Abstract
Corynebacterium efficiens is the closest relative of Corynebacterium glutamicum, a species widely used for the industrial production of amino acids. C. efficiens but not C. glutamicum can grow above 40°C. We sequenced the complete C. efficiens genome to investigate the basis of its thermostability by comparing its genome with that of C. glutamicum. The difference in GC content between the species was reflected in codon usage and nucleotide substitutions. Our comparative genomic study clearly showed that there was tremendous bias in amino acid substitutions in all orthologous ORFs. Analysis of the direction of the amino acid substitutions suggested that three substitutions are important for the stability of the C. efficiens proteins: from lysine to arginine, serine to alanine, and serine to threonine. Our results strongly suggest that the accumulation of these three types of amino acid substitutions correlates with the acquisition of thermostability and is responsible for the greater GC content of C. efficiens.
More than 100 bacterial genomes have already been sequenced (http://www.tigr.org/tdb/mdb/). Although many of these bacteria were pathogens or model organisms, some are of industrial interest (Nelson et al. 2000). Corynebacterium glutamicum is a well-known industrial strain widely used for the production of various amino acids by fermentation, such as glutamate and lysine. Corynebacterium efficiens is a Gram-positive nonpathogenic bacterium previously known as Corynebacterium thermoaminogenes. This strain has recently been shown to be a near relative of C. glutamicum and Corynebacterium callunae, both recognized as glutamic acid-producing species (Fudou et al. 2002). The optimal temperature for glutamate production by C. glutamicum is around 30°C, and this micro-organism can neither grow nor produce glutamate at 40°C or above. On the other hand, C. efficiens can grow and produce glutamate above 40°C. Some comparative experimental results are summarized in Table 1, showing clearly distinct upper temperature limits for growth (E. Kimura, S. Hirano, Y. Matsuzaki, G. Nonaka, H. Itaya, N. Akiyoshi, Y. Kawahara, S. Sugimoto, in prep.). The relative glutamate productivity of C. glutamicum by the biotin limitation method (Kimura et al. 1999) was shown to be severely reduced at 37°C, whereas that of C. efficiens was unaffected (H. Itaya, unpubl.). The thermostability of C. efficiens is a useful trait from an industrial viewpoint as it reduces the considerable cost of cooling needed to dissipate the heat generated during glutamate fermentation.
Table 1.
Summary of Characteristics of Corynebacteria
| C. efficiens | C. glutamicum | C. diphtheriae | |
|---|---|---|---|
| Upper temperature limit for growth (°C) | 45 | 40 | — |
| Glutamate production at 32°C (%)a | 80 | 100 | — |
| Glutamate production at 37°C (%) | 78 | 40 | — |
| Genome size (bp) | 3,147,090 | 3,309,401 | 2,488,635 |
| GC content (%) | 63.4 | 53.8 | 53.5 |
| Number of predicted genes | 2950 | 3099 | — |
Glutamate production in typical experiments using the biotin limitation method as a percent of the production by C. glutamicum at 32°C
Many physiologic, biochemical, and genetic analyses of C. glutamicum have been performed, and the genome sequence of C. glutamicum ATCC 13032 determined by Kyowa Hakko is in the public domain. The finding that C. efficiens can grow at a temperature 10°C higher than C. glutamicum and that its guanine plus cytosine (GC) content is 5% higher (Fudou et al. 2002), provides an attractive topic for study by comparative genomics. Experimental data on the thermal stabilities of 11 metabolic enzymes of the two species suggest that many C. efficiens proteins are more thermostable than those of C. glutamicum (E. Kimura, S. Hirano, Y. Matsuzaki, G. Nonaka, H. Itaya, N. Akiyoshi, Y. Kawahara, S. Sugimoto, in prep.). Furthermore, the two species are closely related phylogenetically, despite the above differences in physiologic characteristics. The genome sequence of Corynebacterium diphtheriae, a well-known pathogenic strain, has been determined by the Sanger Institute. Because C. diphtheriae does not belong to the glutamic acid-producing species, it is useful as a phylogenetic outgroup.
Hyperthermophilic enzymes have been extensively studied (Vieille and Zeikus 2001) and genome-wide comparisons between thermophilic archaea and mesophilic bacteria have been reported (Chakravarty and Varadarajan 2000; Kreil and Ouzounis 2001). Thermophilic enzymes are indeed useful for industrial purposes, and many examples of protein thermostabilization have been reported (Vieille and Zeikus 2001). However, the genome-wide amino acid substitutions responsible for the thermal stability of an organism have not been studied. The genome sequences of C. efficiens and C. glutamicum permit us to compare mesophiles with different optimal temperatures for growth. The greatest advantage is the opportunity to compare more than 1000 orthologous genes one by one, because they are so closely related. We have tried here to elucidate the mechanism underlying the thermal stability of C. efficiens by a genome-wide comparison of amino acid substitutions, in the hope that such a comparison may indicate a general method for protein thermostabilization.
RESULTS
Genome Sequence and GC Content
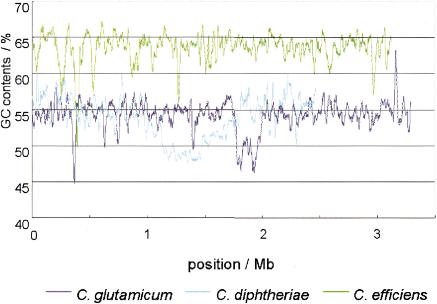
Sequencing was performed by the whole genome shotgun method. Genome size, GC content, and the numbers of predicted genes used in this study are shown in Table 1 for C. efficiens, C. glutamicum, and C. diphtheriae. To gain an overview of Corynebacterial genome structure, we compared the GC content (Fig. 1), GC skew (Fig. 2), and gene order (Suppl. Fig. 2). C. glutamicum had a GC content between 50% and 60% in most regions of the chromosome, and its average GC content was 53.8%. On the other hand, the average GC content of C. efficiens was 63.4%, higher than C. glutamicum over the entire chromosome (Fig. 1). This tendency was also clearly displayed by the predicted ORFs (Suppl. Fig. 1A for C. efficiens; Suppl. Fig. 1B for C. glutamicum). Although the GC content of C. efficiens had previously been reported to be 5% higher than that of C. glutamicum (Fudou et al. 2002), the whole genome analysis revealed that the true figure is 10%.
Figure 1.
GC content of three corynebacterial genomes. Window analysis of GC content performed at 20-kb window size and 1-kb step size. Linear representation of GC content along the chromosome. Green, C. efficiens; dark blue, C. glutamicum; light blue, C. diphtheriae.
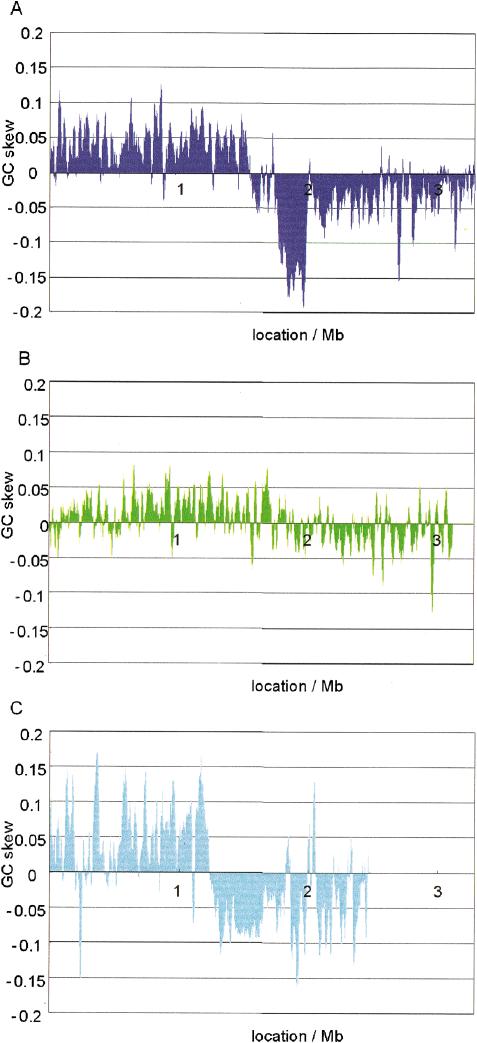
Figure 2.
C. glutamicum GC skew of the three Corynebacteria. Window analysis of GC skew was performed at 20-kb window size and 1-kb step size. C. glutamicum (A), C. efficiens (B), and C. diphtheriae (C).
C. diphtheriae was used as an outgroup of the glutamic acid-producing strains. C. diphtheriae showed a window analysis profile of GC content more similar to C. glutamicum than to C. efficiens (Fig. 1, Suppl. Fig. 1C). This suggests that the ancestral genome structure of Corynebacteria may be closer to that of C. glutamicum than to that of C. efficiens. The GC skew profile supported this hypothesis: whereas C. glutamicum (Fig. 2A) and the outgroup, C. diphtheriae (Fig. 2C), showed clear GC skew profiles with an inversion point that corresponds to the replication terminus (McLean et al. 1998), C. efficiens gave an irregular GC skew profile (Fig. 2B). In addition, gene order was well conserved (Suppl. Fig. 2), while the GC content of C. efficiens was higher than that of C. glutamicum and C. diphtheriae (Fig. 1). We therefore inferred that the genome structure of the common ancestor was more similar to that of C. glutamicum and C. diphtheriae than to C. efficiens, so that C. efficiens must have acquired its thermostability by an increase of GC content after divergence from its sister species.
There was a region of low GC content between 1.8 Mb and 2.0 Mb in C. glutamicum (Suppl. Fig. 1B) and another from 1.2 Mb to 1.7 Mb in C. diphtheriae (Suppl. Fig. 1C). In these regions the values of GC skew in C. glutamicum were under -0.1, whereas in C. diphtheriae, they were above -0.1 (Fig. 2C), pointing to a difference between the two regions. In the comparison of orthologous gene order, prominent gaps between C. glutamicum and C. efficiens (Suppl. Fig. 2A) and C. glutamicum and C. diphtheriae (Suppl. Fig. 2B) corresponded to the region of low GC content of C. glutamicum. We did not find a similar large gap corresponding to the low GC content region of C. diphtheriae (Suppl. Fig. 2B,C). These results suggest that the low GC content region in C. glutamicum was acquired by horizontal gene transfer, and that we found transposase homologs in this region (data not shown). There may be a tendency towards lower GC content in that region in C. diphtheriae. Thus, despite the conserved gene order, there is massive variability in genomic GC content among Corynebacteria that may be a strong driving force for evolution.
Codon Usage and Amino Acid Composition of ORFs
The numbers of ORFs extracted by the Glimmer program as a function of GC content were analyzed (Suppl. Fig. 3). The peak of ORF number in C. efficiens shifts to higher GC than in C. glutamicum. The difference in average GC content between the two micro-organisms is directly reflected in the GC content of the ORFs. To investigate the difference in GC content of the ORFs, codon usage and nucleotide substitutions were examined in the genecoding regions.
Figure 3.
Proposed hydrophobic interaction in C. glutamicum diaminopimelate dehydrogenase. The residue 113Ala is substituted to Ser in C. efficiens. This substitution may destroy hydrophobic interaction and destabilize the protein structure. Flat arrows represent β-sheet.
The codon usage of C. efficiens genes was much more biased than that of C. glutamicum (Table 2). For example, CTC (Leu) and CTG (Leu) were used more frequently in C. efficiens, although the two species had almost the same total number of Leu codons. The most frequently used Asp and Ala codons, GAC (Asp) and GCC (Ala) in C. efficiens differed from those in C. glutamicum, GAT (Asp), and GCA (Ala), respectively. Thirteen codons are rarely used in the highly expressed genes of C. glutamicum (Malumbres et al. 1993). The number of codons per 1000 bases (fraction values) are below 10 in C. glutamicum, whereas the number of GGG (Gly) and CGG (Arg) codons exceeds 10 in C. efficiens (Table 2).
Table 2.
Codon Usage in C. glutamicum and C. efficiens
|
Fraction
valueb
|
||||
|---|---|---|---|---|
| Codon | Amino acid | Rare codona | C. glutamicum | C. efficiens |
| GCA | Ala | 30.66 | 12.29 | |
| GCC | Ala | 27.18 | 54.41 | |
| GCG | Ala | 23.15 | 28.82 | |
| GCT | Ala | 24.96 | 8.75 | |
| TGC | Cys | 4.87 | 6.33 | |
| TGT | Cys | 2.66 | 2.81 | |
| GAC | Asp | 26.11 | 34.68 | |
| GAT | Asp | 32.89 | 29.26 | |
| GAA | Glu | 35.50 | 18.10 | |
| GAG | Glu | 27.41 | 42.27 | |
| TTC | Phe | 22.87 | 26.53 | |
| TTT | Phe | 13.78 | 3.82 | |
| GGA | Gly | 15.43 | 12.47 | |
| GGC | Gly | 33.34 | 37.00 | |
| GGG | Gly | + | 6.97 | 18.19 |
| GGT | Gly | 24.44 | 31.09 | |
| CAC | His | 14.52 | 19.77 | |
| CAT | His | 7.22 | 8.68 | |
| ATA | Ile | + | 2.05 | 1.89 |
| ATC | Ile | 33.55 | 42.38 | |
| ATT | Ile | 21.48 | 5.21 | |
| AAA | Lys | 14.27 | 5.58 | |
| AAG | Lys | 20.75 | 17.38 | |
| CTA | Leu | + | 5.94 | 1.78 |
| CTC | Leu | 22.05 | 33.67 | |
| CTG | Leu | 27.38 | 47.63 | |
| CTT | Leu | 17.01 | 8.15 | |
| TTA | Leu | + | 5.31 | 1.38 |
| TTG | Leu | 19.99 | 6.08 | |
| ATG | Met | 21.90 | 19.52 | |
| AAC | Asn | 21.94 | 18.40 | |
| AAT | Asn | 11.29 | 6.26 | |
| CCA | Pro | 16.80 | 6.76 | |
| CCC | Pro | 9.83 | 20.98 | |
| CCG | Pro | 10.38 | 21.56 | |
| CCT | Pro | 11.26 | 3.94 | |
| CAA | Gln | 13.23 | 3.33 | |
| CAG | Gln | 20.76 | 31.64 | |
| AGA | Arg | + | 2.71 | 1.94 |
| AGG | Arg | + | 3.79 | 4.73 |
| CGA | Arg | + | 6.73 | 4.07 |
| CGC | Arg | 24.54 | 26.91 | |
| CGG | Arg | + | 5.13 | 16.99 |
| CGT | Arg | 13.50 | 12.90 | |
| AGC | Ser | 10.89 | 8.91 | |
| AGT | Ser | + | 5.28 | 3.76 |
| TCA | Ser | + | 8.43 | 4.01 |
| TCC | Ser | 21.01 | 25.23 | |
| TCG | Ser | + | 7.71 | 7.91 |
| TCT | Ser | 10.99 | 2.43 | |
| ACA | Thr | + | 7.90 | 4.23 |
| ACC | Thr | 32.14 | 46.33 | |
| ACG | Thr | 8.96 | 10.18 | |
| ACT | Thr | 12.51 | 3.22 | |
| GTA | Val | + | 8.41 | 5.23 |
| GTC | Val | 22.22 | 34.05 | |
| GTG | Val | 29.01 | 37.03 | |
| GTT | Val | 21.03 | 9.58 | |
| TGG | Trp | 14.13 | 13.21 | |
| TAC | Tyr | 14.40 | 13.35 | |
| TAT | Tyr | 7.45 | 5.00 | |
Rare codons are adapted from Malumbres et al. (1993)
Fraction value represents the number of codons per 1000 bases
Among the 10 most frequently used codons in C. glutamicum, seven have GC in the third position, whereas all 10 codons do so in C. efficiens. Of 10 rarely used codons, none contains GC in the third position in C. efficiens against three in C. glutamicum. It should also be noted that only the fraction value of the GGT (Gly) codon, among the codons with AT in the third position, was higher by more than six points in C. efficiens than in C. glutamicum. These findings seem to reflect clearly the higher GC content of C. efficiens.
The amino acid composition of the protein coding regions is analyzed in Table 3. Lys, Asn, Ser, Ile, and Phe are more frequently used in C. glutamicum than in C. efficiens. The increased usage of Arg, Asp, Gly, His, Pro, and Val in C. efficiens is shown to be statistically significant by the z-test. The high utilization frequency of Asn, Ile, Phe, and Lys in C. glutamicum agrees with the tendency of these amino acids to increase with decreasing GC content reported in a statistical analysis of the complete genomes of six thermophilic archaea, two thermophilic bacteria, 17 mesophilic bacteria, and two eukaryotic species (Kreil and Ouzounis 2001). On the other hand, the high frequency of Gly and Arg in C. efficiens concurs with the view that these amino acid residues increase parallel to rises in GC content.
Table 3.
Amino Acid Composition of Protein Coding Regions
|
Number
|
Ratio
(%)a
|
||||
|---|---|---|---|---|---|
| Amino acid | C. glutamicum | C. efficiens | C. glutamicum | C. efficiens | Pb |
| Ala | 107,484 | 122,084 | 10.58 | 10.44 | |
| Arg | 57,210 | 79,096 | 5.63 | 6.76 | *** |
| Asn | 33,710 | 28,875 | 3.32 | 2.47 | *** |
| Asp | 59,858 | 74,866 | 5.89 | 6.40 | *** |
| Cys | 7643 | 10,706 | 0.75 | 0.92 | |
| Gln | 34,477 | 40,943 | 3.39 | 3.50 | |
| Glu | 63,816 | 70,689 | 6.28 | 6.04 | * |
| Gly | 81,344 | 115,628 | 8.01 | 9.88 | *** |
| His | 22,050 | 33,308 | 2.17 | 2.85 | *** |
| Ile | 57,899 | 57,934 | 5.70 | 4.95 | *** |
| Leu | 99,098 | 115,556 | 9.76 | 9.88 | |
| Lys | 35,527 | 26,882 | 3.50 | 2.30 | *** |
| Met | 22,217 | 22,860 | 2.19 | 1.95 | * |
| Phe | 37,182 | 35,530 | 3.66 | 3.04 | *** |
| Pro | 48,961 | 62,331 | 4.82 | 5.33 | *** |
| Ser | 65,246 | 61,183 | 6.42 | 5.23 | *** |
| Thr | 62,400 | 74,898 | 6.14 | 6.40 | * |
| Trp | 15,465 | 14,339 | 1.52 | 1.23 | * |
| Tyr | 22,164 | 21,488 | 2.18 | 1.84 | ** |
| Val | 81,846 | 100,565 | 8.06 | 8.60 | *** |
| Total | 1,105,597 | 1,169,761 | |||
The ratio is the percentage of the number of a given amino acid to the total number of amino acids
P is the significant difference level by z test
P < 0.05
P < 0.01
P < 0.001
Base Replacement and Amino Acid Substitution
The orthologous ORFs of C. glutamicum and C. efficiens were extracted and sorted according to their degree of identity. They were then divided into three groups, a group with identity of more than 95%, another with identity from 60% to 95%, and a third with identity under 60%. More than 95% of the genes belonging to the first group are ribosomal proteins, and we did not analyze these proteins because of their anticipated conservative nature. The third group, with identity under 60%, was also omitted, because of the large calculated p-distance value of 0.4 and the need to take account of backward and parallel mutations (Nei and Sudhir 2000). One thousand six hundered nineteen orthologous pairs of genes with identity from 60% to 95% (p-distance value 0.2) were used to examine position-specific mutations. Synonymous codon replacement was analyzed (data not shown), and among the 30 most frequent synonymous substitutions, 26 were changes in the third letter from AT in C. glutamicum to GC in C. efficiens. The only substitution that involved GC in C. glutamicum and AT in C. efficiens was from GGC (Gly) in C. glutamicum to GGT (Gly) in C. efficiens. Among the 30 most frequent nonsynonymous substitutions in C. efficiens, 27 increased GC content, and in 21 of these, GC was in the third position. Of these 21, three substitutions, from Lys to Arg (AAA to CGG, AAA to CGC, and AAG to CGC), involved changes in all three letters. The trend of nucleotide substitutions at each codon position in C. efficiens also involved an increase of GC content (data not shown).
The amino acid sequences of 1619 orthologous genes with identity from 60% to 95% were aligned using the pairwise alignment program, Stretcher (Fraczkiewicz and Braun 1998), and the amino acid substitutions obtained were placed in a matrix. By analyzing the differences between the matrix and the transposed matrix, the asymmetric mutations from C. glutamicum to C. efficiens were extracted (Table 4). The results of biased mutations in the two other categories (the groups with identity under 60% and over 95%) differed from those in Table 4 (data not shown). Some of the amino acid substitutions in this table have often been observed before, with Leu, Ile, Val, and Met replacing each other (Henikoff and Henikoff 1992). Because the fourth most frequent substitution, from Ile to Val, is commonly observed in situations unrelated to thermostabilization, the three most frequent substitutions (Lys to Arg, Ser to Thr, Ser to Ala) are the best candidates for stabilizing the proteins. Indeed, many studies have suggested that the Lys to Arg substitution affects thermal stability (Vieille and Zeikus 2001). If the evolutionary development of the thermal stability of proteins is responsible for the thermostability of C. efficiens itself, then the observed amino acid substitutions must be adaptive mutations leading to overall thermostability. In a separate study, the thermal stability of 13 pairs of enzymes on the Glu and Lys biosynthetic pathways in the two species were compared on the basis of the enzymatic activities remaining after heat treatment of crude extracts (E. Kimura et al., in prep.). In Table 5, the numbers of the three kinds of amino acid substitutions within the amino acid sequence are assigned points depending on their directions, and we compare the number of calculated points with the experimental results of enzyme thermal stability. Nine out of 13 enzymes, the thermostabilities of which had been measured, agree with the calculated points, three can not be determined, and only one does not coincide (Table 5). These results suggest that there is a significant correlation between the three kinds of amino acid substitutions and the thermal stability of proteins.
Table 4.
Biased Amino Acid Substitutions in the Orthologous Genes of C. glutamicum and C. efficiens
| C. glutamicum | C. efficiens | Forward | Reverse | Pointa | G + C changing by one-base substitution |
|---|---|---|---|---|---|
| Lys | Arg | 2855 | 664 | 1095.5 | AAA → AGA, AAG → AGG |
| Ser | Ala | 3378 | 2372 | 503 | TCA → GCA, TCC → GCC, TCG → GCG, TCT → GCT |
| Ser | Thr | 2623 | 1723 | 450 | |
| Ile | Val | 4332 | 3585 | 373.5 | ATA → GTA, ATC → GTC, ATT → GTT |
| Asn | Arg | 978 | 372 | 303 | |
| Gln | Glu | 1321 | 747 | 287 | |
| Ile | Leu | 2191 | 1642 | 274.5 | ATA → CTA, ATC → CTC, ATT → CTT |
| Ser | Gly | 1013 | 610 | 201.5 | AGC → GGC, AGT → GGT |
| Lys | Thr | 600 | 235 | 182.5 | AAA → ACA, AAG → ACG |
| Ala | Pro | 1019 | 656 | 181.5 |
Point is defined as the difference between the number of amino acid substitutions from C. glutamicum to C. efficiens and the number of substitutions in the opposite direction, divided by 2. All of the asymmetrical amino acid substitutions showed probability of obtaining the observed deviation from 50:50 by chance less than 0.001
Table 5.
Check of Predictions Against Actual Measurements
| Entry | Enzyme | Thermostable species | Point | Result |
|---|---|---|---|---|
| 1 | 2-Oxoglutarate dehydrogenase | C. efficiens | 0 | — |
| 2 | Glutamate dehydrogenase | C. efficiens | 1 | Yes |
| 3 | Isocitrate lyase | C. efficiens | 2 | Yes |
| 4 | Phosphofructokinase | C. efficiens | -3 | No |
| 5 | Fructose-1-phosphate kinase | C. efficiens | 5 | Yes |
| 6 | Isocitrate dehydrogenase | C. efficiens | 4 | Yes |
| 7 | Aconitase | C. efficiens | 0 | — |
| 8 | Phosphoenolpyruvate carboxylase | C. efficiens | 10 | Yes |
| 9 | Citrate synthase | C. efficiens | 3 | Yes |
| 10 | Aspartate kinase | C. glutamicum | -1 | Yes |
| 11 | Dihydrodipicolinate synthase | C. efficiens | 0 | — |
| 12 | Diaminopimelate dehydrogenase | C. glutamicum | -2 | Yes |
| 13 | Diaminopimelate decarboxylase | C. efficiens | 2 | Yes |
Point is defined as the difference between the sum of the three kinds of substitutions from C. glutamicum to C. efficiens (Lys to Arg, Ser to Ala and Ser to Thr) and the sum of the reverse substitutions (Point = {number of (Lys → Arg + Ser → Ala + Ser → Thr)} - {number of (Arg → Lys + Ala → Ser + Thr → Ser)})
Results are indicated by (1) Yes: when the enzyme from C. efficiens was more thermostable and the point is positive, or when the enzyme from C. glutamicum was more thermostable and point is negative. (2) —: when the point value was zero. (3) No: all other cases
DISCUSSION
There is controversy over whether the first life forms were hyperthermophiles (Woese 1987; Pace 1991; Nisbet and Fowler 1996; Yamagishi et al. 1998) or not (Miller and Lazcano 1995; Forterre 1996; Galtier et al. 1999). As far as we know, among the species belonging to the genus Corynebacterium, C. efficiens can grow at the highest temperature, and is unique in its ability to produce glutamate above 40°C. The main point of interest in relation to the above controversy is whether C. efficiens acquired the ability to grow at higher temperature, or whether C. glutamicum lost it. On the basis of GC content and GC skew analyses, we concluded that C. glutamicum is closer to the common ancestor of the glutamic acid-producing strains, and therefore that C. efficiens has acquired its thermostability and higher GC content during the course of evolution. To understand the basis of this thermostability, we compared the C. efficiens and C. glutamicum genomes.
Studies of protein thermostability using genome sequences have generally compared hyperthermophiles or thermophiles, and mesophiles (Chakravarty and Varadarajan 2000; Kreil and Ouzounis 2001). In such cases, the differences in growth temperature are clear, but the amino acid residues do not correspond one to one because thermophiles and mesophiles are not close phylogenetically. In this report, we have compared two mesophiles with different optimal temperatures for growth, and were able to make a statistical comparison of amino acid residues one by one because of the close phylogenetic relationship. Among asymmetrical amino acid substitutions between C. glutamicum and C. efficiens, the substitution of Lys by Arg was the most frequent (Table 4). This substitution is known to contribute to protein stability. The mechanism of thermostabilization is thought to depend on the resonance stabilisation effect of Arg (Vieille and Zeikus 2001). Thus, Arg is assumed to contribute to protein thermostability because it maintains ion pairs more easily. Nevertheless, the Arg/Lys ratios, 2.94 in C. efficiens and 1.61 in C. glutamicum, are larger than the 2.19 ratio of Aeropyurum pernix, which has the highest Arg/Lys ratio of the hyperthermophiles and a GC content of 53.6% (Kreil and Ouzounis 2001). Thus, the substitutions from lysine in C. glutamicum to Arg in C. efficiens appear to result from the increase of GC content and constitute the basis of protein thermostabilization.
With regard to the substitutions from Ser in C. glutamicum to Ala or Thr in C. efficiens, we consider that Ala and Thr can strengthen hydrophobic interaction inside proteins, because Ala and Thr are more hydrophobic in the environment of a protein than Ser (Taylor 1986). McDonald et al. (1999) have analyzed the asymmetric amino acid substitution patterns in 229 genes of the bacterial genus Bacillus and 99 genes of the archaeal genus Methanococcus. The differences in GC content between Bacillus species are similar (B. stearothermophilus 52% versus B. subtilis 43.5%) to the difference between C. efficiens and C. glutamicum, and the asymmetrical amino acid substitution patterns found in Bacillus are very similar. However, the analysis of Bacillus and other works were based on far fewer genes than our analysis and did not confirm orthologous relationships (Haney et al. 1999; McDonald 2001). The two most frequent substitutions found in Bacillus were the same as in our analysis (Lys to Arg and Ser to Thr), but the Ser to Ala substitution found in genus Corynebacterium was less evident in Bacillus. Nevertheless, Wintrode et al. (2001) have reported substitutions from serine to various amino acids in a thermostable subtilisin made by directed evolution, and their findings suggest that mutation from Ser to Ala or Thr may be one of the effective ways to generate thermostable proteins.
The X-ray structure of the diaminopimelate dehydrogenase (Ddh) of C. glutamicum, one of the enzymes in our analysis, has been determined (Cirilli et al. 2000). Interestingly, C. glutamicum Ddh was found to be more stable than that of C. efficiens, and the mutations responsible are of great interest. We have tried to identify the most effective of the three amino acid substitutions responsible for the thermostability of C. glutamicum Ddh over C. efficiens. The amino acid substitution that acts to lower thermostability of C. glutamicum Ddh is most probably that of 113Ala, which is replaced by Ser in C. efficiens. The Ser residue tends to impair hydrophobic interaction between β-strands, whereas the Ala can be effective in bridging strands (Fig. 3). It is likely that some but not all of the observed substitutions affect protein stability. To identify those that do, actual amino acid substitution experiments and measurements of protein thermostability are needed. Recently, many protein crystal structures have been determined and structure-modeling technology is developing rapidly, so that we may soon be able to predict which mutations among the proposed substitutions increase stability.
An interesting question concerns which event occurred first in evolution: the increase in genomic GC content, or the adaptive amino acid substitutions. Due to the close phylogenetic relationship of C. efficiens and C. glutamicum, this study was focused on only one letter substitutions, and the three substitutions that are not caused by the replacement of the third letter of codons. The one-base substitutions from Lys (AAA and AAG) to Arg (AGA and AGG) and from Ser (TCA, TCC, TCG, and TCT) to Ala (GCA, GCC, GCG, and GCT) are compatible with the increase of GC content in C. efficiens. However, the possible replacements from Ser (TCA, TCC, TCG, and TCT) to Thr (ACA, ACC, ACG, and ACT) are not explained by the GC increase. Thus, the increase in GC content alone cannot predict all three amino acid substitutions suggested to be involved in thermostabilization by the results of the statistical analysis.
A comparative analysis was made of two closely related organisms, C. efficiens and C. glutamicum, which differ in their optimal growth temperatures. C. efficiens has an average GC content that is 10% higher than that of C. glutamicum, with which it shares a conserved gene order. The codon usage and amino acid frequency in predicted coding regions of C. efficiens reflected the increased GC content. From genome structure analyses, C. glutamicum was estimated to be closer to the common ancestor of Corynebacteria. We concluded that C. efficiens would have acquired the thermostablity through the specific amino acid substitutions, which are represented by the increase in GC content that has occurred during the course of evolution. The analysis of amino acid substitutions in the orthologous genes revealed the three asymmetric amino acid substitutions from C. glutamicum to C. efficiens that are involved in thermostabilization in C. efficiens. The involvement of the three kinds of amino acid substitutions identified in this study will be tested experimentally for their roles in achieving protein thermostability.
METHODS
Genome Sequencing
The genome of C. efficiens JCM 44549 (strain YS-314) was sequenced by the shotgun method (Fleischmann et al. 1995). The end sequences from two pUC118 shotgun libraries, one containing short fragments (0.8–1.2 kb) and the other longer fragments (2.0–2.5 kb), were collected. Sequencing reactions were performed on 377 DNA sequencers using dye primer and dye terminator cycle sequencing kits, and M13 universal primers. The data were processed with the Phred/Phrap/Consed package (http://www.phrap.org/) and the assembled sequences, after being split into 30 kb segments, were reassembled and edited by Sequencher (GeneCodes). The details of genome sequencing will be described (Y. Kawarabayasi, Y. Hino, J. Yamazaki, H. Horikawa, H. Nakazawa, T. Tanaka, M. Nishimura, Y. Nishio, H. Shimizu, A. Yamagishi, et al., in prep.). Prediction of protein coding regions was performed with the Glimmer 2.0 program under default conditions (Delcher et al. 1999). The sequence, 5′-AAAGAGG-3′, was used as Shine-Dalgarno sequence (Amador et al. 1999). The genome sequence itself was used for training.
Informatics
The genome sequences of C. glutamicum ATCC 13032 determined by Kyowa Hakko (European Patent No. 1108790, BA000036 in DDBJ/EMBL/GenBank database) and of C. diphtheriae NCTC13129 by the Sanger Institute (http://www.sanger.ac.uk/Projects/C_diphtheriae/), were used as references. The BLASTP program was used (Altschul et al. 1997) to determine orthologous corynebacterial pairs. Codon usage was examined using cusp programs (http://www.uk.embnet.org/Software/EMBOSS/Apps/cusp.html). The GC contents of ORFs were examined using geecee programs (http://www.uk.embnet.org/Software/EMBOSS/Apps/geecee.html). Window analyses for GC content ([G + C]/[G + A + T + C]) and GC skew ([G - C)/[C + G]) were performed by the windowgc.pl script (Y. Nakamura, unpubl.). Stretcher (Myers and Miller 1988) was used for pairwise alignment. Orthologous genes are defined as the best pair of homologs in comparisons between two organisms (Tatusov et al. 1997). GETAREA 1.1 was used to calculate solvent accessible surface areas from Protein Data Bank (PDB) files (Fraczkiewicz and Braun 1998). For calculation of various interactions between amino acid residues in a protein, LPCCSU server was employed (Sobolev et al. 1999).
Acknowledgments
The authors are grateful to Dr. S. Mano for mathematical suggestions, Dr. Y. Niimura for GC content analysis, H. Itaya for providing the data on glutamate production, and Dr. S.J. Van Dien for critical reading of the manuscript. The authors also acknowledge the contributions of Mr. T. Osumi and K. Totsuka, and Drs. O. Tosaka, T. Utagawa, and T. Nishiyama.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1285603.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to DDBJ/EMBL/GenBank under accession nos. BA000035, and AP005214–AP005224.]
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amador, E., Castro, J.M., Correia, A., and Martin, J.F. 1999. Structure and organization of the rrnD operon of “Brevibacterium lactofermentum”: Analysis of the 16S rRNA gene. Microbiology 145: 915-924. [DOI] [PubMed] [Google Scholar]
- Chakravarty, S. and Varadarajan, R. 2000. Elucidation of determinants of protein stability through genome sequence analysis. FEBS Lett. 470: 65-69. [DOI] [PubMed] [Google Scholar]
- Cirilli, M., Scapin, G., Sutherland, A., Vederas, J.C., and Blanchard, J.S. 2000. The three-dimensional structure of the ternary complex of Corynebacterium glutamicum diaminopimelate dehydrogenase-NADPH-L-2-amino-6-methylene-pimelate. Protein Sci. 9: 2034-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher, A.L., Harmon, D., Kasif, S., White, O., and Salzberg, S.L. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27: 4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann, R.D., Adams, M.D., White, O., Clayton, R.A., Kirkness, E.F., Kerlavage, A.R., Bult, C.J., Tomb, J.F., Dougherty, B.A., Merrick, J.M., et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269: 496-512. [DOI] [PubMed] [Google Scholar]
- Forterre, P. 1996. A hot topic: The origin of hyperthermophiles. Cell 85: 789-792. [DOI] [PubMed] [Google Scholar]
- Fraczkiewicz, R. and Braun, W. 1998. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comp. Chem. 19: 319-333. [Google Scholar]
- Fudou, R., Jojima, Y., Seto, A., Yamada, K., Kimura, E., Nakamatsu, T., Hiraishi, A., and Yamanaka, S. 2002. Corynebacterium efficiens sp. nov., a glutamic-acid-producing species from soil and vegetables. Int. J. Syst. Evol. Microbiol. 52: 1127-1131. [DOI] [PubMed] [Google Scholar]
- Galtier, N., Taurasse, N., and Gouy, M. 1999. A nonhyperthermophilic common ancestor to extant life forms. Science 283: 220-221. [DOI] [PubMed] [Google Scholar]
- Haney, P.J., Badger, J.H., Buldak, G.L., Reich, C.I., Woese, C.R., and Olsen, G.J. 1999. Thermal adaptation analyzed by comparison of protein sequences from mesophilic and extremely thermophilic Methanococcus species. Proc. Natl. Acad. Sci. 96: 3578-3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S. and Henikoff, J.G. 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. 89: 10915-10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, E., Yagoshi, Y., Kawahara, Y., Ohsumi, T., Nakamatsu, T., and Tokuda, H. 1999. Corynebacterium glutamicum triggered by a decrease in the level of a complex comprising DtsR and a biotin-containing subunit. Biosci. Biotechnol. Biochem. 63: 1274-1278. [DOI] [PubMed] [Google Scholar]
- Kreil, D.P. and Ouzounis, C.A. 2001. Identification of thermophilic species by the amino acid compositions deduced from their genomes. Nucleic Acids Res. 29: 1608-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres, M., Gil, J.A., and Martin, J.F. 1993. Codon preference in corynebacteria. Gene 134: 15-24. [DOI] [PubMed] [Google Scholar]
- McDonald, J.H. 2001. Patterns of temperature adaptation in proteins form the bacteria Deinococcus radiodurans and Thermus thermophilus. Mol. Biol. Evol. 18: 741-749. [DOI] [PubMed] [Google Scholar]
- McDonald, J.H., Grasso, A.M., and Rejto, L.K. 1999. Patterns of temperature adaptation in proteins from Methanococcus and Bacillus. Mol. Biol. Evol. 16: 1785-1790. [DOI] [PubMed] [Google Scholar]
- McLean, M.J., Wolfe, K.H., and Devine, K.M. 1998. Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. J. Mol. Evol. 47: 691-696. [DOI] [PubMed] [Google Scholar]
- Miller, S.L. and Lazcano, A. 1995. The origin of life—Did it occur at high temperatures? J. Mol. Evol. 41: 689-692. [DOI] [PubMed] [Google Scholar]
- Myers, E.W. and Miller, W. 1988. Optimal alignments in linear space. Comput. Appl. Biosci. 4: 11-17. [DOI] [PubMed] [Google Scholar]
- Nei, M. and Sudhir, K. 2000. Molecular evolution and phylogenetics, pp. 17-31. Oxford University Press, New York.
- Nelson, K.E., Paulsen, I.T., Heidelberg, J.F., and Fraser, C.M. 2000. Status of genome projects for nonpathogenic bacteria and archaea. Nat. Biotechnol. 18: 1049-1054. [DOI] [PubMed] [Google Scholar]
- Nisbet, E.G. and Fowler, C.M.R. 1996. Some linked it hot. Nature 382: 404-405. [Google Scholar]
- Pace, N.R. 1991. Origin of life—Facing up to the physical setting. Cell 65: 531-533. [DOI] [PubMed] [Google Scholar]
- Sobolev, V., Sorokine, A., Prilusky, J., Abola, E.E., and Edelman, M. 1999. Automated analysis of interatomic contacts in proteins. Bioinformatics 15: 327-332. [DOI] [PubMed] [Google Scholar]
- Tatusov, R.L., Koonin, E.V., and Lipman, D.J. 1997. A Genomic perspective on protein families. Science 278: 631-637. [DOI] [PubMed] [Google Scholar]
- Taylor, W.R. 1986. The classification of amino acid conservation. J. Theor. Biol. 119: 205-218. [DOI] [PubMed] [Google Scholar]
- Vieille, C. and Zeikus, G.J. 2001. Hyperthermophilic enzymes: Sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 65: 1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintrode, P.L., Miyazaki, K., and Arnold, F.H. 2001. Patterns of adaptation in a laboratory evolved thermophilic enzyme. Biochim. Biophys. Acta 1549: 1-8. [DOI] [PubMed] [Google Scholar]
- Woese, C.R. 1987. Bacterial evolution. Microbiol. Rev. 51: 221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi, A., Kon, T., Takahashi, G., and Oshima, T. 1998. From the common ancestor of all living organisms to protoeukaryotic cell. In: The keys to molecular evolution and the origin of life? (eds. J. Wiegel, and M.W.W. Adams), pp. 287-295. Taylor & Francis, London.
WEB SITE REFERENCES
- http://www.tigr.org/tdb/mdb/; TIGR Microbial Database home page.
- http://www.phrap.org/; The Phred/Phrap/Consed System home page.
- http://www.sanger.ac.uk/Projects/C_diphtheriae/; The genome project of C. diphtheriae.
- http://www.uk.embnet.org/Software/EMBOSS/Apps/cusp.html; The manual of cusp.
- http://www.uk.embnet.org/Software/EMBOSS/Apps/geecee.html; The manual of geecee.