Abstract
Protein structural diversity encompasses a finite set of architectural designs. Embedded in these topologies are evolutionary histories that we here uncover using cladistic principles and measurements of protein-fold usage and sharing. The reconstructed phylogenies are inherently rooted and depict histories of protein and proteome diversification. Proteome phylogenies showed two monophyletic sister-groups delimiting Bacteria and Archaea, and a topology rooted in Eucarya. This suggests three dramatic evolutionary events and a common ancestor with a eukaryotic-like, gene-rich, and relatively modern organization. Conversely, a general phylogeny of protein architectures showed that structural classes of globular proteins appeared early in evolution and in defined order, the α/β class being the first. Although most ancestral folds shared a common architecture of barrels or interleaved β-sheets and α-helices, many were clearly derived, such as polyhedral folds in the all-α class and β-sandwiches, β-propellers, and β-prisms in all-β proteins. We also describe transformation pathways of architectures that are prevalently used in nature. For example, β-barrels with increased curl and stagger were favored evolutionary outcomes in the all-β class. Interestingly, we found cases where structural change followed the α-to-β tendency uncovered in the tree of architectures. Lastly, we traced the total number of enzymatic functions associated with folds in the trees and show that there is a general link between structure and enzymatic function.
Proteins are numerous, extraordinarily diverse in sequence, and varied in function. They display unique three-dimensional structures and contain protein domains (Doolittle 1995; Ponting and Russell 2002), minimal building blocks (modules) that share common ancestry (Riley and Labedan 1997; Murzin 1998; Apic et al. 2001; Aravind et al. 2002a) and can be unified into a comparatively small set of folding architectures (Murzin et al. 1995; Swindells et al. 1998). The universe of protein topology is therefore finite and structurally redundant, with protein folds being among the most conserved elements in biology (Gerstein and Hegyi 1998). The number of fold categories is at present curbed by progress in structural genomics and acquisition of entire protein complements (proteomes) from organisms spanning all branches of the universal tree of life (Teichmann et al. 1999). Because protein diversity apparently originated from a limited set of architectural designs (Koonin et al. 2000), the challenge is to find these designs imprinted as ancestral signatures in extant molecules. In evolutionary studies, genomes have been organized hierarchically in trees (Wolf et al. 2002) that were derived using measurements of order, sharing or usage of protein sequences (e.g., gene content; Snel et al. 1999) and structures (e.g., fold occurrence; Gerstein 1998). These genome trees describe overall similarities between the proteomes of different organisms, and are therefore phenetic in nature. In the present study, we used cladistic principles to uncover phylogenetic histories of organismal and protein-fold diversification. A general scheme recently applied to the study of evolved RNA structure (Caetano-Anollés 2002a,b) was used to infer phylogenetic relationships on the basis of shared and derived characteristics in protein fold-usage. Using this approach, we reconstructed inherently rooted phylogenetic trees of proteomes and protein folds, identified ancestral fold configurations, and revealed interesting patterns of evolution in protein architecture.
RESULTS
Cladistic Method and Character Argumentation
We used cladistic tools to trace the evolution and compare systematically the architecture of protein molecules that fall within the different fold categories in the Structural Classification of Proteins (SCOP) database. We characterized each fold using attributes (characters) that describe numerically its genomic abundance (G), distribution, and sharing (Ḡ and f). These metrics were here used to reconstruct phylogenetic relationships in three steps. First, values were normalized to compensate for differences in genome size, converted into linearly ordered multistate characters using gap-recoding techniques, and compared in alignment matrices. Second, maximum states were specified as being ancestral (plesiomorphic) to all other, establishing an evolutionary direction of character transformation. And third, the alignment matrices were analyzed with maximum parsimony, reconstructing branching histories of inheritance in the form of phylogenetic trees. The model of character evolution proposed here is supported by the survey and comparison of protein architectures, statistical analyses of genome sequences, and arguments of evolutionary divergence that postulate that new protein structures arise by descent with modification from an ever smaller set of ancestral designs. Character argumentation, the logical process of determining ancestral states in a transformation series, rests on the following considerations.
1. Distribution of Protein Folds Across Domains
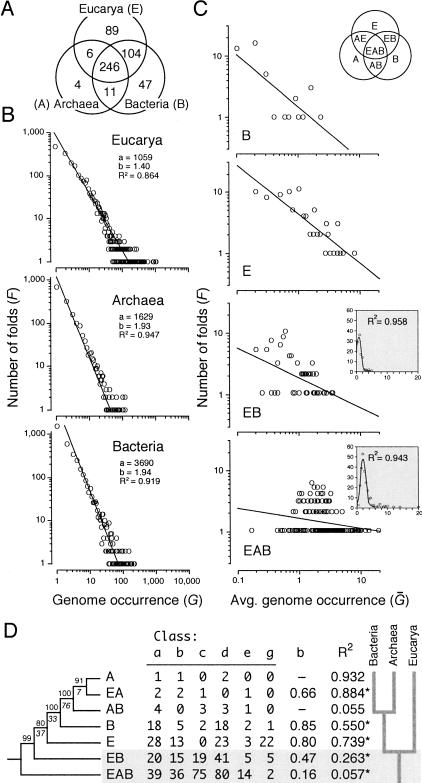
Distribution patterns of fold architectures are good indicators of how proteins have diversified, especially if patterns are studied across a wide organismal transect. Uniform distribution patterns are suggestive of common ancestry and long-term architectural stability, and are spread by vertical descent (Hegyi et al 2002). Uneven distribution patterns are suggestive of horizontal gene transfer (HGT), gene loss, convergence, and rapid divergence (Eisen 2000). We studied the frequency of fold distribution and extent of architectural sharing across domains. Seven major distribution patterns were evident and are depicted in the Venn diagram of Figure 1A. Almost half of the protein folds were common to Eucarya, Archaea, and Bacteria (EAB). Almost all folds in Archaea and the majority of folds containing the large superfamilies were part of this distribution pattern. Eucarya and Bacteria shared about 20% of folds (EB). These probably resulted from the transfer of bacterial genes from primitive organelles to the nucleus, following ancestral processes of endocytosis and other HGT events (Eisen 2000). In contrast, only 2% were shared between the two prokaryotic domains (AB), perhaps reflecting remnants of common ancestry. Finally, about 18%, 9%, and 1% were unique to Eucarya (E), Bacteria (B) and Archaea (A), respectively, and were probably the result of coupled gene transfer and gene loss (nonorthologous displacement), rapid evolution, and HGT events postulated to be pervasive in prokaryotes (Koonin et al. 2000). Interestingly, the frequency of all these patterns of fold distribution did not change significantly despite the continuous increase of known folds in the databases (cf. Wolf et al. 1999).
Figure 1.
Fold distribution, power-law behavior, and history of fold diversification in the three domains of life. (A) The Venn diagram shows the distribution of phylogenetically informative SCOP 1.59 folds in Eucarya, Archaea, and Bacteria (genomes analyzed are described in Fig. 2). (B) The double logarithmic plots show the relationship between the frequency (F) of a protein fold exhibiting a certain attribute and the attribute itself. In this case, the attribute is fold occurrence (G). The relationship between frequency and occurrence was fitted to a straight line (R2 = 0.864–0.947; P < 0.001) that drops off sharply and similarly for each genome (plots not shown) or group of genomes, according to a power law defined by constants a and b. This behavior follows Zipf's law, a description of the frequency of words in natural languages. (C) Double logarithmic plots also show the relationship between the frequency of folds with a particular pattern of distribution and the average number of times these folds occur in genomes within one or more organismal domains, normalized to a 0–20 scale (Ḡ). The nomenclature of patterns of fold distribution is described in the Venn diagram (inset). All plots show significant linear correlations (P < 0.05; see below). However, values in the EAB and EB plots (binned to reduce noise in the data) can be best fitted to a Poisson distribution (P = 0.001) (insets). (D) The table shows the number of folds in the six classes of protein structure (named according to SCOP nomenclature) present in different distribution patterns among organismal domains, together with decay indices and coefficients of linear correlation (R2) describing the fit to a power law (*, P < 0.05). These values were coded (0–26) and weighted (4, 2.5, 3.5 6, 1, and 1, respectively) to compensate for fold representation differences. A single rooted tree of 520 steps (CI = 0.901, RI = 0.925; g1 = -1.460; PTP, P = 0.001) was recovered after an exhaustive search (D). BS values >80% are shown above nodes, and double decay indices below them (CIC = 13.34).
2. Statistical Analysis of Genome Sequences
Genome surveys have shown that fold occurrence follows a power-law distribution (Quian et al. 2001), with only a few popular fold designs occurring amid many that are infrequent. This pattern has been described with a graph that connects evolutionarily related architectures (vertices) in a scale-free network (Apic et al. 2001) that grows when connections establish preferentially with the more highly connected vertices (Jeong et al. 2000). These networks have been used to describe many phenomena, including metabolic pathways, social networks, and the World Wide Web. Linear regression analysis in double-logarithmic plots demonstrates that the frequency of folds (F) displaying a given occurrence decays according to the equation F = aG-b, for both genomes considered individually and those pooled by organismal domain. Figure 1B shows plots for sets of genomes of eucaryal, archaeal, and bacterial origin. Prokaryotic genomes produce steeper decay gradients (i.e., larger b exponents) than eukaryotic genomes, rejecting a null hypothesis of slope homogeneity (ANCOVA, P < 0.0001) and showing that there is a larger level of architectural redundancy in proteomes from complex organisms. This power-law behavior implies a preference for duplication of genes encoding folds that are already common, as recently summarized in an evolutionary model that takes into account both duplication of existing genes and acquisition (and loss) of novel genes by lateral transfer (especially in prokaryotes; Quian et al. 2001). Gene duplication has long been considered a major evolutionary mechanism responsible for genetic diversity and innovation (Ohno 1970). Strong support for this view comes from recent statistical analyses of genome sequences that suggest that the proteome renews every ∼100 million years by gene duplication alone (Lynch and Conery 2000; Bailey et al. 2002; Moundsey et al. 2002). Gene duplication entails innovation in structure and function. Advances in protein structure determination reveal instances where structural innovation appears driven by changes in folding pathways, divergence, and convergence to stable architectures, shuffling of structural components and topologies, and generation of single-chain multidomain arrangements (Apic et al. 2001; Grishin 2001). Sequence and structure comparisons also suggest that innovations result from architectural divergence in paralogous molecules (Aravind et al. 2002b) and exchange of more primitive components by lateral transfer (Riley and Labedan 1997; Lupas et al. 2001).
3. Fold Sharing Across Domains
The extent of fold sharing was measured within each organismal domain or combination of domains with the Ḡ and f metrics. These two statistics rendered similar results; those using Ḡ are shown in Figure 1C. Most EAB and EB fold architectures were widely shared within each domain and followed Poisson-like distributions characteristic of a random graph. In fact the connectivity of folds in these exponential networks was statistically homogeneous (data not shown). All other fold distribution patterns contained relatively few architectures that were widely shared. In most of these cases, fold-sharing patterns resembled a power law characteristic of a scale-free network, which is highly heterogeneous (Fig. 1C). The null hypothesis of slope homogeneity was rejected (ANCOVA, P = 0.0001) in double logarithmic plots, and decay gradients and correlation coefficients were significantly larger for fold distributions other than EAB. The contrasting behavior of patterns of fold distribution (Fig. 1C) suggests that the spread of protein folds across organismal domains is complex and can be described by both homogeneous (random network) and heterogeneous (scale-free network) processes. Both lateral transfer (stochastic) and vertical descent (ordered) mechanisms probably delimit these architectural redundancy patterns.
4. The Evolution of Patterns of Fold Distribution
We used parsimony reasoning to establish phylogenetic relationships between the patterns of fold distribution themselves. In doing so, we tested our model of character evolution. Fold occurrence was averaged across populated domains for each distribution pattern and for each of the six major structural classes of proteins, and the resulting matrix was used to generate a most-parsimonious tree reconstruction (Fig. 1D). Characters were polarized by simply considering ancestral those fold distributions that were more frequent. This reasoning is based on two basic assumptions: (1) lineages can be traced to a common ancestor, and (2) redundancy is a favored evolutionary outcome. The tree was rooted in the group of folds that is shared among the three domains of life, with groups following the sequence EAB>EB>E>B>AB>EA=A, from ancestral to derived. This tree constitutes a hierarchical statement of ontogenetic reasoning, in which molecular features widely shared appear ancestral. If correct, this tree fails to reject the null hypothesis that fold occurrence increases in the course of evolution, and is therefore consistent with the model that supports character argumentation. A corollary of this statement is that Bacteria originated earlier than Archaea when evolving from the primitive cenancestor.
5. Phylogenetic Reconstruction Using Characters of Usage and Sharing of Protein Architectures
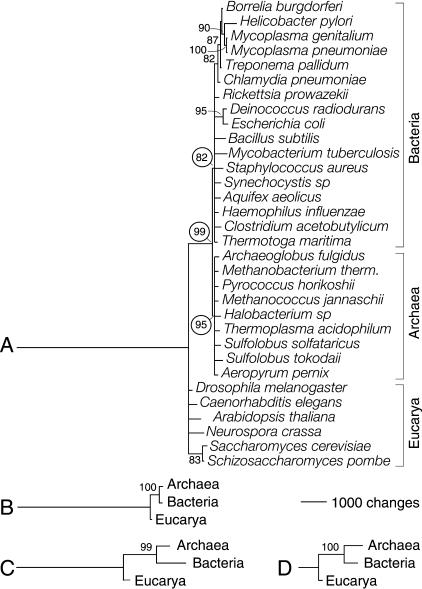
Finally, we tried to falsify our hypothesis of character polarity by comparing phylogenies reconstructed using independent characters sets (Kitching 1992; Maddison and Maddison 1999). Universal genome trees generated using G and f produced identical three-domain statements, all of them rooted in Eucarya (Fig. 2). This indicates that characters defined by the usage and sharing of protein architectures were cladistically compatible.
Figure 2.
Phylogenetic reconstruction of a universal tree. Phylogenetic relationships were inferred from genomic abundance values of SCOP 1.59 fold categories. Bootstrap support (BS) values >80% are shown above nodes. (A) Reduced phylogenetic tree reconstructed from fold occurrence (G) data. A total of 507 informative out of 536 total characters with 20 character states each were analyzed. Two most-parsimonious trees of 16,157 steps (CI = 0.625, RI = 0.486; g1 = -0.659; PTP test, P = 0.001) were retained after a heuristic search with tree-bisection-reconnection (TBR) branch swapping and 50 replicates of random addition sequence. The tree shown is congruent with the 50% majority-rule consensus. The null hypothesis of congruence could not be rejected when folds in the six structural classes were tested for homogeneity of data partitions (P = 0.498). (B) Tree reconstructed from fold occurrence data averaged across genomes in each organismal domain (Ḡ). Characters had 20 states, and 300 informative characters were analyzed. A single tree of 5885 steps (CI = 0.970, RI = 0.660; g1 = -0.702; PTP, P = 0.001) was retained after an exhaustive search. (C) Tree reconstructed from the fraction of genomes in each organismal domain that share individual folds (f). Characters had 17 states; 447 informative out of 507 total characters were analyzed. A single tree of 7603 steps (CI = 0.852, RI = 0.543; g1 = -0.559; PTP, P = 0.001) was retained after an exhaustive search. (D) Tree reconstructed as in C but from the subset of folds that is shared by the three organismal domains. Characters had 17 states, and 149 informative out of 246 total characters were analyzed. A single tree of 1601 steps (CI = 0.895, RI = 0.752; g1 = -0.672; PTP, P = 0.001) was retained after an exhaustive search.
Given all five considerations discussed above, we can assume safely that extant proteins that have originated from a common ancestor retain memory of its ancestral structure and function, and consequently, that protein architectures prevalently used and shared by a wide range of organisms originate from innovations which occurred earlier in evolutionary time.
Proteome Phylogenies
We reconstructed phylogenetic trees from fold occurrence data, and tested the effect of character coding and progress in database entry (see Supplemental Material available at www.genome.org). Polarized multistate characters were used to reconstruct rooted phylogenies from a set of 32 genomes that have been completely sequenced and belong to organisms spanning the three domains of life (Fig. 2A). Distribution of cladogram lengths and PTP tests showed strong cladistic structure in the data (P < 0.01). The global topology of the universal tree was reasonably supported by double decay (data not shown) and bootstrap (BS) analysis. However, many branches were not well resolved, and some groups in Bacteria were polyphyletic (such as the γ-proteobacteria and the Spirochaetales). Two clear monophyletic groups were evident and comprised genomes from Archaea (95% BS) and Bacteria (82% BS). These prokaryotic groups exhibited a strong sister-clade relationship (99% BS) and were derived. In contrast, genomes in Eucarya were basal, suggesting a eukaryotic rooting of the tree of life. The topology and rooting of the genome tree were also recovered when examining patterns of fold distribution and sharing among the three organismal domains (Fig. 2B,C). Identical three-domain statements rooted in Eucarya were reconstructed from fold occurrence data averaged across genomes and from the fraction of genomes in Archaea, Bacteria, and Eucarya that share individual folds. Venn diagrams reveal that almost half of fold architectures were common, and that only about one in four folds were characteristic of individual domains (Fig. 1). Phylogenetic analysis of folds shared by the three domains revealed again the same tree topology rooted in the eukaryotic branch (Fig. 2D).
Phylogenetic Trees of Protein Architectures
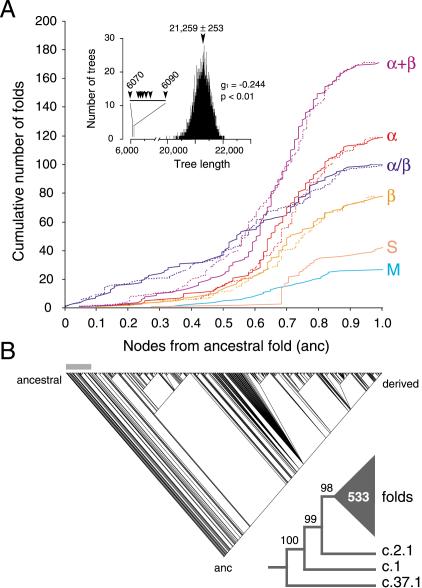
In order to study evolutionary patterns embedded in protein architecture, we generated phylogenetic histories of protein diversification from fold occurrence data. Studies involved small and large subsets of protein folds, and complete data sets matching two releases of SCOP. Figure 3 shows the phylogenetic relationship of 536 folds in SCOP 1.59. Although there was strongcladistic structure in the data (P < 0.01), only 32 characters (with 20 states) were used to describe fold occurrence in different genomes, and consequently, most branches of the tree were poorly supported by bootstrap analysis. Despite this limitation, general evolutionary patterns in protein structure were clearly recovered. Cumulative frequency plots revealed order and rate of appearance of folds falling within each of the six major structural classes of globular proteins (Fig. 3A). Considering that folds are finite, the total number of nodes (ranging from 418–424 cladogenic events) and node distances (range 84–90) in the trees defined a relative but total time frame of protein diversification. All classes appeared very early in the tree of architectures, within the first 42 cladogenic events (9.7%) and within a distance of 24 nodes from the root (0.28 in a relative 0–1 scale). Folds in the α/β protein class arose first and were followed by those in the α+β, all-α, all-β, small, and multidomain classes, in that order. These folds accumulated at different levels. The α/β folds occurred at relatively constant rates and were prevalent in the bottom half of the tree. In contrast, the α+β folds started to accumulate significantly later but with increasing rates until these folds became the most prevalent class. Folds in all other classes followed this same pattern of accumulation but with lower rates. Maximum rates diminished in the order of fold appearance in the tree; that is, all-α, all-β, small, and multidomain proteins. These general evolutionary patterns were robust and were similarly inferred from trees reconstructed using Ḡ (see below) and folds defined by SCOP 1.49 and 20 genomes (data not shown).
Figure 3.
Phylogenetic reconstruction of a universal tree of protein architecture. (A) Cumulative frequency plots illustrate the accumulation of folds in the six major classes of protein architecture along optimal (continuous lines) and suboptimal phylogenetic trees (dashed lines). Cumulative fold number is given as a function of distance in nodes from the hypothetical ancestral fold (anc) in a relative scale. Suboptimal tree reconstructions (spanning 6070 and 6090 steps) show that systematic and random error did not substantially affect the rates of fold accumulation. The inset shows tree distribution profiles and metrics of skewness. (B) One optimal most-parsimonious tree (6070 steps; CI = 0.105, RI = 0.773; PTP test, P = 0.001) was recovered from a heuristic search with TBR branch swapping and 10 replicates of random addition sequence. To decrease search times during branch swapping of suboptimal trees, only 10 trees of length ≥D + 1 were kept in each replicate, with D being the minimum tree length found in multiple iterative searches. The bar defines when protein classes occurred for the first time. The reduced cladogram shows branches with BS supports <98% collapsed into a multifurcation (triangle with number of multifurcating branches).
Evolutionary Patterns and Pathways of Protein Architecture
The general tree of protein architectures identified three α/β folds as the most ancestral (Fig. 3B). These were the P-loop hydrolase (c.37), the TIM β/α-barrel (c.1), and the Rossmann (c.2) fold, in order from ancestral to derived. Phylogenies reconstructed separately from folds belonging to each protein class were more informative and contained better-supported branches than the general tree. These trees revealed ancestral fold configurations in each class and clear evolutionary patterns (Fig. 4). Ancestral folds were generally folds with top genomic representation and included barrel folds (e.g., c.1, b.40 [OB-fold], b.43 [reductase/elongation factor], and b.84 [barrel-sandwich hybrid]) and classic folds with helices packed on either side (e.g., c.37, c.2, c.23 [flavodoxin-like], and d.104 [SH2-like fold]) or onto a single face (e.g., d.58 [ferredoxin-like], c.3 [FAD/NAD(P)-binding], and d.142 [ATP-grasp fold]) of a central β-sheet arrangement. Ancestral folds in the α/β class were generally superfolds widely distributed amonggenomes (Gerstein 1998; Wolf et al. 1999). Remarkably and with the exception of the TIM β/α-barrel, they all shared a common architecture of interleaved β-sheets and α-helices. The ferredoxin-like fold (d.58) was the most ancestral architecture in the α+β class. This fold contains simple and irregular protein architectures and packs an α+β sandwich with an antiparallel β-sheet. Ancestral folds in the all-α class were mostly composed of bundles of longhelices, sometimes constituting a layer packing arrangement. This suggests that all-α folds that fit a polyhedron model (roughly half of SCOP entries in this protein class; Chothia et al. 1997) are evolutionarily derived architectures. Hence, evolution of the all-α protein class appears driven by a search for order in protein packing. Ancestral folds in the all-β class were β-barrels (of n = 5,6 and S = 8–10 with Greek-key) (b.40 and b.43), a barrel sandwich hybrid (b.84), and a left-handed β-helix with turns composed of three short β-strands (b.81). These ancestral folds have β-sheets staggered into closed, partly open, or open β-barrel architectures, or are packed in prism-like fashion into a 3-sheet β-helix arrangement (see UDP N-acetylglucosamine acyltransferase [1lxa]). This suggests that β-sandwiches, β-propellers, and β-prisms are all derived fold architectures, and that β-helices (which are mostly right-handed) derive from left-handed and closely packed superhelical structures. Interestingly, an evolutionary tendency towards right-handedness in β-helices is supported thermodynamically by folding pathways of β-β-β units in β-sheets that favor right-handed connections (Chothia et al. 1997).
Figure 4.
Reduced cladograms representing the phylogenetic relationships of folds belonging to individual protein classes. Branches with BS values <50% were collapsed into multifurcations (triangles with areas proportional to the number of folds unified by the polytomy). Trees were retained after heuristic searches with TBR branch swapping and 10 replicates of random addition sequence. Their lengths ranged from 786 steps (CI = 0.814, RI = 0.709; g1 = -2.215; PTP test, P = 0.001) for small proteins to 2375 steps (CI = 0.270, RI = 0.761; g1 = -0.528; PTP, P = 0.001) for the α/β protein class. Cladograms depicting trees with alternative reconstructions were congruent with the 50% majority-rule consensus.
Protein transformation pathways that describe likely scenarios of structural evolution (Murzin 1998; Grishin 2001) could be traced in our tree of architectures (see Supplemental Material). Similarly, evolutionary patterns of architectural design were also evident in individual protein classes. For example, we selected all-β folds that had barrel-like structures and were phylogenetically informative, pooled them in groups according to β-sheet topology (Greek-key, meander, and complex; Chothia et al. 1997), reconstructed phylogenetic trees, and searched for patterns in structure (Fig. 5). In the three groups, there was a clear increase in barrel strand (n) and shear (S) number. Moreover, open and partly open barrel structures were derived characteristics. Furthermore, analysis of ancestral folds from each group showed an evolutionary progression in β-sheet topology from the highly ordered Greek-key and the simple up-and-down meander pattern to more complex topological arrangements.
Figure 5.
Phylogenetic trees of all-β protein folds with β-barrel-like architecture. Maximum parsimony was used to reconstruct a general tree of β-barrel-like folds with different β-sheet topologies and barrel mimic folds (A) and trees of β-barrel folds with Greek-key, meander, and complex β-sheet topologies (B). Barrel mimic folds include architectures such as the barrel-sandwich hybrid, with two β-sheets in the shape of a half-barrel packed in a sandwich-like arrangement, and the β-clip, with two-stranded β-sheets that fold upon themselves. Folds are described by general characteristics such as barrel architecture [closed (C), partly open (P), or open barrel (O)], number of strands (n), and shear number (S), and special features (SF) such as cross-over psi loops (p), over-side connections (oc), capping by α-helices (c), and internal pseudo-threefoil symmetry (i). Trees with lengths ranging 659–768 steps [CI = 0.833–0.941, RI = 0.729–0.904; g1 = -(0.554–0.904); PTP tests, P = 0.001] were retained after branch-and-bound or exhaustive searches.
Evolution of Enzymatic Function
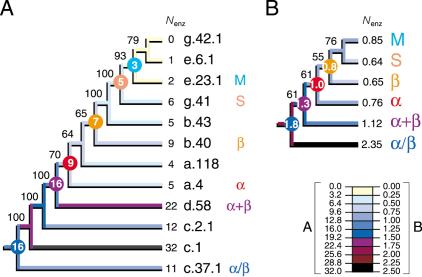
Lastly, we explored the relationship between protein architecture and function by tracing the total number of enzymatic functions associated with folds in the trees (Fig. 6). As expected, the most ancestral folds had generally the most enzymatic functions associated with them. This evolutionary tendency was seen in ancestral folds sampled throughout the tree of architectures and in a tree of protein classes. Squared-change parsimony allowed inference of the number of functions associated with the hypothetical ancestors of each protein class, which also decreased in time.
Figure 6.
Tracing the evolutionary association of enzymatic function and protein architecture. Cladograms show the phylogenetic relationship of primitive folds (A) and protein classes (B) and were derived from a single tree of 907 steps (CI = 0.690, RI = 0.809; g1 = -0.911; PTP, P = 0.001) and 5209 steps (CI = 0.683, RI = 0.598; g1 = -0.792; PTP, P = 0.001) retained after branch-and-bound and exhaustive searches, respectively. The tree of protein classes was derived from fold occurrence data averaged across populated domains for each distribution pattern (Fig. 1) and for each of the six protein classes. The number of enzymatic functions (Nenz) was similarly averaged. Square-change parsimony was used to reconstruct ancestral Nenz states as continuous characters in the trees using McCLADE with the rooted option. These values are shown encircled for selected internal nodes.
DISCUSSION
Character attributes represent transformation pathways and hypotheses of relationship (amenable to Popperian falsification) that link character states to each other by specific evolutionary processes (Kitching 1992; Maddison and Maddison 1999). Our phylogenetic study rests on the central assumption that protein folds are more prevalent and more widely shared the more ancestral is their origin, with characters transforming from one state to another in pathways which are linear (restrictive statement that prohibits branched or reticulate arrangements), directed (statement of asymmetry in transformation costs), and polarized (statement that invokes ancestral states). This simple (and perhaps simplistic) model of character evolution is based on the parsimony principle of preferring simple explanations to complex ones (Ockham's razor: “Pluralitas non est ponenda sine neccesitate”) and is supported by patterns in the distribution and sharing of protein folds across domains, statistical analyses and scale-free network behavior of protein fold occurrence, and a phylogenetic study of the evolution of patterns of fold distribution (Fig. 1). The model and supporting results are compatible with the findings and evolutionary model of Quian et al. (2001). More complex models may be warranted however in the future to account for possible factors such as variation in evolutionary rates across characters and branches of the trees, and changes in the size of the protein universe expected to have occurred during evolution.
The reconstruction of histories of proteome diversification showed two monophyletic sister-groups delimiting Bacteria and Archaea, and a topology rooted in Eucarya (Fig. 2). The rooting of the universal tree constitutes a highly debated and controversial issue. Genomic analysis has shown that lateral gene transfer and lineage-specific gene loss are common phenomena (at least in prokaryotes; Koonin et al. 2000), casting doubt on the existence of a universal common ancestor (the `cenancestor'; Fitch and Upper 1987), and complicating parsimony-based reasoning(Woese 1998; Doolittle 1999). On the other hand, the analysis of complete genomes has rescued the notion of the universal tree, as sufficient phylogenetic signal in the sequence, content, and order of gene complements enabled tree reconstruction (Wolf et al. 2002). These unrooted trees continue to support the three-domain classification of life (Woese et al. 1990) but failed to define deep phylogenetic relationships. Other approaches had to be sought (Doolittle 2000). For example, a rooted universal tree was recently recovered from rRNA structure using cladistic principles and considerations in statistical mechanics (Caetano-Anollés 2002a,b). These phylogenies were suggestive of three dramatic evolutionary events and an equally parsimonious eukaryotic or prokaryotic origin of diversified life. Our results complement these findings, by reflecting global evolutionary relationships at a genomic scale. However, they conflict with the accepted view of a prokaryotic ancestor (Woese et al. 1990), supporting instead recent proposals that describe the genome of the cenancestor as an eukaryotic-like, gene-rich, and relatively modern architecture (Forterre and Philippe 1999; Penny and Poole 1999). We favor the view that molecular evolution is driven by a search for innovation that increases molecular complexity and modularity in all lineages of the universal tree (see below).
A general phylogeny of protein architectures showed that protein classes appeared early in evolution and in defined order (Figs. 3,5). Our results suggest that the most primitive protein forms contained interspersed α-helical and β-sheet elements (as in the α/β class) that in the course of evolution were first segregated within their structure (α+β class) and then confined to separate molecules (all-α and all-β classes). It is likely that during this time, structural simplification and re-arrangement occurred pervasively and at low levels (Lupas et al. 2001), resulting in the slow accumulation of small proteins and multidomain folds. This hypothetical scenario is consistent with patterns of modularity and simplification in molecular design (Ancel and Fontana 2000; Hartwell et al. 1999), recently revealed in rRNA structure (Caetano-Anollés 2002b), and in the suggestion that diversity in protein architecture originated by stochastic processes expressed in both protein sequence and structure (the random origin hypothesis; White 1994).
Proteins belonging to a fold category maintain a core of three-dimensional packing delimited by topological connections of α-helices and β-sheets, but they harbor peripheral elements of secondary structure and turn and coil regions that can be substantially variable, in both size and conformation. In SCOP, fold categories are also delimited by evolutionary considerations (Murzin et al. 1995; Lo Conte et al. 2002). In the absence of significant sequence similarity, functional features such as catalytic or binding sites, and structural characteristics such as unusual motifs or loops are used as evidence of common ancestry. It is therefore of interest to understand how topological connections defining fold categories have changed during protein diversification from a structural perspective. Phylogenies reconstructed from small and large subsets of protein folds allowed reliable identification of the most ancestral fold categories, and uncovered general patterns of evolution in fold architecture. We found that most ancestral folds shared a common architecture of barrels or interleaved β-sheets and α-helices (Fig. 4). We also uncovered interesting evolutionary patterns when studying the evolution of certain fold architectures that are prevalently used in nature. For example, β-barrels are simple highly geometrical structures that represent about a third of folds in the all-β protein class and appear quite early in evolution (Figs. 4,5). The study of these β-barrel folds showed an evolutionary tendency to increase: (1) the tilt of the β-strands in relation to the barrel axis in the context of the “n, S model” (by increasing n and S; McLachlan 1979), (2) the frequency of partly-open or open barrel structures, and (3) the complexity of strand topology in the curled β-sheets (Fig. 5). These tendencies suggest that barrel architectures with increased curl and stagger of β-sheets (sensu Taylor 2002) should be regarded as favored evolutionary outcomes.
It was also possible to trace changes in fold structure that have an impact on fold architecture. Grishin (2001) recently provided examples of architectural transformations in evolutionarily related proteins. Several of these examples involved insertions/deletions (indels), circular permutations, and strand re-arrangements that induce changes in structure capable of modifying general architecture. One example is the conversion of an α-helix into a three-stranded β-meander that replaces the α-β-α layered fold architecture characteristic of NAD(P)-binding Rossmann-fold domain (c.2) in lactate dehydrogenase (1ldn) by the β-β-α architecture characteristic of the FAD/NAD(P)-binding domain (c.3) in NADH peroxidase (1npx; Aravind et al. 2002b). This conversion is of significance as it is rather common in Rossmann fold proteins. Phylogenetic analysis (Figs. 3,4) showed that the classical Rossmann c.2 fold is ancestral to the c.3 architecture, suggesting that structural change follows the general α-to-β tendency revealed in the universal tree of protein architectures (Fig. 3). This same tendency can be observed in other pathways involving insertions, deletions, substitutions, circular permutations, and re-arrangements in β-sheet topologies (see Supplemental Material).
Lastly, we traced the total number of enzymatic functions associated with folds and found that their number increased towards the base of the trees (Fig. 6). These results suggest that architectural multifunctionality in proteins was replaced by specialized function. It is tempting to speculate that this evolutionary tendency is also associated with the rise of modular design in proteins.
Taken together, statistical and phylogenetic studies suggest a likely evolutionary scenario. At an early evolutionary stage, divergence in structure and function resulted in massive proliferation of proteins that were functionally versatile, had numerous lineage-specific variants, and clustered in large structural ensembles. The resulting architectures were common and widely distributed across taxa and domains, forming random exponential networks of relationships, and ultimately large structural superfamilies. With time, certain architectures behaved as modules, combined by “domain shuffling”, and gave rise to structural innovations associated with specific functions. Driven by strong selection pressures, each innovation was rare but spread across lineages, perhaps during horizontal transfer events that occurred progressively and at different levels in informational and operational gene sets (Jain et al. 1999). This ultimately produced scale-free networks of relationships that were more robust (error tolerant) than the originating networks.
However, results argue against mass lateral transfer of genes among the domains of life, as this would have obscured phylogenetic signal in the data. Results also show that diversity in architecture appears prevalent in Eucarya. This could stem from the early appearance of eukaryotic-like ancestors or from natural selection forces acting on gene duplication. Eucaryotic organisms are believed to be the subject of K-selection, taking advantage of the carrying capacity of the environment rather than rapid growth in times of nutrient availability (Carlile 1982). They also harbor multiple centers of replication that could enhance possibilities of gene duplication, genomic redundancy, and architectural diversification. Given that genetic redundancy is common and can be stable (Nowak et al. 1997), mutation and selection on `structural' and `functional' replicates would ultimately result in increased architectural innovation.
The evolutionary patterns revealed here are important but rely on how thorough and extensive are the protein databases analyzed. Results may be affected by biases such as over- and underrepresentation of certain sequences and structures, incorrect structural assignment of proteins to fold categories, and genome sampling (Gerstein and Hegyi 1998). PDB databanks are biased by research preferences for targets and organisms and physical constraints imposed by crystallography and NMR spectroscopy. At present, repositories classify only a small subset (averaging ∼35%) of sequences into fold categories, and fold classification remains an empirical endeavor. Fortunately, the number of `orphan' sequences without a structure will diminish with progress in structural genomics. Our approach is general and will benefit by the constant increase in number and breadth of genomes that are being sequenced. We do not expect significant changes in the SCOP classification, especially because SCOP organizes protein architectures robustly according to both structural similarity and evolutionary origin (Swindells et al. 1998). Notwithstanding, discovery of new folds and accretion of fold categories will continue. The analysis of proteomes defined by two releases of SCOP recovered similar phylogenies of genomes and folds (data not shown). Consequently, the use of larger character sets in future analyses will only enhance the confidence of our phylogenetic statements.
METHODS
The Structural Classification of Proteins (SCOP) database describes the evolutionary and structural relationship of proteins with known atomic structure (Murzin et al. 1995). Release 1.59 (May 2002) clusters 15,979 PDB structural entries and 39,893 domains into 686 fold categories, encompassing 1073 superfamilies and 1827 families (Lo Conte el al. 2002). Protein entries matching fold categories in SCOP were retrieved from the PEDANT 1.0.2 database (Frishman et al 2001) in a set of six eukaryotic, nine archaeal, and 17 bacterial genomes sampled over 100 finished genomic sequences. Out of 138,377 entries, an average of 38.4±1.5 (SE) % (range 9.7%–48.6%) matched SCOP domains, and 19.5±0.8 % (range 9.5%–28.3%) had enzymatic activities associated with them. For comparison purposes, we also used a data set that matched 420 fold categories in SCOP 1.39 and was generated by PSI-BLAST comparisons between PDB and genome sequence entries in the first 20 genomes ever to be sequenced (Hegyi et al. 2002). We considered soluble proteins that grouped into major structural classes: all-α proteins with structures composed mostly of α-helices (α), all-β proteins with mostly β-sheets (β), α/β proteins with interspersed α-helices and β-sheets (α/β), α+β proteins containing segregated α-helices and β-sheet regions (α+β), multidomain proteins containing domains belonging to different classes and without known homologs (M), and small proteins (S). The usage and sharing of protein folds was characterized with three metrics: fold occurrence (Gij), average genome occurrence (Ḡi), and fraction of genomes harboring a fold (fi). Gij defines how often a protein fold (i) occurs in a given proteome (j). Ḡi represents averages of Gij values. Ḡi and fi measure the extent of fold sharing within each domain or combination of domains. Values were converted into linearly ordered multistate characters using the gap-recoding technique of Thiele (1993), normalized using an arbitrary scale (generally 0–20) to compensate for differences in genome size, and compared in alignment matrices. These matrices constitute frequency ensembles of protein architectures. Character states were represented by a discrete alphanumerical format with numbers 0–9 and letters A–Q, and matrices encoded in the NEXUS format. Phylogenetic relationships were inferred using PAUP* v.4.0 (Swofford 1999). Characters were polarized with the ANCSTATES command. Trees were reconstructed using maximum parsimony as the optimality criterion, and were automatically rooted at the point where the hypothetical ancestor connected to the tree. Phylogenetic reliability was evaluated by the nonparametric bootstrap method (Felsenstein 1985; implemented using 2 × 103 pseudoreplicates) and by double decay analysis (Wilkinson et al. 2000) using RadCon (Thorley and Page 2000). The structure of phylogenetic signal in the data was tested by the skewness (g1) of the length distribution of 104 random trees, and permutation tail probability (PTP) tests of cladistic covariation using 103 replicates. Ensemble consistency (CI) and retention (RI) indices were used to measure homoplasy and synapomorphy. The homogeneity of partitions was analyzed using a modified Michevich-Farris index of incongruence among data sets and 103 heuristic search replicates (Farris et al. 1995). Topological congruence was measured with several tree comparison metrics and randomization tools using Component (Page 1993). Cumulative frequency plots were used to illustrate the accumulation of folds belonging to a protein class alonga phylogenetic tree. Cumulative fold number was given as a function of distance in nodes from the root. These plots can be considered time plots of lineages (Nee et al. 1994) with a time axis defined in relative units (e.g., cladogenic events). Enzymatic functions were retrieved from the Kyoto Encyclopedia of Genes and Genomes (KEGG) v. 24 (Wixon and Kell 2000). They were defined up to the third level of the Enzyme Commission (EC) classification (IUPAC-IUBMB), and their number was traced on the trees using MacClade v. 3.08 (Maddison and Maddison 1999). Square-change parsimony was used to reconstruct the ancestral states of continuous-valued characters (Maddison 1991). Data matrices can be retrieved from the TreeBase repository (http://herbaria.harvard.edu/treebase/).
Acknowledgments
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1161903.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Ancel, L.W. and Fontana, W. 2000. Plasticity, evolvability, and modularity in RNA. J. Exp. Zool. Part B. Mol. Dev. Evol. 288: 242-283. [DOI] [PubMed] [Google Scholar]
- Apic, G., Gough, J., and Teichmann, S.A. 2001. An insight into domain combinations. Bioinformatics 17: S83-S89. [DOI] [PubMed] [Google Scholar]
- Aravind, L., Mazumder, R., Vasudevan, S., and Koonin, E.V. 2002a. Trends in protein evolution inferred from sequence and structure analysis. Curr. Opin. Struct. Biol. 12: 392-399. [DOI] [PubMed] [Google Scholar]
- Aravind, L., Anantharaman, V., and Koonin, E.V. 2002b. Monophily of class I aminoacyl tRNA synthetase, USPA, ETFP, photolyase, and PP-ATPase nucleotide-binding domains: Implications for protein evolution in the RNA world. Proteins 48: 1-14. [DOI] [PubMed] [Google Scholar]
- Bailey, J.A., Gu, Z., Clark, R.A., Reinert, K., Samonte, R.V., Schwartz, S., Adams, M.D., Myers, E.W., Li, P.W., and Eichler, E.E. 2002. Recent segmental duplications in the human genome. Science 297: 1003-1007. [DOI] [PubMed] [Google Scholar]
- Caetano-Anollés, G. 2002a. Evolved RNA structure and the rooting of the universal tree of life. J. Mol. Evol. 54: 333-345. [DOI] [PubMed] [Google Scholar]
- Caetano-Anollés, G. 2002b. Tracing the evolution of RNA structure in ribosomes. Nucleic Acids Res. 30: 2575-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile, M. 1982. Prokaryotes and eukaryotes: Strategies and successes. Trends. Biochem. 7: 128-130. [Google Scholar]
- Chothia, C., Hubard, T., Brenner, S., Barns, H., and Murzin, A. 1997. Protein folds in the all-β and all-α classes. Annu. Rev. Biophys. Biomol. Struct. 26: 597-627. [DOI] [PubMed] [Google Scholar]
- Doolittle, R.F. 1995. The multiplicity of domains in proteins. Annu. Rev. Biochem. 64: 287-314. [DOI] [PubMed] [Google Scholar]
- Doolittle, W.F. 1999. Phylogenetic classification and the universal tree. Science 284: 2124-2128. [DOI] [PubMed] [Google Scholar]
- Doolittle, W.F. 2000. The nature of the universal ancestor and the evolution of the proteome. Curr. Opin. Struct. Biol. 10: 355-358. [DOI] [PubMed] [Google Scholar]
- Eisen, J.A. 2000. Horizontal gene transfer among microbial genomes: New insights from complete genome analysis. Curr. Opin. Genet. Dev. 10: 606-611. [DOI] [PubMed] [Google Scholar]
- Farris, J.S., Kållersö, M., Kluge, A.G., and Bult, C. 1995. Testing significance of incongruence. Cladistics 10: 315-319. [Google Scholar]
- Felsenstein, J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783-791. [DOI] [PubMed] [Google Scholar]
- Fitch, W.M. and Upper, K. 1987. The phylogeny of tRNA sequences provides evidence for ambiguity reduction in the origin of the genetic code. Cold Spring Harbor Symp. Quant. Biol. 52: 759-767. [DOI] [PubMed] [Google Scholar]
- Forterre, P. and Philippe, H. 1999. Where is the root of the universal tree of life? BioEssays 21: 871-879. [DOI] [PubMed] [Google Scholar]
- Frishman, D., Albermann, K., Hani, J., Heumann, K., Metanomski, A., Zollner, A., and Mewes, H.-W. 2001. Functional and structural genomics using PEDANT. Bioinformatics 17: 44-57. [DOI] [PubMed] [Google Scholar]
- Gerstein, M. 1998. Patterns of protein-fold usage in eight microbial genomes: A comprehensive structural census. Proteins 33: 518-534. [DOI] [PubMed] [Google Scholar]
- Gerstein, M. and Hegyi, H. 1998. Comparing genomes in terms of protein structure: Surveys of a finite parts list. FEMS Microbiol. Rev. 22: 277-304. [DOI] [PubMed] [Google Scholar]
- Grishin, N.V. 2001. Fold change in evolution of protein structures. J. Struct. Biol. 134: 167-185. [DOI] [PubMed] [Google Scholar]
- Hartwell, L.H., Hopfield, J.J., Leibler, S., and Murray, A.W. 1999. From molecular to modular cell biology. Nature 402: C47-C52. [DOI] [PubMed] [Google Scholar]
- Hegyi, H., Lin, J., Greenbaum, D., and Gerstein, M. 2002. Structural genomics analysis: Characteristics of atypical, common, and horizontally transferred folds. Proteins 47: 126-141. [DOI] [PubMed] [Google Scholar]
- Jain, R., Rivera, M.C., and Lake, J.A. 1999. Horizontal gene transfer amonggenomes: The complexity hypothesis. Proc. Natl. Acad. Sci. 96: 3801-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, H., Tombor, B., Albert, R., Ottvai, Z.N., and Barabási, A.-L. 2000. The large scale organization of metabolic networks. Nature 407: 651-654. [DOI] [PubMed] [Google Scholar]
- Kitching, I.J. 1992. The determination of character polarity. In Cladistics, (eds. P.L. Forey, C.J. Humphries, I.J. Kitching, R.W. Scotland, D.J. Siebert, and D.M. Williams), pp. 22-43. Clarendon Press, Oxford.
- Koonin, E.V., Aravind, L., and Kondrashov, A.S. 2000. The impact of comparative genomics on our understanding of evolution. Cell 101: 573-576. [DOI] [PubMed] [Google Scholar]
- Lo Conte, L., Brenner, S.E., Hubbard, T.J.P., Chothia, C., and Murzin, A. 2002. SCOP database in 2002: Refinements accommodate structural genomics. Nucleic Acids Res. 30: 264-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas, A.N., Ponting, C.P., and Russell, R.B. 2001. On the evolution of protein folds: Are similar motifs in different protein folds the result of convergence, insertion, or relics of an ancient peptide world? J. Struct. Biology 134: 191-203. [DOI] [PubMed] [Google Scholar]
- Lynch, M. and Conery, J.S. 2000. The evolutionary fate and consequences of duplicate genes. Science 10: 1151-1155. [DOI] [PubMed] [Google Scholar]
- Maddison, W.P. 1991. Squared-change parsimony reconstructions of ancestral states for continuous-valued characters on a phylogenetic tree. Syst. Zool. 40: 304-314. [Google Scholar]
- Maddison, W.P. and Maddison, D.R. 1999. MacClade: Analysis of phylogeny and character evolution, version 3.08. Sinauer Assoc., Sunderland, MA.
- McLachlan, A.D. 1979. Gene duplications in the structural evolution of chymotripsin. J. Mol. Biol. 12: 49-79. [DOI] [PubMed] [Google Scholar]
- Moundsey, A., Bauer, P., and Hope, I.A. 2002. Evidence suggesting that a fifth of annotated Caenorhabditis elegans genes may be pseudogenes. Genome Res. 12: 770-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin, A. 1998. How far divergent evolution goes in proteins. Curr. Opin. Struct. Biol. 8: 380-387. [DOI] [PubMed] [Google Scholar]
- Murzin, A., Brenner, S.E., Hubbard, T., and Clothia, C. 1995. SCOP: A structural classification of proteins for the investigation of sequences and structures. J. Mol. Biol. 247: 536-540. [DOI] [PubMed] [Google Scholar]
- Nee, S., Holmes, E.C., May, R.M., and Harvey, P.H. 1994. Extinction rates can be estimated from molecular phylogenies. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 344: 77-82. [DOI] [PubMed] [Google Scholar]
- Nowak, M.A., Boerlijst, M.C., Cooke, J., and Smith, J.M. 1997. Evolution of genetic redundancy. Nature 388: 167-171. [DOI] [PubMed] [Google Scholar]
- Ohno, S. 1970. Evolution by gene duplication. Springer-Verlag, Berlin.
- Page, R.D.M. 1993. COMPONENT, tree comparison software for Microsoft Windows, v. 2.0. The Natural History Museum, London.
- Penny, D. and Poole, A. 1999. The nature of the universal common ancestor. Curr. Opin. Genet. Dev. 9: 672-677. [DOI] [PubMed] [Google Scholar]
- Ponting, C.P. and Russell, R.R. 2002. The natural history of protein domains. Annu. Rev. Biophys. Biomol. Struct. 31: 45-71. [DOI] [PubMed] [Google Scholar]
- Quian, J., Luscombe, N.M., and Gerstein, M. 2001. Protein family and fold occurrence in genomes: Power-law behavior and evolutionary model. J. Mol. Biol. 313: 673-681. [DOI] [PubMed] [Google Scholar]
- Riley, M. and Labedan, B. 1997. Protein evolution viewed through Escherichia coli protein sequences: Introducing the notion of a structural segment of homology, the module. J. Mol. Biol. 268: 857-868. [DOI] [PubMed] [Google Scholar]
- Snel, B., Bork, P., and Huynen, M.A. 1999. Genome phylogeny based on gene content. Nat. Genet. 21: 108-110. [DOI] [PubMed] [Google Scholar]
- Swindells, M.B., Orengo, C.A. Jones, D.T., Hutchinson, E.G., and Thornton, J.M. 1998. Contemporary approaches to protein structure classification. BioEssays 20: 884-891. [DOI] [PubMed] [Google Scholar]
- Swofford, D.L. 1999. Phylogenetic analysis using parsimony and other programs (PAUP*), version 4. Sinauer Assoc., Sunderland, MA.
- Taylor, W.R. 2002. A `periodic table' for protein structures. Nature 416: 657-660. [DOI] [PubMed] [Google Scholar]
- Teichmann, S.A., Chothia, C., and Gerstein, M. 1999. Advances in structural genomics. Curr. Opin. Struct. Biol. 9: 390-399. [DOI] [PubMed] [Google Scholar]
- Thiele, K. 1993. The holy grail of the perfect character: The cladistic treatment of morphometric data. Cladistics 9: 275-304. [DOI] [PubMed] [Google Scholar]
- Thorley, J.L. and Page, R.D.M. 2000. RadCon: Phylogenetic tree comparison and consensus. Bioinformatics 16: 486-487. [DOI] [PubMed] [Google Scholar]
- White, S.H. 1994. Global statistics of protein sequences: Implications for the origin, evolution, and prediction of structure. Annu. Rev. Biophys. Biomol. Struct. 23: 407-439. [DOI] [PubMed] [Google Scholar]
- Wilkinson, M., Thorley, J.L., and Upchurch, P. 2000. A chain is no longer than its weakest link: Double decay analysis of phylogenetic hypotheses. Syst. Biol. 49: 754-776. [DOI] [PubMed] [Google Scholar]
- Wixon, J. and Kell, D. 2000. The Kyoto encyclopedia of genes and genomes—KEGG. Yeast 17: 48-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese, C. 1998. The universal ancestor. Proc. Natl. Acad. Sci. 95: 6854-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese, C.R., Kandler, O., and Wheelis, M.L. 1990. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. 87: 4576-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, Y.I., Brenner, S.E., Bash, P.A., and Koonin, E.V. 1999. Distribution of protein folds in the three superkingdoms of life. Genome Res. 9: 17-26. [PubMed] [Google Scholar]
- Wolf, Y.I., Rogozin, I.B., Grishin, N.V., and Koonin, E.V. 2002. Genome trees and the tree of life. Trends Genet. 18: 472-479. [DOI] [PubMed] [Google Scholar]