-Ost1–2HA-NubI.
pADH-RGS4–2HA-NubG Deletion Mutants
These constructs were generated by PCR amplification of different portions of RGS4 and subsequent cloning by in vivo recombination in yeast in the Nde I-digested pADH-PREY-2HA-NubG.
Construction of the NubG-Fused Random Human Brain cDNA Library
The random human brain cDNAs were generated using the SuperScriptII RNase H-Reverse Transcriptase (INVITROGEN) from 5 μg of total human fetal brain RNA (AMBION). The primary library was introduced in pADH-PREY-2HA-NubG vector (Y-NubG orientation, where Y is an insert cDNA) using the Gateway system and had a complexity of 1 × 107 clones. The subsequent random analysis of 23 E. coli colonies showed that the percentage of recombinants is 87% and that the average insert size is 1.1 kb. Requests for the Y-NubG-fused random human brain cDNA library should be directed to info@dualsystems.com.
ErbB3 Split-Ubiquitin Membrane Yeast Two-Hybrid Screen
Two hundred micrograms of the human brain cDNA library fused N terminally to NubG (Y-NubG orientation) were transformed into the yeast reporter strain L40(MATa trp1 leu2 his3 LYS2::lexA-HIS3 URA3::lexA-lacZ) expressing ErbB3-Cub-TF bait using the lithium acetate protocol. Approximately 1.5 × 107 TRP+ LEU+ transformants were selected on SD Leu-Trp-His- medium containing 10 mM 3-aminotriazole (3-AT). Library plasmids were isolated from 170 initial positive HIS3+/lacZ+ yeast colonies and rescued into E. coli XL1-Blue according to standard procedures. Isolated library plasmids were retransformed into L40 expressing either ErbB3-Cub-TF or the yeast Wbp1-Cub-TF control bait. Three individual colonies of each transformant were tested for the activation of the lacZ reporter by X-gal filter lift-off assay after incubation for 5 h at room temperature. Only the plasmids that activated the HIS3 and lacZ reporters in combination with the ErbB3-Cub-TF bait, but not with the Wbp1-Cub-TF control bait, were selected for sequencing and further studies.
Protein Purification and Immunoblotting
Protein extracts and membrane fractionations were performed as described previously (David et al. 1997). Briefly, cells equivalent to A600 = 100 were resuspended in 50 mM Tris-HCl (pH 7.9) supplemented with protease inhibitors (Roche Molecular Biochemicals) and lysed by vortexing with glass beads (Sigma). Unlysed cells were removed by centrifugation at 700g, and the membrane fraction was collected by centrifugation at 150,000g. To solubilize the membrane fraction, we resuspended the pellet in the lysis buffer containing 1% Triton X-100, followed by centrifugation at 150,000g. Rabbit anti-VP16 polyclonal antibody (1:1000; Clontech), mouse anti-HA monoclonal antibody (1:1000; Babco), mouse monoclonal anti-ErbB3 antibody (1:5000; Neomarkers), rabbit polyclonal anti-RGS4 antibody (1:1000; kindly provided by Kirk Druey), rabbit polyclonal Sec61p antibody, and Yrb1p antibody (respectively 1:2000 and 1:5000; kindly provided by Claude Jakob) were used.
Immunoprecipitation
The full-length ErbB3 cDNA was amplified by PCR and inserted into the XbaI and HindIII sites of pcDNA3 (Invitrogen). HA-tagged full-length human RGS4 in pcDNA3 was kindly provided by Kirk Druey. HEK 293T cells were grown and transiently transfected as previously described (Derrien and Druey 2001). Two days after transfection, cells were washed with PBS and lysed in a buffer containing 100 or 150 mM Nacl, 50 mM Tris (pH 7.5), 5 mM EDTA, 1% Nonidet-P-40, 2 mM sodium vanadate, 1 mM phenylmethylsulfonyl fluoride, and a mixture of protease inhibitors (Roche Molecular Biochemicals). Lysed cells were incubated for 1 h at 4 °C and centrifuged at 13,000g for 15 min. The supernatant was designated as cell lysate. RGS4 was immunoprecipitated with 10μg goat polyclonal anti-RGS4 N-16 antibody (1:1000; Santa Cruz Biotechnology) or control IgG and then incubated with Protein G sepharose beads (Amersham Pharmacia Biotechnology). The beads were washed three times with lysis buffer containing 0.5% Nonidet-P-40 prior to addition of Laemmli buffer and SDS-PAGE. Immunoblottings were performed using the appropriate antibodies.
Acknowledgments
We thank Michael Fetchko, Stan Fields, Michael Hottiger, and John Miller for helpful discussions; John Koland, Kirk Druey, and Claude Jakob for reagents; and Ulrich Hübscher for his support. We also thank Stephan te Heesen for his generous help during the initial phase of the split-ubiquitin membrane YTH project. The I.S. group is supported by Zürcher Krebsliga, Gebert-Rüf Foundation, Walter Honegger Foundation, Bonizzi-Theler Foundation, EMDO Foundation, Novartis Foundation, Olga Mayenfisch Foundation, Sassella Foundation, Fonds für medizinische Forschung, Kommission für Technische Inovation (KTI, Nr. 5343.2 SUS), and the Swiss National Science Foundation (31–58798.99 and 3100A0–100256/1).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1276503.
Footnotes
[Supplementary material is available online at www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: J. Koland, K. Druey, and C. Jakob.]
References
- Amorino, G.P., Hamilton, V.M., Valerie, K., Dent, P., Lammering, G., and Schmidt-Ullrich, R.K. 2002. Epidermal growth factor receptor dependence of radiation-induced transcription factor activation in human breast carcinoma cells. Mol. Biol. Cell 13: 2233-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronheim, A., Engelberg, D., Li, N., al-Alawi, N., Schlessinger, J., and Karin, M. 1994. Membrane targeting of the nucleotide exchange factor Sos is sufficient for activating the Ras signaling pathway. Cell 78: 949-961. [DOI] [PubMed] [Google Scholar]
- Auerbach, D., Galeuchet-Schenk, B., Hottiger, M.O., and Stagljar, I. 2002a. Genetic approaches to the identification of interactions between membrane proteins in yeast. J. Recept. Signal Transduct. Res. 22: 471-481. [DOI] [PubMed] [Google Scholar]
- Auerbach, D., Thaminy, S., Hottiger, M.O., and Stagljar, I. 2002b. The post-genomic era of interactive proteomics: Facts and perspectives. Proteomics 2: 611-623. [DOI] [PubMed] [Google Scholar]
- Beckmann, A.M. and Wilce, P.A. 1997. Egr transcription factors in the nervous system. Neurochem. Int. 31: 477-510. [DOI] [PubMed] [Google Scholar]
- Broder, Y.C., Katz, S., and Aronheim, A. 1998. The ras recruitment system, a novel approach to the study of protein–protein interactions. Curr. Biol. 8: 1121-1124. [DOI] [PubMed] [Google Scholar]
- Cervantes, S., Gonzalez-Duarte, R., and Marfany, G. 2001. Homodimerization of presenilin N-terminal fragments is affected by mutations linked to Alzheimer's disease. FEBS Lett. 505: 81-86. [DOI] [PubMed] [Google Scholar]
- David, N.E., Gee, M., Andersen, B., Naider, F., Thorner, J., and Stevens, R.C. 1997. Expression and purification of the Saccharomyces cerevisiae α-factor receptor (Ste2p), a 7-transmembrane-segment G protein-coupled receptor. J. Biol. Chem. 272: 15553-15561. [DOI] [PubMed] [Google Scholar]
- de Bono, J.S. and Rowinsky, E.K. 2002. The ErbB receptor-family: A therapeutic target for cancer. Trends Mol. Med. 8: S19-26. [DOI] [PubMed] [Google Scholar]
- Derrien, A. and Druey, K.M. 2001. RGS16 function is regulated by epidermal growth factor receptor-mediated tyrosine phosphorylation. J. Biol. Chem. 276: 48532-48538. [DOI] [PubMed] [Google Scholar]
- Diamonti, A.J., Guy, P.M., Ivanof, C., Wong, K., Sweeney, C., and Carraway, 3rd, K.L. 2002. An RBCC protein implicated in maintenance of steady-state neuregulin receptor levels. Proc. Natl. Acad. Sci 99: 2866-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druey, K.M., Sullivan, B.M., Brown, D., Fischer, E.R., Watson, N., Blumer, K.J., Gerfen, C.R., Scheschonka, A., and Kehrl, J.H. 1998. Expression of GTPase-deficient Giα2 results in translocation of cytoplasmic RGS4 to the plasma membrane. J. Biol. Chem. 273: 18405-18410. [DOI] [PubMed] [Google Scholar]
- Dunnwald, M., Varshavsky, A., and Johnsson, N. 1999. Detection of transient in vivo interactions between substrate and transporter during protein translocation into the endoplasmic reticulum. Mol. Biol. Cell 10: 329-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhard, K.N., Jacoby, J.J., Fu, X.Y., Jahn, R., and Dohlman, H.G. 2000. Use of G-protein fusions to monitor integral membrane protein–protein interactions in yeast. Nat. Biotechnol. 18: 1075-1079. [DOI] [PubMed] [Google Scholar]
- Fields, S. and Song, O. 1989. A novel genetic system to detect protein–protein interactions. Nature 340: 245-246. [DOI] [PubMed] [Google Scholar]
- Hollinger, S. and Hepler, J.R. 2002. Cellular regulation of RGS proteins: Modulators and integrators of G protein signaling. Pharmacol. Rev. 54: 527-559. [DOI] [PubMed] [Google Scholar]
- Hubsman, M., Yudkovsky, G., and Aronheim, A. 2001. A novel approach for the identification of protein–protein interaction with integral membrane proteins. Nucleic Acids Res. 29: E18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., and Sakaki, Y. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. 98: 4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson, N. and Varshavsky, A. 1994. Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl. Acad. Sci. 91: 10340-10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khachigian, L.M., Williams, A.J., and Collins, T. 1995. Interplay of Sp1 and Egr-1 in the proximal platelet-derived growth factor A-chain promoter in cultured vascular endothelial cells. J. Biol. Chem. 270: 27679-27686. [DOI] [PubMed] [Google Scholar]
- Laser, H., Bongards, C., Schuller, J., Heck, S., Johnsson, N., and Lehming, N. 2000. A new screen for protein interactions reveals that the Saccharomyces cerevisiae high mobility group proteins Nhp6A/B are involved in the regulation of the GAL1 promoter. Proc. Natl. Acad. Sci. 97: 13732-13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massaad, M.J. and Herscovics, A. 2001. Interaction of the endoplasmic reticulum α 1,2-mannosidase Mns1p with Rer1p using the split-ubiquitin system. J. Cell Sci. 114: 4629-4635. [DOI] [PubMed] [Google Scholar]
- Matheny, C., Day, M.L., and Milbrandt, J. 1994. The nuclear localization signal of NGFI-A is located within the zinc finger DNA binding domain. J. Biol. Chem. 269: 8176-8181. [PubMed] [Google Scholar]
- Mumberg, D., Muller, R., and Funk, M. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119-122. [DOI] [PubMed] [Google Scholar]
- Olayioye, M.A., Neve, R.M., Lane, H.A., and Hynes, N.E. 2000. The ErbB signaling network: Receptor heterodimerization in development and cancer. EMBO J. 19: 3159-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahl, P.M., Hodges, Y.K., Meltesen, L., Perryman, M.B., Horwitz, K.B., and Horwitz, L.D. 1998. ZNF207, a ubiquitously expressed zinc finger gene on chromosome 6p21.3. Genomics 53: 410-412. [DOI] [PubMed] [Google Scholar]
- Qiu, X.B. and Goldberg, A.L. 2002. Nrdp1/FLRF is a ubiquitin ligase promoting ubiquitination and degradation of the epidermal growth factor receptor family member, ErbB3. Proc. Natl. Acad. Sci. 99: 14843-14848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders, A., Schulze, W., Thaminy, S., Stagljar, I., Frommer, W.B., and Ward, J.M. 2002. Intra- and intermolecular interactions in sucrose transporters at the plasma membrane detected by the split-ubiquitin system and functional assays. Structure 10: 763-772. [DOI] [PubMed] [Google Scholar]
- Riese II, D.J. and Stern, D.F. 1998. Specificity within the EGF family/ErbB receptor family signaling network. Bioessays 20: 41-48. [DOI] [PubMed] [Google Scholar]
- Serebriiskii, I., Estojak, J., Berman, M., and Golemis, E.A. 2000. Approaches to detecting false positives in yeast two-hybrid systems. Biotechniques 28: 328-330, 332-326. [DOI] [PubMed] [Google Scholar]
- Srinivasa, S.P., Bernstein, L.S., Blumer, K.J., and Linder, M.E. 1998. Plasma membrane localization is required for RGS4 function in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 95: 5584-5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagljar, I. and Fields, S. 2002. Analysis of membrane protein interactions using yeast-based technologies. Trends Biochem. Sci. 27: 559-563. [DOI] [PubMed] [Google Scholar]
- Stagljar, I. and te Heesen, S. 2000. Detecting interactions between membrane proteins in vivo using chimeras. Methods Enzymol. 327: 190-198. [DOI] [PubMed] [Google Scholar]
- Stagljar, I., Korostensky, C., Johnsson, N., and te Heesen, S. 1998. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc. Natl. Acad. Sci. 95: 5187-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaminy, S. and Stagljar, I. 2002. The membrane-based yeast two-hybrid system. In Protein–protein interactions (ed. E. Golemis), pp. 395-405. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Tsujimoto, Y., Numaga, T., Ohshima, K., Yano, M.A., Ohsawa, R., Goto, D., Naito, S., and Ishikawa, M. 2003. Arabidopsis tobomavirus multiplication (TOM) 2 locus encodes a transmembrane protein that interacts with TOM1. EMBO J. 22: 335-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz, P., Giot, L., Cagney, G., Mansfield, T.A., Judson, R.S., Knight, J.R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., et al. 2000. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403: 623-627. [DOI] [PubMed] [Google Scholar]
- Varshavsky, A. 1996. The N-end rule: Functions, mysteries, uses. Proc. Natl. Acad. Sci. 93: 12142-12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtek, A.B., Hollenberg, S.M., and Cooper, J.A. 1993. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74: 205-214. [DOI] [PubMed] [Google Scholar]
- von Mering, C., Krause, R., Snel, B., Cornell, M., Oliver, S.G., Fields, S., and Bork, P. 2002. Comparative assessment of large-scale data sets of protein–protein interactions. Nature 417: 399-403. [DOI] [PubMed] [Google Scholar]
- Wittke, S., Lewke, N., Muller, S., and Johnsson, N. 1999. Probing the molecular environment of membrane proteins in vivo. Mol. Biol. Cell 10: 2519-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden, Y. 2001. Biology of HER2 and its importance in breast cancer. Oncology 61: 1-13. [DOI] [PubMed] [Google Scholar]