Abstract
Asthma was the most common comorbidity in hospitalized patients during the 2009 influenza pandemic. For unknown reasons, hospitalized asthmatics had less severe outcomes and were less likely to die from pandemic influenza. Our data with primary human bronchial cells indicate that changes intrinsic to epithelial cells in asthma may protect against cytopathology induced by influenza virus. To further study influenza virus pathogenesis in allergic hosts, we aimed to develop and characterize murine models of asthma and influenza comorbidity to determine structural, physiological and immunological changes induced by influenza in the context of asthma. Aspergillus fumigatus-sensitized and -challenged C57BL/6 mice were infected with pandemic H1N1 influenza virus, either during peak allergic inflammation or during airway remodeling to gain insight into disease pathogenesis. Mice infected with the influenza virus during peak allergic inflammation did not lose body weight and cleared the virus rapidly. These mice exhibited high eosinophilia, preserved airway epithelial cell integrity, increased mucus, reduced interferon response and increased insulin-like growth factor-1. In contrast, weight loss and viral replication kinetics in the mice that were infected during the late airway remodeling phase were equivalent to flu-only controls. These mice had neutrophils in the airways, damaged airway epithelial cells, less mucus production, increased interferons and decreased insulin-like growth factor-1. The state of the allergic airways at the time of influenza virus infection alters host responses against the virus. These murine models of asthma and influenza comorbidity may improve our understanding of the epidemiology and pathogenesis of viral infections in humans with asthma.
Keywords: acute asthma, airway epithelium, chronic asthma, comorbidity, mouse model
Asthma is a multifactorial disease of the airways that precipitates from genetic predisposition and environmental triggers. The World Health Organization estimates that 235 million people have asthma and an additional 100 million will develop the disease over the next 15 years.1 In allergic asthma, exposure to environmental stimuli results in elevated serum immunoglobulin E, acute airway hyperresponsiveness (AHR), eosinophilic peribronchovascular inflammation and goblet cell (GC) metaplasia with associated mucus hypersecretion together with chronic airway remodeling, which includes angiogenesis, subepithelial fibrosis and smooth muscle cell hyperplasia.
Asthma has long been appreciated as a risk factor for hospitalization because of influenza, and asthmatics were a key target group for administration of influenza vaccines in the United States even before the 2010 recommendation for universal vaccination.2 This association was highlighted during the 2009 H1N1 influenza pandemic.3 Several recent studies reported asthma prevalence in 6–18% of children and adults hospitalized with seasonal flu and 14–30% in those hospitalized with pandemic influenza.4, 5, 6 Surprisingly, despite the high rate of hospitalization, asthma was associated with lower rates of pneumonia, intensive care unit admission, mechanical ventilation and death.3, 4, 5, 7, 8, 9 Reasons for these seemingly contradictory outcomes in asthmatics that develop symptomatic influenza are unclear, but they are of obvious scientific and public health interest.
Animal models of comorbidity are crucial in understanding host–pathogen interactions, which may improve patient care and increase successful medical outcomes. Existing murine model systems of asthma and respiratory virus comorbidity use influenza virus as an inducing agent for asthma development,10 but have not examined the pathogenesis of influenza in established models of allergic asthma. As acute asthma (AA) and chronic asthma (CA) are considerably different syndromes requiring different interventions, there is a need for models that recapitulate the manner in which these types of allergic airways respond to respiratory pathogens. This void in model systems provided the impetus for our work. We previously reported a mouse model that mimics the characteristics of acute inflammatory asthma soon after fungal challenge; the mice later undergo architectural changes that are characteristic of CA with the resolution of inflammation over time.11, 12 Using these models of allergic asthma as platforms, herein, we questioned how influenza virus impacts hosts with characteristics of either AA (marked by AHR, inflammation and GC metaplasia) or CA (marked by airway remodeling). These models serve as a basis to understand the immunological responses of the asthmatic host and may aid in the development of strategies to protect asthmatics from respiratory viral infections.
Results
Asthmatic bronchial epithelium is protected from influenza virus-induced damage
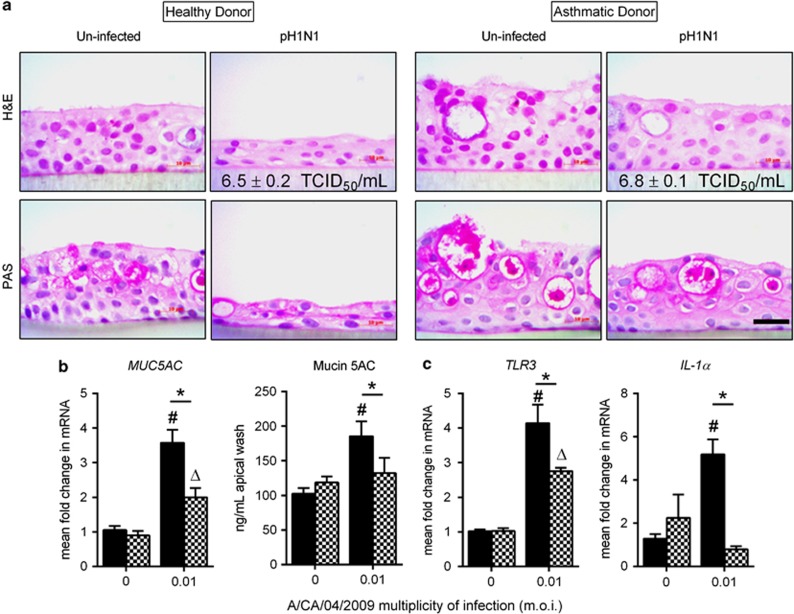
To explore potential differences that allow for better outcomes in asthmatics hospitalized with influenza, we used primary human bronchial epithelial cells from healthy and asthmatic donors grown in an air–liquid interface to determine whether differential susceptibility was observed and, if so, whether it was due to differences in viral replication and/or cell sloughing. Cells from both donors were fully differentiated with pseudostratified morphology and mucus-producing cells (Figure 1a). After pandemic H1N1 (pH1N1) infection, cells from the healthy donor appeared squamous, while the morphology of cells from the asthmatic donor were unaffected by virus infection (Figure 1a and Table 1). We found no difference in viral titers between donors (6.5±0.2 vs 6.8±0.1 median dose infectious dose for 50% of tissue culture wells (TCID50) ml−1), although viral infection resulted in higher mucus gene expression and protein production in healthy cells (Figure 1b).
Figure 1.
Human bronchial epithelial cells from healthy and asthmatic donors grown in the air–liquid interface infected with pH1N1 influenza virus at 0.01 m.o.i. Cells from both donors maintained a pseudostratified morphology and produced mucus under steady state. The thickness of the epithelial layer from the healthy donor was reduced and appeared squamous after pH1N1 infection, while cells from the asthmatic donor maintained normal morphology (a). Viral replication was comparable between cells (inset values, b). Cells from the healthy donor (solid bars) produced more mucus (b) and expressed more TLR3 and IL-1α after virus infection compared to asthmatic donor cells (checkered bars) (c). Data are presented as the mean and s.d. from a representative experiment, Δ,#P<0.05 compared with uninfected control and *P<0.05 by Student's t-test. Top panel hematoxylin and eosin (H&E) staining; bottom panel, periodic acid Schiff's (PAS) staining. Scale bar=10 μm.
Table 1. Histological analyses of primary human bronchial epithelial cells from healthy and asthmatic donors grown in the air–liquid interface.
| Donor | Group | Effects on tissue structure |
|---|---|---|
| Healthy | Uninfected | Apical surface intact with visible cilia. Basal cells intact with healthy morphology. Pseudostratified cells with some PAS+ cells and acinar-like structures |
| Infected with A/CA/04/2009 (pH1N1) | Apical cells detached with cell sloughing. Tissue thinned and more squamous morphology. Very few PAS+ cells present | |
| Asthmatic | Uninfected | Apical surface intact with visible cilia. Basal cells intact with healthy morphology. Pseudostratified cells with many PAS+ cells and acinar-like structures |
| Infected with A/CA/04/2009 (pH1N1) | Apical surface intact with visible cilia. Basal cells intact with healthy morphology. Pseudostratified cells with many PAS+ cells and acinar-like structures. Tissue appears slightly more compacted than uninfected controls |
Abbreviations: PAS, periodic acid Schiff; pH1N1, pandemic H1N1.
As the innate immune mediators of the epithelial barrier are crucial to the host's antiviral defense, we examined whether the cells from the asthmatic donor were protected from damage due to enhanced expression of antiviral factors. Expression of both TLR-3 and IL-1α were higher in healthy donor cells following viral infection (Figure 1c). Taken together, these data suggest that epithelial cells from the asthmatic donor are equally susceptible to pH1N1 infection but are resistant to virus-induced damage, possibly through reduced production of inflammatory cytokines such as IL-1α. These findings provided a rationale for more in-depth studies in mice.
Stage in allergic asthma pathogenesis alters influenza morbidity
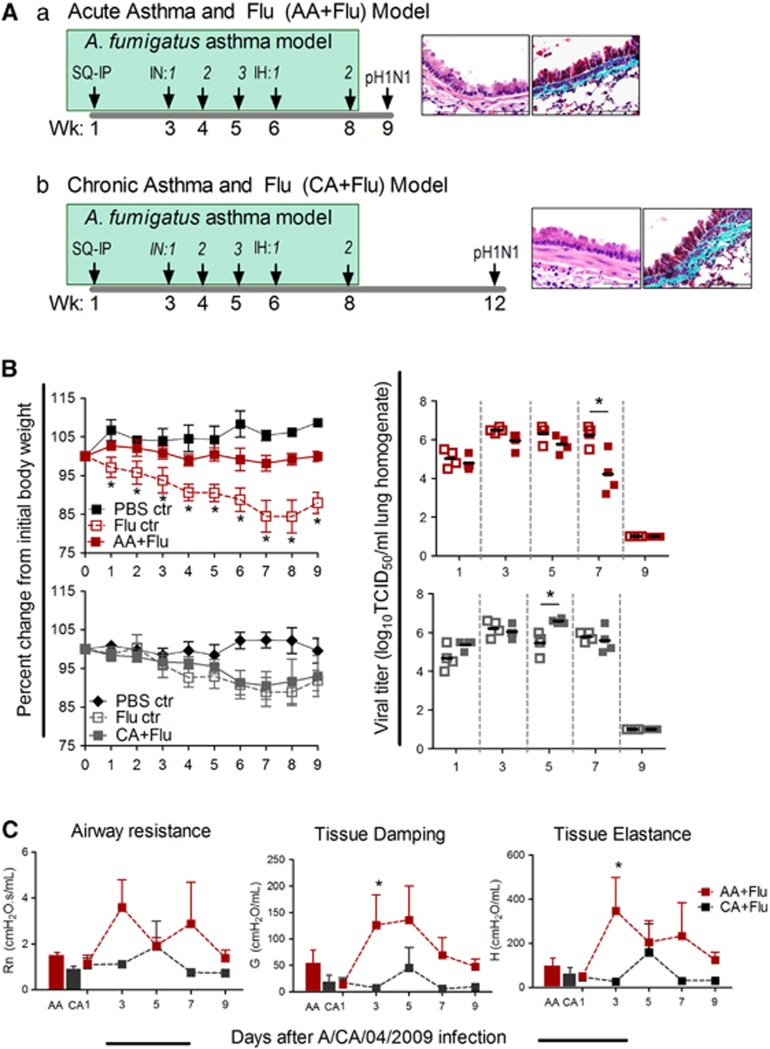
Two murine models were created by infecting mice during different stages of allergic asthma pathogenesis (Figure 2A): a fungal asthma model where mice were infected with pH1N1 during peak acute inflammation (AA+Flu; Figure 2Aa) and a fungal asthma model where mice were infected with pH1N1 during chronic remodeling (CA+Flu; Figure 2Ab). Histological images are representative of the state of the large airways in the allergen-challenged mice at the time of virus infection (Figure 2A). Weight loss was used as an indicator of influenza morbidity. Naive mice did not lose weight in either model, while flu-only controls gradually lost weight until day 8 with peak weight loss of 12–15% before recovery (Figure 2B). AA+Flu mice maintained weight throughout the model, while CA+Flu mice lost weight after infection, mimicking their flu-only counterparts (Figure 2B). Viral load in lungs peaked at day 3 and remained high through day 7 in flu-only controls, correlating with peak weight loss (Figure 2B). Viral titers in the AA+Flu group were decreased at this time point, indicating accelerated viral clearance; this was not observed in the CA+Flu model (Figure 2B). These data indicate that the allergic state of the airways at the time of influenza virus infection affects viral pathogenesis marked by body weight loss and viral replication.
Figure 2.
The developmental stage of allergic asthma impacts influenza morbidity. Schema of comorbidity models (A): (a) AA+Flu and (b) CA+Flu. Images represent the level of airway remodeling in AA and CA lungs at the time of infection. Weight loss and viral titers were affected by the state of allergy in lungs (B, n=8–9 per group). Airway physiological parameters including Newtonian resistance (Rn), tissue damping (G) and tissue elastance (H) were reduced in the CA+Flu model compared with that of AA+Flu (C, n=4 per group). Data are representative of two independent experiments and presented as the mean and s.d., *P<0.05 by analysis of variance. Ctr, control; IH, inhalation; IN, intranasal; PBS, phosphate-buffered saline; SQ/IP, subcutaneous/intraperitoneal; Wk, weeks.
As viral infection causes symptomology that may mimic asthma episodes,13 and respiratory viruses such as rhinovirus and respiratory syncytial virus have been shown to induce asthma, we measured the resistance in the conducting airways (Rn) and changes in tissue parameters, tissue damping (G) and tissue elastance (H), in the models. Mice in the flu-only control groups of both models responded to methacholine challenge with values and trends equivalent to the AA+Flu groups (data not shown). However, responses in the CA+Flu groups were lower than in the AA+Flu group (Figure 2C). Thus, the allergic state of the airways at the time of virus introduction alters the physiological response of the lungs to virus infection. We investigated airway immune profiles next, because inflammatory cells and their products can cause pathophysiological changes.
Cell recruitment patterns differed between acute and chronic models after influenza
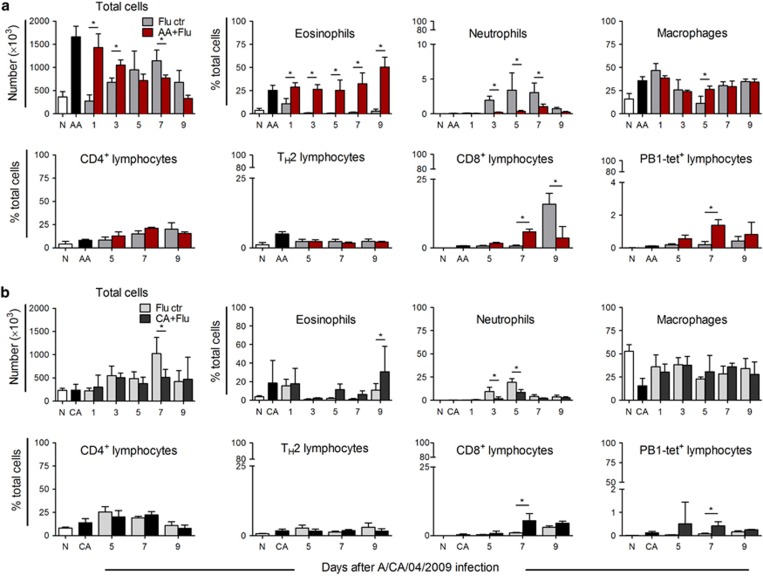
Lung inflammation occurs in asthma and respiratory infections, albeit with different types of immune cells taking precedence. There were threefold more cells in the airways at day 0 in AA compared with that of the CA model and naive controls (Figure 3). A reduction in cell infiltration occurred over time after virus in the AA+Flu model but not in the CA+Flu model (Figure 3). Peak airway inflammation of flu-only controls occurred at day 7 (Figure 3), coinciding with sustained viral replication in these mice (Figure 2).
Figure 3.
Inflammatory cell recruitment into the airways after pH1N1 infection. There were more cells recruited into the AA+Flu airways, particularly eosinophils and flu-specific CD8+ T cells (a). The influx of cells was reduced in the CA+Flu airways, wherein macrophages made up a large proportion (b). Data are representative of two independent experiments and presented as the mean and s.d., n=5 mice per group and *P<0.05 by analysis of variance. Ctr, control; TH2, T helper type 2. N, naive.
Influenza virus is known to induce neutrophil and T-cell recruitment, and this was seen in both ‘flu-only' control groups (Figure 3). Eosinophils, which were prominent in AA airways, continued to increase after viral infection (Figure 3a), but did not change in the CA+Flu (Figure 3b). The AA+Flu model had fewer neutrophils and equal compositions of macrophages and CD4+ lymphocytes compared with the CA+Flu model. Although the total number of CD8+ lymphocytes in the airways of both models was similar, pH1N1-specific CD8+ T cells were more abundant in AA+Flu (Figure 3). These data suggest that the existing allergic state of the airways has an impact on immune cell recruitment following pH1N1 infection.
Mice in the chronic model, including the flu-only controls, exhibited a reduced phenotype for weight loss, although viral titers were roughly equal to that in the acute model (Figure 2B). The reduction in inflammatory cells was observed in the flu controls age matched to the CA+Flu compared with those age matched to the AA+Flu (Figure 3). Although the difference between the two groups at the time of viral infection was 3 weeks, immune function was likely different because these ages fell at the exponential decline phase of thymic involution,14 highlighting the importance of mouse age in studies with infectious agents.
Histopathology of lungs differed between the acute and chronic models of comorbidity
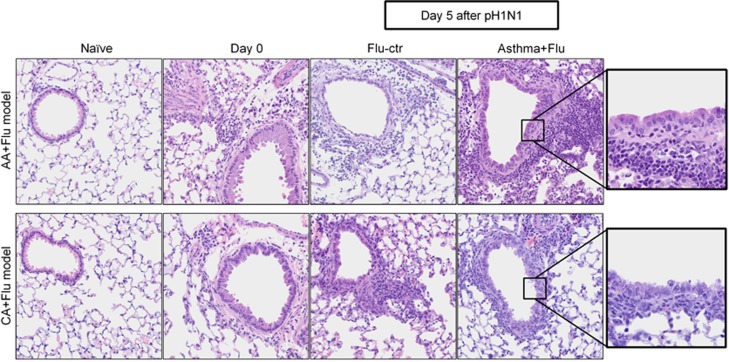
Structural damage to the airways is a characteristic of influenza.15 Virus infection resulted in peribronchovascular inflammation in both models of comorbidity. Inflammatory foci in the peribronchovascular areas peaked 5 days after pH1N1 infection in AA+Flu lungs (Figure 4 and Table 2A) coinciding with the viral plateau (Figure 2). There was a delay in the recruitment of inflammatory cells into the CA+Flu airways (Figure 4 and Table 2B). Although influenza virus is cytopathic, we noted that morphological changes in bronchial epithelia were only apparent in the flu controls and CA+Flu but not in AA+Flu airways (Figure 4 and Table 2).
Figure 4.
Peribronchovascular inflammation after pH1N1 infection. Inflammation increased after viral infection in both models of comorbidity. There was less/no damage to the bronchial epithelium of AA+Flu compared with that of CA+Flu. Data are representative of two independent experiments. Representative images are at × 400 (first four columns) and × 1000 (last column) after hematoxylin and eosin staining. Ctr, control.
Table 2. Mean histological scoring of lungs from comorbidity models and controls.
| (A) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Parameter |
AA model |
|||||||||
| AA+Flu | Flu ctr | |||||||||
| Days after pH1N1 | 1 | 3 | 5 | 7 | 9 | 1 | 3 | 5 | 7 | 9 |
| PB inflammation | + | + | +++(+) | +++ | ++(+) | − | + | + | +++ | ++ |
| PV inflammation | ++(+) | +++ | +++ | ++(+) | ++(+) | − | + | + | +++ | ++(+) |
| Epi. hyperplasia | − | − | − | − | − | − | + | + | ++ | ++ |
| Epi. necrosis | − | − | − | − | − | − | ++ | ++ | ++ | − |
| (B) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Parameter |
CA model |
|||||||||
| CA+Flu | Flu ctr | |||||||||
| Days after pH1N1 | 1 | 3 | 5 | 7 | 9 | 1 | 3 | 5 | 7 | 9 |
| PB inflammation | + | ++ | ++ | +++ | +++ | − | (+) | + | +(+) | ++ |
| PV inflammation | +(+) | ++ | ++ | +++ | +++ | (+) | + | +(+) | ++ | ++ |
| Epi. hyperplasia | − | + | + | ++ | ++ | − | − | + | +(+) | + |
| Epi. necrosis | − | − | − | − | − | − | − | (+) | (+) | − |
Abbreviations: AA, acute asthma; CA, chronic asthma; ctr, controls; Epi, epithelial; Flu, influenza; PB, peribronchial; PV, perivascular.
Mean staining intensity score for analyzed parameters from 0 to 4, where ‘−' indicates 0 and (+) indicates a half-point.
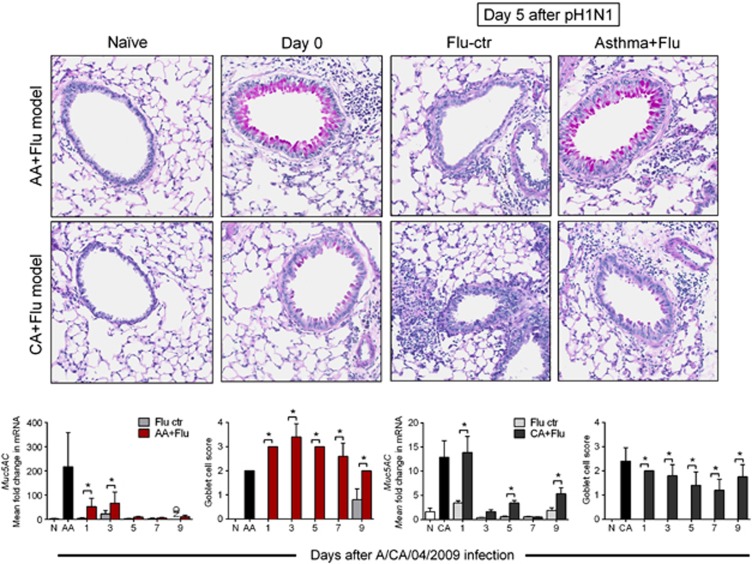
Mucus production is an important innate defense in the respiratory tract. Virus alone did not induce GC metaplasia, while both GC metaplasia and Muc5AC were increased in the comorbid states (Figure 5). Although Muc5AC levels decreased after viral infection in both groups, the number of GCs remained elevated, suggesting that other mucin genes may have a supplementary role in comorbidity. The abundance of GCs and induction of Muc5AC expression in AA+Flu may have served as an additional innate defense that protected these mice from influenza. Taken together, these data infer that intact structural defenses of lungs may have contributed to protection from influenza in the AA+Flu model.
Figure 5.
Presence of GCs in the airways of mice after pH1N1 infection. Virus infection alone did not induce GC metaplasia. More GCs were noted in the AA+Flu compared with CA+Flu. Representative images are at × 400 after periodic acid Schiff's staining. Although Muc5AC gene expression was reduced, the number of GCs that interspersed the airways remained elevated in the comorbidity groups. Data are representative of two independent experiments and presented as the mean and s.d., n=5 mice per group and *P<0.05 by analysis of variance. Ctr, control.
Host interferon response may contribute to influenza morbidity
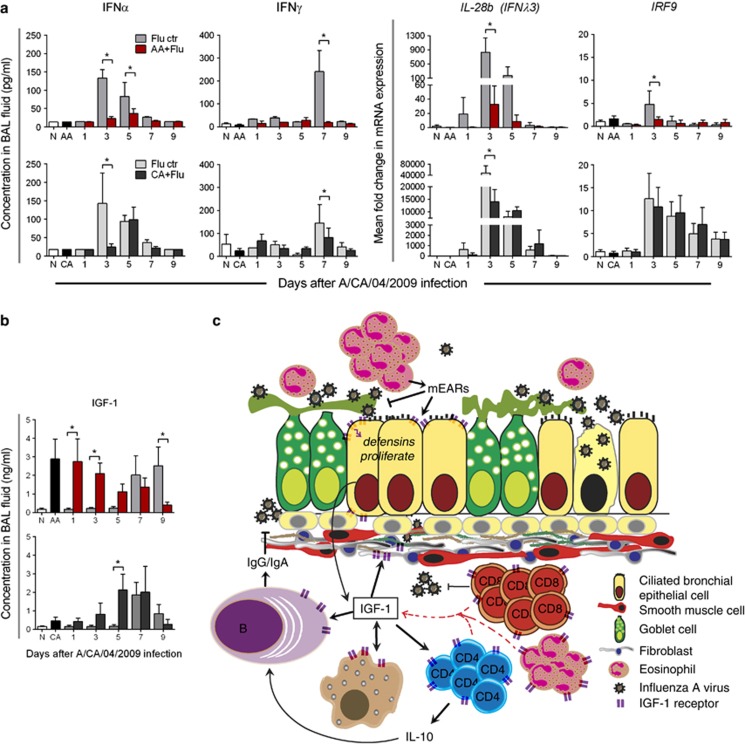
As the interferon (IFN) system is implicated in antiviral defenses and virus-mediated tissue damage in the lungs,16 we measured the host's local IFN response. The antiviral IFN response, as characterized by Type I (IFNα), Type II (IFNγ), Type III (IFNλ3/IL-28b) and IRF9 (IFN regulatory factor-9), was activated in flu-only controls. The peak in available IFNα and IFNγ in the bronchoalveolar lavage (BAL) fluid occurred at day 5 after infection in the CA+Flu model, while peak expression of IL-28b and IRF9 genes occurred at day 3 (Figure 6a). The Type I and Type III IFN responses are initiated sooner during the viral growth curve in the flu-only control groups than in the asthma and flu groups, while the Type II system is initiated in the flu-only control groups at day 7 (Figure 6a). Nonetheless, unlike the other parameters measured, pH1N1 infection triggered an IFN response in the CA+Flu groups compared with the AA+Flu groups (Figure 6a). An opposing trend in levels of the proinflammatory molecule insulin-like growth factor-1 (IGF-1) was observed between the two models (Figure 6b). IGF-1 was prominent before viral infection in allergic mice during the acute inflammatory phase, but not the chronic remodeling phase, and was induced late in the course of infection by influenza virus in both allergen-naive and CA+Flu models. These data suggest that canonical IFN pathways are not responsible for improved outcomes in acute, allergic asthma, but highlight a potential relationship between IGF-1 and asthma that may mediate protection from influenza.
Figure 6.
Host immune factors were altered in the lungs following pH1N1 infection. Viral infection results in reduced IFNs and their downstream factors in AA+Flu, while levels in CA+Flu were more equivalent to flu-only controls (a). An opposing trend was observed for IGF-1 between the AA+Flu model and flu-only controls. IGF-1 levels in the BAL fluid followed the same trend between the CA+Flu and flu-only controls (b). Data are representative of two independent experiments and presented as the mean and s.d., n=5 mice per group and *P<0.05 by analysis of variance. Schematic representation of our proposed mechanism of enhanced viral clearance and reduced lung damage in the AA+Flu model based on IGF-1 (c). Ctr, control.
Discussion
Asthma, a complicated disease involving multiple intrinsic and extrinsic parameters, is a major underlying condition associated with respiratory infections. Clear clinical and epidemiological data demonstrate that asthmatics are at high risk of hospitalization during influenza epidemics; however, recent studies show that asthmatics were less likely to die from pH1N1 and suffer less severe symptoms and outcomes once hospitalized.3, 4, 5, 7, 8, 9 As the existing models typically utilize influenza viruses as initiators or exacerbators of asthma in mice,17, 18 the question of how the asthmatic hosts' immune response to influenza differs from that of the normal host has not been previously addressed. In this study, we developed models of comorbidity by combining a murine model that recapitulates the hallmarks of human asthma11, 12, 19 with a model of influenza A virus infection.20 Using these, we have found that the inflammatory state of the host's allergic airways has a fundamental role in influenza pathogenesis. Our study shows that acute allergic airways are protected from influenza, supporting a hypothesis proposed by Chang et al.17 to explain why in some cases respiratory viruses inhibited asthma, while in other cases asthma development was promoted. This is also strengthened by a recent report showing improved outcomes in ovalbumin-sensitized mice challenged with a mouse-adapted lab strain of influenza.21
Respiratory viruses are the leading cause of asthma exacerbations in children and adults.22 A prospective study during the 2009 influenza pandemic showed that pH1N1 preferentially infected asthmatics more than non-asthmatics, which the authors speculated may be due to impairments of the bronchial epithelium.23 However, our findings show that there was no difference in viral replication capacity in epithelia from healthy/asthmatic donors. In fact, cells from the healthy donor were more susceptible to pH1N1-mediated cytopathology, findings that were recapitulated in our murine models, wherein the airway epithelia of AA+Flu were intact, while that of the flu controls and CA+Flu were damaged. The respiratory epithelium performs crucial physical and innate immune defenses against environmental pathogens through mucus, reactive oxygen species, defensin and cytokine production.24, 25 Although asthmatic lungs have mucus-producing cells and mucus hypersecretion, our data with ex vivo human tissues suggest that mucus production in response to virus may require a higher viral burden in asthmatics. As virus-induced activation of TLR3 on epithelial cells promotes proinflammatory cytokine production leading to tissue injury,26 and IL-1α can promote cytopathology in epithelial cells,27 the induction of TLR3 and IL-1α in cells from the healthy donor may have contributed to the enhanced susceptibility to pH1N1-induced damage. Because we noted a similar protection in the airway epithelia of the AA+Flu group, a specific mechanism may be in place to signal cell survival in spite of viral replication in asthma. Further analyses into the barrier functions of epithelial cells will help elucidate the mechanism in which epithelia from asthmatics maintain structural integrity in the presence of a cytolytic pathogen such as influenza A virus.
In addition to the structural integrity of airway epithelium and inflammatory cells, respiratory viruses also induce changes in lung physiology.28 Our findings support the growing literature that these parameters are not merely reflective of airway smooth muscle stiffness/shortening, but that they are complex phenomena that are indicative of the state of the airways such as airway narrowing (which may be due to mucus or cells), increased epithelial thickness and vascular leakage.29 The role of eosinophils in AHR has been debated in the past, wherein their presence has been demonstrated both as necessary30 and unnecessary31 for AHR. Our data demonstrate that changes in airway physiological parameters, Rn, G and H, can occur in the absence or limited number of eosinophils as in the flu-only control groups and CA+Flu, respectively, as well as in their presence as in AA+Flu. The increase in Rn that resulted because of pH1N1 alone possibly involved other pathways that increase airway resistance such as IL-1β and IL-13,32, 33 which were elevated in these mice (data not shown). As the flu-only control animals had airway responses equivalent to AA+Flu groups in spite of having less immune cell recruitment, the reduction in lung physiological parameter of the CA+Flu group cannot be due to reduced immune infiltrates. This non-responsiveness in the CA+Flu groups is interesting, and may be explained by the ‘inflammatory twitch' theory recently proposed by Pothen et al.34 In the CA+Flu model where the influenza virus infection occurred during the resolution/refractory phase of the allergic inflammatory response, we propose that the lung was not primed to mount a second response to the invading pathogen. This ‘unprepared' state of the chronic allergic lung, marked by reduced inflammation, and lack of pathophysiologic changes, may have then given way to the increased pathogen burden (Figure 2) and host pathology (Figure 4), which were observed in the CA+Flu model.
Antiviral immunity is associated with the recruitment of neutrophils35 and virus-specific CD8+ T cells.36 Our findings that AA+Flu mice were protected from influenza likely reflect the importance of virus-specific CD8+ T cells in viral clearance. As neutrophils were absent in the AA+Flu lungs, their potential antiviral role may have been performed by another granulocyte that was abundant, the eosinophil. Functions of eosinophils that transcend that of the traditional antiparasitic role37 such as antigen presentation,38 phagocytosis,39 antiviral40 and antibacterial41 defense advocate for a direct or indirect antiviral role of eosinophils in allergic asthma and influenza comorbidity. Influenza virus has been previously shown to hinder the recruitment of eosinophils and T helper type 2 cells into airways after allergen challenge.42 This reduction of hallmark T helper type 2 immunity is linked increased IFNγ and CD8+ T cells following influenza virus infection.43 We did not observe an inhibition of T-cell or eosinophil recruitment following viral infection in our models, further suggesting that the manner in which influenza virus impacts the allergic airways may be linked to the pre-existing immunological state of the lung and the microenvironment of the airways.
Infiltrating cells produce cytokines that direct the defense mechanisms in the lungs against influenza. Type I and Type II IFNs have a vital role in viral immunity, as well as the immunopathology associated with influenza.44, 45 Type III IFNs (IFNλ) are recently discovered conserved genes with three members.46 For these studies, we focused on IFNλ3 (IL-28b) because the IFNλ1 is absent in mice47 and IFNλ2 and -3 are homologous. As certain proteins are shared between the Type I and Type III IFN systems, we looked at IRF-9 as a common upstream indicator of viral-induced IFN gene expression. Herein, we noted that IFN Types I–III were not induced by influenza A virus infection in the AA+Flu model, while mice in the CA+Flu groups responded to viral infection with IFN Type I and III production. As T helper type 2 cytokines in BAL fluid and lungs were equivalent between AA+Flu and CA+Flu (data not shown), the downregulation of IFNs in the AA+Flu model could not be attributed to an overexpression of T helper type 2 cytokines. Although the IFN response is mounted by the host as a primary antiviral defense mechanism, IFNs have a myriad of effects on host cells including increasing expression of major histocompatibility complexes and inducing cell death,16, 48 which can be detrimental to the host. The CA+Flu model was more susceptible to pH1N1-mediated morbidity and had a higher viral burden in spite of inducing an IFN response. Therefore, we speculate that in the CA+Flu model, the induction of the IFN cascade may have resulted in the observed immunopathology in the airways (Figure 4) compared with the AA+Flu model in which IFNs were reduced.
Largely considered a hormone, IGF-1 has proinflammatory properties; its neutralization in allergen-challenged lungs led to decreased immune cell infiltration and adhesion molecules and blocked AHR49 and prosurvival properties of cells.50 Yamashita et al.49 demonstrated that airway epithelial cells were prominent producers of IGF-1 in the mouse lung. The reduced availability of IGF-1 in the CA+Flu model may have resulted from the damage induced in these cells by the invading virus, and may have partially contributed to the reduction in inflammation and AHR. Receptors for IGF-1 expressed on most immune cells signal proliferation or cytokine release.51, 52 Therefore, high availability of IGF-1 in the microenvironment of AA+Flu may have promoted survival and activation of recruited immune cells compared with CA+Flu model, which had reduced cell infiltration. Although not a direct goal of the present work described here, we are currently investigating the novel hypothesis that IGF-1 produced by the airway epithelial cells of mice in the AA model enhances antiviral defenses against influenza A virus infection in this mouse model (Figure 6c).
Asthmatics may be infected with respiratory viruses during the inflammatory or remodeling phases of the disease. The present study, aimed at generating and characterizing a mouse model of asthma and influenza comorbidity, has shown that the timing of influenza infection on pre-existing asthmatic airways affects the immunological, virological and histopathological outcomes. Our main finding that influenza morbidity differed based on the state of the allergic airways corroborates a human study in which rhinovirus infection did not worsen asthma symptomology.53 Although precise information on the allergic state of the airways of the hospitalized asthmatics during the 2009 influenza pandemic is not readily available, clinical reports highlight that the highest mortality occurred in adults between 18 and 64 years of age.54, 55 Phenotypic differences between childhood and adult asthma as discussed in depth by Bush and Menzies-Gow56 indicate that adult patients are more likely to have remodeled airways that may be more susceptible to respiratory pathogens. Model systems that can recapitulate the microenvironment of these patient subsets are necessary and can be used to further our understanding on infectious disease pathogenesis in allergic asthma. Those developed herein are ideally suited to investigate the pathways that protect asthmatics with acute airways inflammation from influenza virus and increase susceptibility in chronic asthmatics.
Methods
Ethics statement
All experiments were approved by the Institutional Animal Use and Care Committee at St Jude Children's Research Hospital (SJCRH, Memphis, TN, USA).
Virus
Pandemic influenza virus A/CA/04/2009 (pH1N1) obtained from Dr Richard Webby at SJCRH was propagated in Madin–Darby canine kidney (MDCK.2; ATCC, Manassas, VA, USA) cells.
Primary bronchial epithelial cells
Fully differentiated primary bronchial cells from a healthy African-American 18-year-old female donor and an African-American 56-year-old female with asthma (MatTek Corporation, Ashland, MA, USA) were infected with low multiplicity of infection (m.o.i.) of 0.01 pH1N1 to mimic in vivo infections or left uninfected as controls.
Mice
Six-week-old C57BL/6 female mice from Jackson Laboratories (Bar Harbor, ME, USA) acclimatized for 1 week in SJCRH's specific pathogen-free ABSL2 facility with ad libitum food and water in a 12-h light:dark cycle and examined and weighed daily after infection.
Allergic asthma was induced in mice using Aspergillus fumigatus as previously described by our group.11, 19 Briefly, animals were sensitized to whole antigen from A. fumigatus extract from Greer Labs (Lenoir, NC, USA) adsorbed in Imject Alum (Pierce, Rockford, IL, USA) via subcutaneous and intraperitoneal injections. After a 2-week resting period, mice were administered 20 μg of A. fumigatus antigen in 20 μl intranasally once a week for 3 weeks. One week following the final intranasal delivery of antigen, anesthetized mice were exposed to live, unmanipulated, mature A. fumigatus conidia for 10 min via natural inhalation route. This fungal challenge was repeated 2 weeks later. The AA model is characterized by eosinophilic inflammation in the BAL spaces and peribronchovascular inflammatory foci, GC metaplasia, epithelial thickening, AHR to methacholine challenge and increased serum immunoglobulin E, all of which peak at 7 days after the fungal challenge. The CA model is characterized by airway remodeling events that begin to occur after 14 days following the fungal challenge, such as peribronchial smooth muscle hyperplasia and hypertrophy, and increased subepithelial fibrosis along with remnant peribronchovascular eosinophilic inflammation and serum immunoglobulin E. The characterization of this model system has been previously published by our group.11
Mice in the ‘flu' groups were infected with pH1N1 virus at 1000 TCID50 in a 50 μl inoculum intranasally. The infectious dose induced morbidity but no mortality. Mice in the ‘naive' groups were administered saline in place of A. fumigatus antigen and pH1N1 virus.
Comorbidity models
AA+Flu groups were infected with pH1N1 virus 1 week after the second fungal inhalation challenge at the peak of allergic inflammation. CA+Flu groups were infected with pH1N1 virus 4 weeks after the second fungal challenge, at which point inflammation had subsided and airway remodeling events had progressed. As we anticipated nonsignificant changes in the inflammatory profile in the asthma-only groups over the 9-day period at the chosen time after the second fungal challenge, naive, ‘AA'-only and ‘CA'-only controls in each study were harvested on the day of viral infections. Mice in the AA+Flu and CA+Flu groups were 15 and 18 weeks of age, respectively, at the time of infection.
BAL and tissue harvest
BAL was performed on animals that were killed. Caudal and accessory lobes of the right lung were harvested and snap frozen for RNA quantification, while left lobes were harvested and infused ex vivo with 1 ml of 10% neutral-buffered formalin for histopathology.
Airway physiology
Anesthetized mice (n=4 per group) were surgically intubated and attached to a computer-controlled small animal ventilator (flexiVent FX1; SCIREQ, Montreal, QC, Canada) data gathered by flexiWare software (SCIREQ) at a mouse default ventilation rate of 150 breaths min−1 using the ‘mouse inhaled dose-response' script. After baseline, 25 mg ml−1 of acetyl-β-methacholine (Sigma, St Louis, MO, USA) was nebulized into airways for 10 s. Newtonian resistance (Rn), tissue damping (G) and elastance (H) were recorded. The mean of software-generated values after methacholine was calculated for each animal before obtaining the mean for each group.
Flow cytometric analysis of BAL cells
BAL cells were washed with staining media (sterile phosphate-buffered saline with 5% fetal bovine serum) and enumerated with the Countess (Invitrogen, Grand Island, NY, USA). All incubations were performed in ice in the dark, and the staining media were used to dilute antibodies and washings. One to two million cells were incubated for 30 min with human γ-globulin (blocks Fc receptors) and live–dead aqua (Invitrogen) at 1 μl per million cells to differentiate live cells from dead. Cells were washed and incubated in fluorescent-tagged antibodies from BD Biosciences (San Jose, CA, USA) at 1:50 for 30 min: CD8-FITC, CD3-PE-Cy7, CD107b (Mac3) -PE, CD193 (CCR3) -APC, Ly6G-V450, CD4-A700 and CD19-PerCP-Cy5.5. Cells were washed and fixed with BD stabilizing fixative (4% paraformaldehyde), and strained through a 40 μm nylon mesh into labeled polystyrene tubes and stored at 4 °C overnight before flow cytometry analysis the next day. Tetramer staining was performed on a separate set of mice (n=4 per group) as described above except staining cocktail included antibodies against CD3-PE-Cy7, CD4-A700, CD8-FITC and PB1-tetramer-PE (diluted 1:8000 per titration by the Thomas Lab at SJCRH). Each experimental sample group was accompanied with an unstained cell population, isotype controls and single-color controls for instrument and compensation setup. Cells from each mouse were analyzed individually, and the mean and standard deviation (s.d.) were calculated for each group (n=5 per group). Data were acquired with a LSRFortessa (BD Biosciences) with 350, 405, 488, 561 and 641 nm lasers.
Samples were acquired and analyzed within the St Jude Children's Research Hospital Flow Cytometry Core. FMO (fluorescence minus one) controls are not required for staining protocols. Isotype controls are used to negate nonspecific binding and background noise. The fluorescence is compensated manually on both bone marrow cells and the tissue of investigation. The LSRFortessa in which these samples were acquired is subjected to a rigid quality control test each day to ensure that proper voltages are used for every experiment. The limiting factor within the experiments is the availability of the tetramer antibody. The tetramer with the highest stain index was selected for the subsequent experiments. Raw FCS files can be submitted at any time for further evaluation. The gating strategies used, determined based on the literature and known markers for various cell types, are demonstrated in the Supplementary Figures 1 and 2.
Histopathology
Paraffin-embedded tissue inserts or lungs were sectioned at 4 μm and stained with hematoxylin and eosin and periodic acid Schiff's stains. Slides were coded and read by a pathologist blinded to the study design (KLB).
RNA analysis
Harvested RNA from human bronchial cells and lungs was converted to cDNA (iScript; BioRad, Hercules, CA, USA) and was analyzed by quantitative polymerase chain reaction with RNA-specific Quantitect primer assays (Qiagen, Valencia, CA, USA) against human HPRT-1, TLR-3 and IL-1α and mouse HPRT-1, Muc5AC, IL-28b and IRF-9. Data were acquired with an ABI-7500 real-time PCR machine (Applied Biosystems, Carlsbad, CA, USA) and relative fold change in gene expression was determined using the 2−ΔΔCt method normalized to the internal housekeeping gene, HPRT-1.
Quantification of specific proteins
To determine the impact viral infection has on the airway milieu, we measured proteins by enzyme-linked immunosorbent assays. We measured the amount of Mucin 5AC in the apical wash of primary bronchial cells with a precoated ELISA kit from CUSABIO (Wuhan, China). IFNα (pbl IFN Source, Piscataway, NJ, USA), IFNγ and IGF-1 (BD Biosciences) were measured in undiluted BAL fluid following the manufacturer's protocols.
Viral titration
Viral content in apical wash of cells or whole-lung homogenates was determined as the TCID50 on Madin–Darby canine kidney cells.
Statistical analysis
Animal studies were repeated for reproducibility. Data are represented as the mean±s.d. Data after virus infection were compared with naive controls with Student's t-test on GraphPad Prism (GraphPad Software, La Jolla, CA, USA) and P<0.05 was considered significant and represented by ‘#' for cells from the healthy donor and ‘Δ' for cells from the asthmatic donor. Data from the flu-only control groups and asthma+flu groups were compared by two-way analysis of variance and P<0.05 was considered as significant and represented by an asterisk (*).
Acknowledgments
We would like to acknowledge Arthur Covington of Animal Resource Center and Drs Scott Perry and Ann-Marie Hamilton-Easton of the Shared Flow Cytometry Resource Center at SJCRH for animal husbandry and assistance in planning the cell staining and acquisition, respectively. We also thank Dr Paul Thomas in the Department of Immunology at SJCRH for PB1-PE tetramer, Dr Jason Bates (University of Vermont) for his time and help in airway physiology data analysis and Dr Helene Rosenberg (NIH/NIAID) for critical review of the manuscript. This work was funded by the National Institute of Allergy and Infectious Diseases, the National Institutes of Health, under Contract number HHSN266200700005C, Centers of Excellence in Influenza Research and Surveillance (CEIRS), N01 7005C (JAM).
Footnotes
The Supplementary Information that accompanies this paper is available on the Immunology and Cell Biology website (http://www.nature.com/icb)
Author contributions
AES: Study conception and design, data acquisition, analysis and interpretation, and drafting and editing the manuscript; SNW and AES: FCS data acquisition and FlowJo analysis; KLB and AES: histopathological data acquisition, analysis and interpretation; SAH and JMS: fungal asthma model and editing the manuscript; JAM: study conception and design, and editing the manuscript. All authors read and approved the final version of the manuscript.
Supplementary Material
References
- WHO Asthma. 2011 ASthma Fact Sheet. Available at http://www.who.int/mediacentre/factsheets/fs307/en/index.html (cited 29 July 2013, last accessed on 29 July 2013)..
- Fiore AE, Shay DK, Broder K, Iskander JK, Uyeki TM, Mootrey G, et al. Prevention and control of seasonal influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2009. MMWR. 2009;58:1–52. [PubMed] [Google Scholar]
- Jain S, Kamimoto L, Bramley AM, Schmitz AM, Benoit SR, Louie J, et al. Hospitalized patients with 2009 H1N1 influenza in the United States, April–June 2009. N Engl J Med. 2009;361:1935–1944. doi: 10.1056/NEJMoa0906695. [DOI] [PubMed] [Google Scholar]
- Nguyen-Van-Tam JS, Openshaw PJ, Hashim A, Gadd EM, Lim WS, Semple MG, et al. Risk factors for hospitalisation and poor outcome with pandemic A/H1N1 influenza: United Kingdom first wave (May–September 2009) Thorax. 2010;65:645–651. doi: 10.1136/thx.2010.135210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson CO, Graham DA, Uyeki TM, Randolph AG. Risk factors for mechanical ventilation in US children hospitalized with seasonal influenza and 2009 pandemic influenza A*. Pediatr Crit Care Med. 2012;13:625–631. doi: 10.1097/PCC.0b013e318260114e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran D, Vaudry W, Moore DL, Bettinger JA, Halperin SA, Scheifele DW, et al. Comparison of children hospitalized with seasonal versus pandemic influenza A, 2004–2009. Pediatrics. 2012;130:397–406. doi: 10.1542/peds.2011-3216. [DOI] [PubMed] [Google Scholar]
- Gilca R, De Serres G, Boulianne N, Ouhoummane N, Papenburg J, Douville-Fradet M, et al. Risk factors for hospitalization and severe outcomes of 2009 pandemic H1N1 influenza in Quebec, Canada. Influenza Other Respir Viruses. 2011;5:247–255. doi: 10.1111/j.1750-2659.2011.00204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna JJ, Bramley AM, Skarbinski J, Fry AM, Finelli L, Jain S. Asthma in patients hospitalized with pandemic influenza A(H1N1)pdm09 virus infection—United States, 2009. BMC Infect Dis. 2013;13:57. doi: 10.1186/1471-2334-13-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple M, Myles P, Enstone J, Openshaw P, Read R, Taylor B, et al. The Relationship of Asthma to Outcome in Influenza A/H1N1 2009 Infections: FLU-CIN Cohort Study. European Respiratory Society: Amsterdam, The Netherlands; 2011. [Google Scholar]
- Chang YJ, Kim HY, Albacker LA, Baumgarth N, McKenzie AN, Smith DE, et al. Innate lymphoid cells mediate influenza-induced airway hyper-reactivity independently of adaptive immunity. Nat Immunol. 2011;12:631–638. doi: 10.1038/ni.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoselton SA, Samarasinghe AE, Seydel JM, Schuh JM. An inhalation model of airway allergic response to inhalation of environmental Aspergillus fumigatus conidia in sensitized BALB/c mice. Med Mycol. 2010;48:1056–1065. doi: 10.3109/13693786.2010.485582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarasinghe AE, Hoselton SA, Schuh JM. A comparison between intratracheal and inhalation delivery of Aspergillus fumigatus conidia in the development of fungal allergic asthma in C57BL/6 mice. Fungal Biol. 2011;115:21–29. doi: 10.1016/j.funbio.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostransky D, Blais FX.Postviral bronchial hyperreactivity syndrome: recognizing asthma's great mimic J Am Osteopath Assoc 199191465–468.471-465. [PubMed] [Google Scholar]
- Hsu HC, Zhang HG, Li L, Yi N, Yang PA, Wu Q, et al. Age-related thymic involution in C57BL/6J × DBA/2J recombinant-inbred mice maps to mouse chromosomes 9 and 10. Genes Immun. 2003;4:402–410. doi: 10.1038/sj.gene.6363982. [DOI] [PubMed] [Google Scholar]
- Yin L, Xu S, Cheng J, Zheng D, Limmon GV, Leung NH, et al. Spatiotemporal quantification of cell dynamics in the lung following influenza virus infection. J Biomed Opt. 2013;18:046001. doi: 10.1117/1.JBO.18.4.046001. [DOI] [PubMed] [Google Scholar]
- Julkunen I, Sareneva T, Pirhonen J, Ronni T, Melen K, Matikainen S. Molecular pathogenesis of influenza A virus infection and virus-induced regulation of cytokine gene expression. Cytokine Growth Factor Rev. 2001;12:171–180. doi: 10.1016/s1359-6101(00)00026-5. [DOI] [PubMed] [Google Scholar]
- Chang YJ, Kim HY, Albacker LA, Lee HH, Baumgarth N, Akira S, et al. Influenza infection in suckling mice expands an NKT cell subset that protects against airway hyperreactivity. J Clin Invest. 2011;121:57–69. doi: 10.1172/JCI44845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N, Suzuki S, Suzuki Y, Shirai A, Nakazawa M, Suzuki M, et al. Immune response induced by airway sensitization after influenza A virus infection depends on timing of antigen exposure in mice. J Virol. 2001;75:499–505. doi: 10.1128/JVI.75.1.499-505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarasinghe AE, Hoselton SA, Schuh JM. The absence of the VPAC(2) receptor does not protect mice from Aspergillus induced allergic asthma. Peptides. 2010;31:1068–1075. doi: 10.1016/j.peptides.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanzeck K, Boyd KL, McCullers JA. Glycan shielding of the influenza virus hemagglutinin contributes to immunopathology in mice. Am J Respir Crit Care Med. 2011;183:767–773. doi: 10.1164/rccm.201007-1184OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Sasaki H, Fukui T, Fujita K, Kutsukake E, Matsumoto T. Mice with asthma are more resistant to influenza virus infection and NK cells activated by the induction of asthma have potentially protective effects. J Clin Immunol. 2012;32:256–267. doi: 10.1007/s10875-011-9619-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos NG, Christodoulou I, Rohde G, Agache I, Almqvist C, Bruno A, et al. Viruses and bacteria in acute asthma exacerbations—a GA(2) LEN-DARE systematic review. Allergy. 2011;66:458–468. doi: 10.1111/j.1398-9995.2010.02505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloepfer KM, Olenec JP, Lee WM, Liu G, Vrtis RF, Roberg KA, et al. Increased H1N1 infection rate in children with asthma. Am J Respir Crit Care Med. 2012;185:1275–1279. doi: 10.1164/rccm.201109-1635OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D, Prince A. Innate immunity in the respiratory epithelium. Am J Respir Cell Mol Biol. 2011;45:189–201. doi: 10.1165/rcmb.2011-0011RT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bals R, Hiemstra PS. Innate immunity in the lung: how epithelial cells fight against respiratory pathogens. Eur Respir J. 2004;23:327–333. doi: 10.1183/09031936.03.00098803. [DOI] [PubMed] [Google Scholar]
- Chan RW, Yuen KM, Yu WC, Ho CC, Nicholls JM, Peiris JS, et al. Influenza H5N1 and H1N1 virus replication and innate immune responses in bronchial epithelial cells are influenced by the state of differentiation. PLoS One. 2010;5:e8713. doi: 10.1371/journal.pone.0008713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LN H, SA M, ER U, WE G. Interleikin-1 is linked to the respiratory epithelial cytopathology pf pertussis. Infect Immun. 1993;61:3123–3128. doi: 10.1128/iai.61.8.3123-3128.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becnel D, You D, Erskin J, Dimina DM, Cormier SA. A role for airway remodeling during respiratory syncytial virus infection. Respir Res. 2005;6:122. doi: 10.1186/1465-9921-6-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesenfeld E, Allen GB, Bates JH, Poynter ME, Wu M, Aimiand S, et al. The temporal evolution of airways hyperresponsiveness and inflammation. J Allergy Ther. 2012;1:1–7. doi: 10.4172/2155-6121.S1-005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, McGarry MP, Farmer SC, Denzler KL, Larson KA, Carrigan PE, et al. Interleukin-5 expression in the lung epithelium of transgenic mice leads to pulmonary changes pathognomonic of asthma. J Exp Med. 1997;185:2143–2156. doi: 10.1084/jem.185.12.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Iijima K, Kita H. Marked airway eosinophilia prevents development of airway hyper-responsiveness during an allergic response in IL-5 transgenic mice. J Immunol. 2003;170:5756–5763. doi: 10.4049/jimmunol.170.11.5756. [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H, Sakamoto T, Xu W, Barnes PJ, Chung KF. Effect of interleukin-1 beta on airway hyperresponsiveness and inflammation in sensitized and nonsensitized Brown–Norway rats. J Allergy Clin Immunol. 1994;93:464–469. doi: 10.1016/0091-6749(94)90355-7. [DOI] [PubMed] [Google Scholar]
- Tomlinson KL, Davies GC, Sutton DJ, Palframan RT. Neutralisation of interleukin-13 in mice prevents airway pathology caused by chronic exposure to house dust mite. PLoS One. 2010;5:10. doi: 10.1371/journal.pone.0013136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pothen JJ, Poynter ME, Bates JH. The inflammatory twitch as a general strategy for controlling the host response. J Immunol. 2013;190:3510–3516. doi: 10.4049/jimmunol.1202595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate MD, Deng YM, Jones JE, Anderson GP, Brooks AG, Reading PC. Neutrophils ameliorate lung injury and the development of severe disease during influenza infection. J Immunol. 2009;183:7441–7450. doi: 10.4049/jimmunol.0902497. [DOI] [PubMed] [Google Scholar]
- Flynn KJ, Belz GT, Altman JD, Ahmed R, Woodland DL, Doherty PC. Virus-specific CD8+ T cells in primary and secondary influenza pneumonia. Immunity. 1998;8:683–691. doi: 10.1016/s1074-7613(00)80573-7. [DOI] [PubMed] [Google Scholar]
- Abraham D, Rotman HL, Haberstroh HF, Yutanawiboonchai W, Brigandi RA, Leon O, et al. Strongyloides stercoralis: protective immunity to third-stage larvae inBALB/cByJ mice. Exp Parasitol. 1995;80:297–307. doi: 10.1006/expr.1995.1036. [DOI] [PubMed] [Google Scholar]
- Shi HZ, Humbles A, Gerard C, Jin Z, Weller PF. Lymph node trafficking and antigen presentation by endobronchial eosinophils. J Clin Invest. 2000;105:945–953. doi: 10.1172/JCI8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SG, Sapp TM. Phagocytosis of bacteria by eosinophils in infectious-related asthma. J Allergy. 1969;44:113–117. doi: 10.1016/0021-8707(69)90007-0. [DOI] [PubMed] [Google Scholar]
- Phipps S, Lam CE, Mahalingam S, Newhouse M, Ramirez R, Rosenberg HF, et al. Eosinophils contribute to innate antiviral immunity and promote clearance of respiratory syncytial virus. Blood. 2007;110:1578–1586. doi: 10.1182/blood-2007-01-071340. [DOI] [PubMed] [Google Scholar]
- Ueki S, Melo RC, Ghiran I, Spencer LA, Dvorak AM, Weller PF. Eosinophil extracellular DNA trap cell death mediates lytic release of free secretion-competent eosinophil granules in humans. Blood. 2013;121:2074–2083. doi: 10.1182/blood-2012-05-432088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlleben G, Muller J, Tatsch U, Hambrecht C, Herz U, Renz H, et al. Influenza A virus infection inhibits the efficient recruitment of Th2 cells into the airways and the development of airway eosinophilia. J Immunol. 2003;170:4601–4611. doi: 10.4049/jimmunol.170.9.4601. [DOI] [PubMed] [Google Scholar]
- Wohlleben G, Erb KJ. The absence of IFN-gamma leads to increased Th2 responses after influenza A virus infection characterized by an increase in CD4+ but not CD8+ T cells producing IL-4 or IL-5 in the lung. Immunol Lett. 2004;95:161–166. doi: 10.1016/j.imlet.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Lin KL, Suzuki Y, Nakano H, Ramsburg E, Gunn MD. CCR2+ monocyte-derived dendritic cells and exudate macrophages produce influenza-induced pulmonary immune pathology and mortality. J Immunol. 2008;180:2562–2572. doi: 10.4049/jimmunol.180.4.2562. [DOI] [PubMed] [Google Scholar]
- Trinchieri G. Type I interferon: friend or foe. J Exp Med. 2010;207:2053–2063. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levraud JP, Boudinot P, Colin I, Benmansour A, Peyrieras N, Herbomel P, et al. Identification of the zebrafish IFN receptor: implications for the origin of the vertebrate IFN system. J Immunol. 2007;178:4385–4394. doi: 10.4049/jimmunol.178.7.4385. [DOI] [PubMed] [Google Scholar]
- Pott J, Mahlakoiv T, Mordstein M, Duerr CU, Michiels T, Stockinger S, et al. IFN-lambda determines the intestinal epithelial antiviral host defense. Proc Natl Acad Sci USA. 2011;108:7944–7949. doi: 10.1073/pnas.1100552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J Leuk Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- Yamashita N, Tashimo H, Ishida H, Matsuo Y, Arai H, Nagase H, et al. Role of insulin-like growth factor-I in allergen-induced airway inflammation and remodeling. Cell Immunol. 2005;235:85–91. doi: 10.1016/j.cellimm.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Butt AJ, Firth SM, Baxter RC. The IGF axis and programmed cell death. Immunol Cell Biol. 1999;77:256–262. doi: 10.1046/j.1440-1711.1999.00822.x. [DOI] [PubMed] [Google Scholar]
- Smith TJ. Insulin-like growth factor-I regulation of immune function: a potential therapeutic target in autoimmune diseases. Pharmacol Rev. 2010;62:199–236. doi: 10.1124/pr.109.002469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudler S, Baumgartl J, Hampel B, Buch T, Waisman A, Snapper CM, et al. Insulin-like growth factor-1 controls type 2 T cell-independent B cell response. J Immunol. 2005;174:5516–5525. doi: 10.4049/jimmunol.174.9.5516. [DOI] [PubMed] [Google Scholar]
- Grunberg K, Sharon RF, Sont JK, In 't Veen JC, Van Schadewijk WA, De Klerk EP, et al. Rhinovirus-induced airway inflammation in asthma: effect of treatment with inhaled corticosteroids before and during experimental infection. Am J Respir Critic Care Med. 2001;164 (Part 1:1816–1822. doi: 10.1164/ajrccm.164.10.2102118. [DOI] [PubMed] [Google Scholar]
- LaRussa P. Pandemic novel 2009 H1N1 influenza: what have we learned. Semin Respir Crit Care Med. 2011;32:393–399. doi: 10.1055/s-0031-1283279. [DOI] [PubMed] [Google Scholar]
- Fowlkes AL, Arguin P, Biggerstaff MS, Gindler J, Blau D, Jain S, et al. Epidemiology of 2009 pandemic influenza A (H1N1) deaths in the United States, April–July 2009. Clin Infect Dis. 2011;52 (Suppl 1:S60–S68. doi: 10.1093/cid/ciq022. [DOI] [PubMed] [Google Scholar]
- Bush A, Menzies-Gow A. Phenotypic differences between pediatric and adult asthma. Proc Am Thorac Soc. 2009;6:712–719. doi: 10.1513/pats.200906-046DP. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.