Abstract
Purpose
Persistent inflammation and impaired neovascularization in type 2 diabetes mellitus (T2DM) patients may lead to development of macro- and microvascular complications. Diabetic retinopathy (DR) is one of the secondary microvascular complications of T2DM. Improper activation of the innate immune system may be an important contributor in the pathophysiology of DR. Toll-like receptor 4 (TLR4) is an important mediator of innate immunity, and genetic alterations in TLR4 support inflammation in the hyperglycemic condition. The present work was designed to investigate whether the TLR4 single nucleotide polymorphisms (SNPs) rs4986790, rs4986791, rs10759931, rs1927911, and rs1927914 are associated with DR in a north Indian population.
Methods
The study group of 698 individuals (128 DR, 250 T2DM, 320 controls) was genotyped by PCR-RFLP. Haplotype and linkage disequilibrium between SNPs were determined using Haploview software.
Results
Combined risk genotypes of TLR4 SNPs rs10759931 (odds ratio [OR] 1.50, p = 0.05) and rs1927914 (OR 1.48, p = 0.05) were found to be significantly associated with pathogenesis of DR. A total of 14 haplotypes with frequency >1% were obtained using Haploview software. Haplotypes ACATC (37.5%) and ACATT (14.8%) were the two most common haplotypes obtained.
Conclusions
Results of the present case-control study that included 698 north Indian subjects suggested that TLR4 SNPs rs10759931 and rs1927914 modulate the risk of DR in T2DM cases. Association analysis using haplotypes showed none of the haplotypes were associated with either susceptibility or resistance to DR in a north Indian population.
Introduction
Type 2 diabetes mellitus (T2DM) is a multifactorial disorder characterized by elevation of blood glucose levels due to peripheral insulin resistance. Chronic hyperglycemia [1], over-production of reactive oxygen species, persistent low-grade inflammation and impaired neovascularization [2] are the characteristic features of T2DM that are critically involved in the development of microvascular complications in diabetic patients. Diabetic retinopathy (DR) is one such microvascular complication. DR has an overall prevalence of 22–37% in individuals with known diabetes, and it may lead to blindness due to continuous blood leakage from retinal pericytes and endothelial cells if left untreated [3]. A chronic low-grade subclinical inflammation is an important contributor in the pathogenesis of DR [4]. In early stages of DR, loss of microvascular cells creates a hypoxic condition that in turn induces the expression of angiogenic factors [5]. This ischemia-induced retinal neovascularization along with the outgrowth of other retinal membranes gives rise to advanced proliferative diabetic retinopathy (PDR) characterized by loss of vision due to hemorrhage and/or tractional retinal detachment [6]. Other systemic abnormalities common in T2DM, such as hyper-reactivity of platelets, greater prevalence of adhesion molecules, hypercoagulability, and lesser fibrinolysis activity, also contribute to the pathophysiology of DR [7,8].
Several studies have linked a genetic abnormality in the mediators of the innate immune response to the secondary complications of T2DM [9,10]. Toll-like receptors (TLRs), a pattern recognition receptor (PRR) family, are a group of transmembrane receptors and are involved in regulating innate immunity by pathogen recognition [11]. Genetic variations within genes encoding these PRRs have been shown to be involved in several inflammatory diseases [12]. TLR4 is an important member of the TLR family, and its expression has been reported on a variety of cell types, including cardiomyocytes, macrophages, airway epithelium, endothelial, and smooth muscle cells [13]. TLR4 as a PRR has been shown to predominantly interact with microorganism-derived lipopolysaccharides, but other interacting molecules, such as heat shock proteins 60 and 70, fibrinogen, and fibronectin, are also known [14]. After ligand binding, TLR4 takes part in the activation of a pro-inflammatory response by the activation of the nuclear factor-κB pathway. Any deregulation of TLR4 signaling due to the single nucleotide polymorphisms (SNPs) in the extracellular domain of TLR4 may alter the ligand binding capacity and hence disturb the pro- and anti-inflammatory cytokines. Our group has recently shown five TLR4 SNPs, viz rs4986790, rs4986791, rs10759931, rs1927911, and rs1927914, to be associated with one of the secondary complications of T2DM [10]. Two of these SNPs, rs4986790 and rs4986791, are located at exon three of TLR4 in tight linkage disequilibrium. These SNPs have been shown to modulate TLR4 effector functions either by interfering with the binding capacity of TLR4 with its ligands or by controlling the extracellular deposition of functional TLR4 [15,16]. Three other SNPs of TLR4, rs10759931, rs1927911, and rs1927914, have been also reported to be associated with inflammatory diseases, including cancer [10,17].
Buraczynska et al. studied the association of rs4986790 and rs4986791 SNPs with early onset of DR in a Polish population and found the G allele of rs4986790 SNP to be associated with DR [18]. Zareparsi et al. analyzed the association of these SNPs with age-related macular degeneration, another progressive eye disease leading to loss of vision in elderly patients [19]. The present study was designed to confirm the association of rs4986790 and rs4986791 with DR in a north Indian cohort. Moreover, we checked the association of three other SNPs rs10759931, rs1927911, and rs1927914 with the pathogenesis and progression of DR in a north Indian population for the first time. Since the five studied SNPs belong to the same gene and are located near to each other, there is a possibility of linkage disequilibrium (LD) among them. We checked this possibility by using haplotype analysis.
Methods
Subjects
In this case-control study, a total of 698 individuals, including 378 T2DM patients (M:F = 244:134; Mean age = 51.24±9.62) and 320 age-matched controls (M:F= 182:138; Mean age = 50.13±7.35 years), were consecutively enrolled between July 2010 and December 2013 (Table 1). The T2DM patients were subclassified as patients with diabetic retinopathy (128) and DM without retinopathy (250) as additional controls. Recruitment of patients was done by the outpatient department (OPD) clinics of the University Hospital, Institute of Medical Sciences, Banaras Hindu University, Varanasi, India. Diabetes was diagnosed according to World Health Organization criteria. Retinopathy was diagnosed according to the Early Treatment Diabetic Retinopathy Study criteria [20]. A total of 320 healthy controls were recruited from the general north Indian population residing in Varanasi. These controls had the same ethnicity, had controlled fasting or postprandial sugar levels, had no family history of T2DM, and were without any other inflammatory or chronic disease. Patients underwent a standardized clinical and laboratory evaluation. Each patient’s family history, habits (e.g., smoking, alcoholism), and disease were recorded through a questionnaire. The study was approved by the Institutional Human Ethics Committee of the Institute of Medical Sciences, Banaras Hindu University, Varanasi, India. Informed written consent was obtained from every participant.
Table 1. Biochemical and demographic details of subjects.
| Parameters | DR (n=128) | T2DM (n=250) | P value |
|---|---|---|---|
| Average age |
54.87±8.72 years |
51.87±10.57 years |
0.05 |
| Average BMI (kg/m2) |
25±5.1 |
23.32±4.68 |
0.13 |
| Average duration of type 2 diabetes (years) |
8.50±6.76 |
6.08±5.32 |
0.006 |
| Male |
86 (67.2%) |
158 (63.2%) |
0.19 |
| Female |
42 (32.8%) |
92 (53.2%) |
0.43 |
| Poor glycemic Control (according to WHO guidelines) |
102 (79.7%) |
133 (%) |
0.32 |
| Family history present |
49(38.3%) |
45 (18%) |
0.64 |
| Hypertension present (as per WHO guidelines) | 32 (25%) | 72 (28.8%) | 0.5 |
Biochemical and Demographic parameters of DR patients (n=128) and T2DM (n=250). Data are presented as mean ± SD or as number (percentage).
Genotyping of SNPs by PCR-restriction fragment length polymorphism
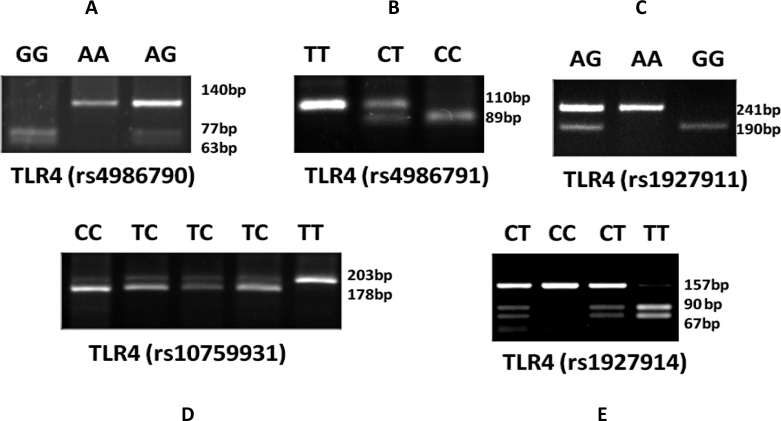
Genomic DNA was extracted from 5 ml peripheral blood collected aseptically from median cubital vein in a sodium heparinized syringe using a standard salting-out procedure as per our previous report [1]. The SNPs of the TLR4 gene, rs4986790, rs4986791, rs10759931, rs1927911, and rs1927914, were analyzed using PCR restriction fragment length polymorphism (PCR-RFLP) as reported previously [10]. The PCR reaction was set in a total reaction volume of 20 µl containing 50 ng genomic DNA, 5 pmol each primer (Sigma-Aldrich, St Louis, MO), 10 µl 2X DreamTaq PCR Master Mix (Fermentas, Hanover, MD) containing DreamTaq DNA Polymerase, 2X DreamTaq buffer, deoxynucleotide triphosphate (dNTPs) and 4 mmol/L MgCl2). PCR thermal cycling was done on Applied Biosystems Veriti 96-well Thermal Cycler. The sequence of the implicated primers used in the study and the methodology adopted is summarized in Table 2. The PCR set-up was composed of an initial denaturation step of 5 min followed by 35 cycles of 40 s at 94 °C, 45 s at 58 °C, and 40 s at 72 °C. This was followed by a final extension step of 10 min. The amplified products of SNPs rs4986790, rs4986791, rs10759931, rs1927911, and rs1927914 were digested with the restriction enzymes BccI, BslI, KpnI, StyI, and SphI, respectively. The digested products were separated on 3% agarose gel (Figure 1).
Table 2. PCR conditions for TLR4 SNPs genotyping.
| SNP ID | Primers (5’-3’) | Restriction enzyme used | Product size and genotypes |
|---|---|---|---|
|
rs4986790 |
F: CTGCTCTAGAGGGCCTGTG |
BccI |
140=AA
140, 77, 63=AG
77, 63=GG |
| R: TTCAATAGTCACACTCACCAG | |||
| rs4986791 |
F: CTACCAAGCCTTGAGTTTCTG |
BslI |
110=TT
110, 89, 22=TC
89, 22=CC |
| R: AAGCTCAGATCTAAATACT | |||
|
rs10759931 |
F: ATAACCTCAGTGGGCTCTGG |
KpnI |
241=AA
241, 190, 51=AG |
| R: ATGTTCTGGCATCTGGGAAG | |||
|
rs1927911 |
F: TCACTTTGCTCAAGGGTCAA |
StyI |
203=TT
203, 178, 25=TC
178, 25=CC |
| R: AAACCTGCATGCTCTGCAC | |||
| rs1927914 | F: ACAAAATGGTCCCTCACAGC |
SphI | 150=TT 157, 90, 67=TC 90, 67=CC |
| R: TGGAAAGTAGCAAGTGCAATG |
Primers for PCR-RFLP of the TLR4, Restriction Enzymes used and base pair products for genotypes.
Figure 1.
Gels showing PCR-RFLP analysis of different SNPs of the TLR4 gene. The amplified products of the SNPs rs4986790, rs4986791, rs10759931, rs1927911, and rs1927914 were digested with restriction enzymes BccI, BslI, KpnI, StyI and SphI, respectively. The restricted products were separated on 3% agarose gel, according to our previous report [10]. A: For genotyping rs4986790, the 140-bp PCR product was digested with BccI. The A allele is not cut by the enzyme, whereas the G allele yields 77 and 63 bp products. B: For genotyping rs4986791, the 110-bp PCR product was digested with BslI. The T allele is not cut by the enzyme, whereas the C allele yields 89 and 21 bp products. C: For genotyping rs1927911, the 203-bp PCR product was digested with StyI. The T allele is not cut by the enzyme, whereas the C allele yields 178 and 25 bp products. D: For genotyping rs10759931, the 241-bp PCR product was digested with KpnI. The A allele is not cut by the enzyme, whereas the G allele yields 190 and 51 bp products. E: For genotyping rs1927914, the 157-bp PCR product was digested with SphI. The T allele is not cut by the enzyme, whereas the C allele yields 90 and 67 bp products.
Statistical analysis for genotype comparison
The allele and genotypic distributions among the groups were evaluated using the χ2 test. The differences in the frequencies between the case and the control groups were analyzed for statistical significance at the 95% confidence interval (CI) using the χ2 test. The allele frequencies of all SNPs were in Hardy–Weinberg equilibrium. Odds ratios (ORs) were calculated and reported within the 95% confidence limits using a calculator for CIs of ORs based on the null hypothesis (Confidhypo). A two-tailed p value of ≤0.05 was considered statistically significant.
Linkage disequilibrium and haplotype analysis
Haplotype frequencies and LD were calculated using Haploview software (version 4.2; The Broad Institute) based on the expectation–maximization (EM) algorithm. This program takes one haplotype at a time and compares its frequency between cases and controls. The standardized disequilibrium coefficient (D’) and correlation coefficient (r2) between these SNPs were also analyzed using the LD plot function of this software to find certain allelic combinations of SNPs that might alter the risk of DR. A p value <0.05 was considered statistically significant for the observed haplotypes.
Results
A comparison of clinical and biochemical characteristics of the study subjects revealed that subjects with DR were older in age compared to T2DM subjects (54.87±8.72 and 51.87±10.57 years, respectively; p = 0.05; Table 1). Duration of diabetes was significantly higher in DR cases compared to T2DM cases (8.50±6.76 years for DR, 6.08±5.32 years for T2DM; p = 0.006). The remaining parameters did not vary significantly between the two groups. The genotypes of TLR4 SNPs rs4986790, rs4986791, rs10759931, rs1927911, and rs1927914 were analyzed in 378 T2DM patients and 320 age-matched controls. Genotypic frequencies of all TLR4 SNPs studied were similar to those reported for other populations [21,22].
The association of TLR4 SNPs was analyzed in the combined T2DM group and compared to the controls. For TLR4 rs4986790 SNP, the prevalence of homozygous risk genotype GG was 0.5% in the combined T2DM group while it was absent in healthy controls (Table 3). Interestingly, for TLR4 rs4986791 SNP, the risk genotype CC showed a higher prevalence in controls (1.2%) compared to the combined T2DM group (0.8%; p value >0.05). The genotypic frequencies of risk genotypes of rs10759931, rs1927911, and rs1927914 were 0.3%, 4.2%, and 8.5%, respectively, in the combined T2DM cases compared to 0.9%, 0.6%, and 13.8% in controls. Genotypic frequencies of all SNPs were found to be in Hardy–Weinberg equilibrium for both study groups.
Table 3. Observed genotype frequencies for studied SNPs among T2DM cases.
| SNP and genotype | Controls, no., (%) | T2DM, no., (%) | OR | 95% CI | p- value |
|---|---|---|---|---|---|
|
rs4986790 (TLR4_Asp299Gly) 119515123 | |||||
| AA |
247 (77.2) |
287 (76.0) |
– |
– |
– |
| AG |
73 (22.8) |
89 (23.5) |
1.01 |
0.71 to 1.44 |
0.93 |
| GG |
0 (0) |
2 (0.5) |
6.2 |
0.38 to 101.4 |
0.19 |
| AG + GG |
73 (22.8) |
91 (24.1) |
1.01 |
0.71 to 1.44 |
0.84 |
|
rs4986791 (TLR4_Thr399Ile) 119515423 | |||||
| CC |
262 (81.9) |
302 (79.9) |
– |
– |
– |
| CT |
54 (16.9) |
73 (19.3) |
1.17 |
0.79 to 1.72 |
0.42 |
| TT |
4 (1.2) |
3 (0.8) |
0.65 |
0.14 to 2.89 |
0.57 |
| CT + TT |
58 (18.1) |
76 (20.1) |
1.13 |
0.77 to 1.65 |
0.35 |
|
rs10759931 (TLR4_1859) 120464147 | |||||
| AA |
173 (54.1) |
183 (48.4) |
– |
– |
– |
| AG |
144 (45.0) |
194 (51.3) |
1.26 |
0.93 to 1.70 |
0.12 |
| GG |
3 (0.9) |
1 (0.3) |
0.34 |
0.04 to 2.48 |
0.29 |
| AG + GG |
147 (46.0) |
195 (51.6) |
1.25 |
0.92 to 1.67 |
0.14 |
|
rs1927914 (TLR4_2437) 120464725 | |||||
| TT |
184 (57.5) |
181 (47.9) |
– |
– |
– |
| TC |
134 (41.9) |
181 (47.9) |
1.37 |
1.01 to 1.85 |
0.04 |
| CC |
2 (0.6) |
16 (4.2) |
4.8 |
1.86 to 12.36 |
0.001 |
| TC + CC |
136 (42.5) |
197 (52.1) |
1.46 |
1.09 to 1.97 |
0.001 |
|
rs1927911 (TLR4_7764) 120470054 | |||||
| CC |
112 (35.0) |
177 (46.8) |
– |
– |
– |
| CT |
164 (51.2) |
169 (44.7) |
0.65 |
0.47 to 0.89 |
0.008 |
| TT |
44 (13.8) |
32 (8.5) |
0.45 |
0.27 to 0.76 |
0.002 |
| CT + TT | 212 (66.3) | 201 (53.2) | 0.6 | 0.44 to 0.81 | 0.001 |
Genotype frequencies of single nucleotide polymorphisms (SNPs) rs4986790, rs4986791, rs10759931, rs1927911 and rs1927914 of TLR4 gene among T2DM and controls
In the T2DM group, 128 patients were diagnosed with DR. The distribution of genotypes of TLR4 SNPs in these DR patients compared to control subjects is documented in Table 4. The frequency of risk genotype GG of rs4986790 was found to be 0.8% in DR cases. The combined risk genotypes (GG + AG) of rs4986790 were found to be evenly distributed in the DR cases (22.7%) compared to controls (22.8%; OR 0.99, 95% CI 0.61–1.61). The risk genotype TT of rs4986791 was absent in DR. The combined risk genotype CT + TT of the TLR4 SNP rs4986791 polymorphism was also higher in DR cases (20.3%) than in controls (18.1%; OR 1.15, 95% CI 0.68–1.95). For TLR4 SNP rs10759931, both the heterozygous genotype AG and the combined risk genotypes AG + GG were significantly associated with DR with respect to controls (OR 1.53, 95% CI 1.01–2.31, p = 0.04 for AG; OR 1.50, 95% CI 0.99–2.26, p = 0.05 for AG + GG). For TLR4 rs1927914, the heterozygous genotype CT along with the combined risk genotype CT + TT were significantly associated with DR compared to controls (OR 1.48, 95% CI 1.00–2.25, p = 0.05 for CT; OR 1.48, 95% CI 1.0–2.24, p = 0.05 for CT + TT). For rs1927911, the frequency of risk genotype TT was slightly but not significantly lower in DR (7.8%) compared to controls (13.8%; OR 0.77, 95% CI 0.26–1.03).
Table 4. Observed Genotype frequencies for studied SNPs among DR cases.
| SNP and genotype | Controls, no., (%) | DR, no., (%) | OR | 95% CI | p- value |
|---|---|---|---|---|---|
|
rs4986790 (TLR4_Asp299Gly) 119515123 | |||||
| AA |
247 (77.2) |
99 (77.4) |
– |
– |
– |
| AG |
73 (22.8) |
28 (21.8) |
0.95 |
0.58 to 1.56 |
0.86 |
| GG |
0 (0) |
1 (0.8) |
– |
– |
– |
| AG + GG |
73 (22.8) |
29 (22.7) |
0.99 |
0.61 to 1.61 |
0.97 |
|
rs4986791 (TLR4_Thr399Ile) 119515423 | |||||
| CC |
262 (81.9) |
102 (79.7) |
– |
– |
– |
| CT |
54 (16.9) |
26 (20.3) |
1.24 |
0.72 to 2.12 |
0.42 |
| TT |
4 (1.2) |
0 (0) |
0.24 |
0.02 to 2.23 |
0.21 |
| CT + TT |
58 (18.1) |
26 (20.3) |
1.15 |
0.68 to 1.95 |
0.59 |
|
rs10759931 (TLR4_1859) 120464147 | |||||
| AA |
173 (54.1) |
56 (43.7) |
– |
– |
– |
| AG |
144 (45.0) |
72 (56.3) |
1.53 |
1.01 to 2.31 |
0.04 |
| GG |
3 (0.9) |
0 (0) |
0.26 |
0.02 to 3.74 |
0.32 |
| AG + GG |
147 (46.0) |
72 (56.3) |
1.5 |
0.99 to 2.26 |
0.05 |
|
rs1927914 (TLR4_2437) 120464725 | |||||
| TT |
184 (57.5) |
61 (47.7) |
– |
– |
– |
| TC |
134 (41.9) |
66 (51.5) |
1.48 |
1.00 to 2.25 |
0.05 |
| CC |
2 (0.6) |
1 (0.8) |
1.56 |
0.11 to 21.58 |
0.73 |
| TC + CC |
136 (42.5) |
67 (52.3) |
1.48 |
1.00 to 2.24 |
0.05 |
|
rs1927911 (TLR4_7764) 120470054 |
|||||
| CC |
112 (35.0) |
52 (40.6) |
– |
– |
– |
| CT |
164 (51.2) |
66 (51.5) |
0.86 |
0.55 to 1.34 |
0.51 |
| TT |
44 (13.8) |
10 (7.8) |
0.52 |
0.26 to 1.03 |
0.06 |
| CT + TT | 212 (66.3) | 76 (59.4) | 0.77 | 0.50 to 1.17 | 0.22 |
Genotype frequencies of single nucleotide polymorphisms (SNPs) rs4986790, rs4986791, rs10759931, rs1927911 and rs1927914 of TLR4 gene among DR and controls.
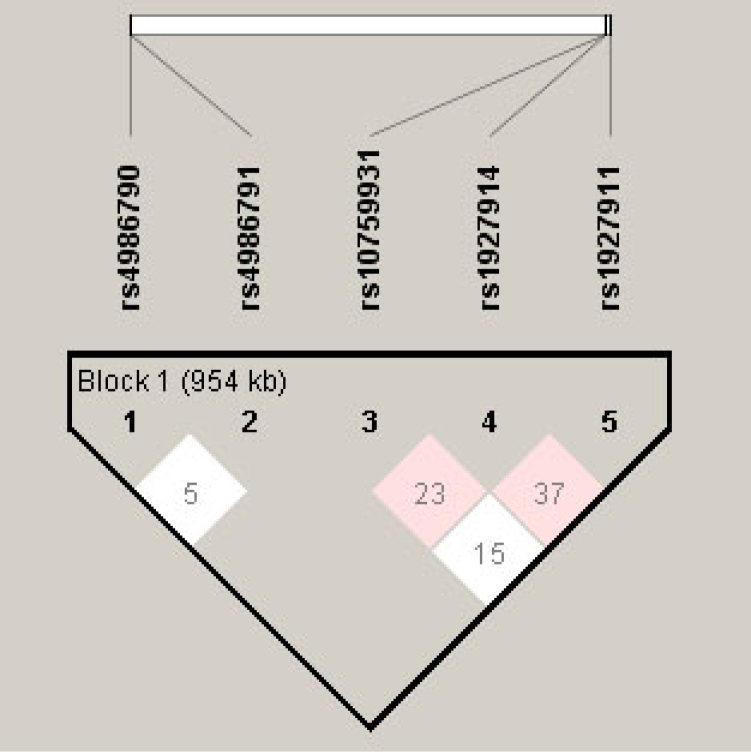
It has previously been shown that some of the TLR4 SNPs reside in strong LD in normal tension glaucoma, another eye-related disease [23]. To estimate haplotype frequencies and analyze the haplotype association with DR, we selected five SNPs—rs4986790, rs4986791, rs10759931, rs1927911, and rs1927914—for LD and haplotype analysis (Table 5, Figure 2). The SNPs rs4986790 and rs4986791were found not to be in significant LD (D’ = 0.05, LOD = 0.0, confidence bound 0.01–0.82, r2 = 0.0). Evidence of intermediate LD was found between loci rs10759931 and rs1927914 (D’ = 0.24, LOD = 2.67, confidence bound 0.12–0.33, r2 = 0.051) and between loci rs1927914 and rs1927911 (D’ = 0.37, LOD = 4.75, confidence bound 0.24–0.49, r2 = 0.069). No significant LD was found between loci rs10759931 and rs1927911 (D’ = 0.155, LOD = 0.73, confidence bound 0.02–0.29, r2 = 0.013). Fourteen haplotypes were identified having a frequency of more than 1% (Table 6). Haplotypes ACATC (37.5%) and ACATT (14.8%) were the two most common haplotypes obtained.
Table 5. Haplotype analysis for the studied SNP markers.
| L1 | L2 | D | LOD | R2 | CI low | CI high | Dist | T-int |
|---|---|---|---|---|---|---|---|---|
|
rs4986790 |
rs4986791 |
0.051 |
0 |
0 |
0.01 |
0.82 |
300 |
0 |
|
rs10759931 |
rs1927914 |
0.236 |
2.67 |
0.051 |
0.12 |
0.33 |
578 |
3.4 |
|
rs10759931 |
rs1927911 |
0.155 |
0.73 |
0.013 |
0.02 |
0.29 |
5907 |
- |
| rs1927914 | rs1927911 | 0.371 | 4.75 | 0.069 | 0.24 | 0.49 | 5329 | 5.48 |
The table describes the LD value calculated for the all present SNPs of TLR4 gene. L1 and L2 are loci in question, D' is the value of D prime between the two loci, LOD is the log of the likelihood odds ratio, r2 is the correlation coefficient between the two loci, CI low is 95% confidence lower bound on D', CI high is the 95% confidence upper bound on D', Dist is the distance (in bases) between the loci, and is only displayed if a marker info file has been loaded, T-int is a statistic used by the HapMap Project to measure the completeness of information represented by a set of markers in a region.
Figure 2.
Linkage disequilibrium plot. Haplotype frequencies and LD were calculated using Haploview software (version 4.2). The LD parameter D is represented by the specific value in each cell. The cells are color graduated representing the strength of LD between the two markers. The rs numbers are SNP IDs extracted from the Ensembl database. The loci rs10759931, rs1927911, and rs1927914 are in intermediate LD.
Table 6. Association status of common haplotypes of TLR4 gene with DR.
| Haplotype | Frequency | Case, control frequencies | Chi square | p value |
|---|---|---|---|---|
| ACATC |
0.375 |
0.374, 0.379 |
0.019 |
0.89 |
| ACATT |
0.148 |
0.155, 0.120 |
1.363 |
0.243 |
| ACGTC |
0.074 |
0.073, 0.081 |
0.135 |
0.713 |
| ACACC |
0.039 |
0.038, 0.042 |
0.06 |
0.806 |
| ACGCT |
0.045 |
0.041, 0.060 |
1.235 |
0.266 |
| ATATC |
0.044 |
0.043, 0.045 |
0.012 |
0.912 |
| GCATC |
0.037 |
0.041, 0.023 |
1.34 |
0.247 |
| GCGTT |
0.018 |
0.019, 0.013 |
0.29 |
0.589 |
| ACACT |
0.052 |
0.052, 0.053 |
0.004 |
0.953 |
| ATACT |
0.014 |
0.013, 0.019 |
0.349 |
0.555 |
| ACGTT |
0.043 |
0.041, 0.049 |
0.218 |
0.641 |
| ACGCC |
0.024 |
0.023, 0.029 |
0.254 |
0.614 |
| ATGCT |
0.01 |
0.009, 0.016 |
0.717 |
0.397 |
| GCACT | 0.02 | 0.019, 0.024 | 0.164 | 0.685 |
Fourteen haplotypes with a frequency of more than 1% was found.
Discussion
Persistent hyperglycemia and oxidative stress in T2DM are shown to elicit the innate immune system and chronic low-grade inflammation in patients [9]. DR is also an inflammatory disease having a multigenic etiology and is shown to be regulated by inadequate activation of members of the immune system [18]. TLRs are important contributors to the innate immune system, and some members of the TLR family, especially TLR4, have been implicated in several inflammatory and immune disorders [24]. Therefore, SNPs, even with comparatively small ORs in the TLR4 gene, when combined can cause alterations in protein functioning that may contribute, to a moderate degree, to the development of DR.
The association of two SNPs included in this study, rs4986790 and rs4986791, have been widely studied in inflammatory diseases, such as Crohn’s disease, and gastric cancer and gastric lymphoma in different cohorts, including Indians [25,26]. Recently, Buraczynska et al. also associated these two SNPs with early onset of DR in a Polish population [18]. The present study was designed to observe the genotypic frequencies of rs4986790 and rs4986791 in a north Indian population to observe the ethnic and population variations of TLR4 polymorphism. The risk genotype GG of rs4986790 was found to be present only in T2DM cases while it was absent in the control group, similar to the results of Buraczynska et al. [18]. Although the combined risk genotype (CT + TT) of TLR4 rs4986791 was greater in the DR group compared to controls, we were unable to find any risk genotype TT in the DR group. Our data suggested neither of these TLR4 SNPs were associated with DR in an Indian population. This observation differs from that of Buraczynska et al. [18] who found the risk allele of TLR4 SNP rs4986790 to be associated with early onset of DR in a Polish population. The difference in ethnic and racial backgrounds of Polish and Indian populations or disease heterogeneity may be the reason for this observed variation. Moreover, stratification of subjects on the basis of age of onset of DR by Buraczynska et al. might be another reason for this observed variance between these two studies. The study of Buraczynska et al. focused on cases with a comparatively early onset, suggesting stronger involvement of genetic factors. However, our study was focused on the general risk for DR patients, regardless of age. Moreover, differences in the interactive effects of environment and lifestyle of Polish and Indian populations can’t be ruled out.
We then analyzed the association of rs10759931, rs1927911, and rs1927914 variants of TLR4 with DR in our population. We found the heterozygous genotype AG of rs10759931 and the combined risk genotype (AG + GG) to be significantly associated with the development of DR in an Indian population. The risk allele A of this variant exhibits altered binding affinity with transcription factors, thereby resulting in a lower expression of TLR4 [27]. Hence, low expression of TLR4 mediated by this variant could explain the different susceptibility of various diseases, including cancer, among different ethnic groups [10,17]. A similar trend in association was found for variant rs1927914 as both heterozygous genotype TC and combined risk genotype (TC + CC) were significantly associated with the pathogenesis of DR. This finding was supported by our previous report that heterozygous genotypes of TLR4 variant rs1927914 are common in the Indian population [10]. Suh et al. (2011) found no significant association of rs1927914 with normal tension glaucoma in a South Korean population [28]. A possible explanation for this contradictory result is the ethnic differences between study subjects and disease heterogeneity. Another difference was in the number of study participants. The Korean study comprised 527 individuals, while our study analyzed 690 Indian subjects. For TLR4 variant rs1927911, we found the risk genotype TT to be marginally but not significantly lower in the DR group compared to control subjects, a finding in agreement with Suh et al. (2011) for a South Korean population [28]. Haplotype analysis for the calculation of LD for the five SNPs of the TLR4 gene yielded 14 haplotypes having a frequency of more than 1%. The two loci combinations rs10759931 and rs1927914, and rs1927914 and rs1927911 were found to be in intermediate LD. The other TLR4 SNP loci combinations in our population showed no sign of LD among them. Association analysis using haplotypes showed none of the haplotypes were associated with either susceptibility or resistance to DR in a north Indian population.
Data regarding the role of TLR4 gene polymorphism in diabetic complication are scarce. Only a few reports document an association of TLR4 with a secondary complication of diabetes. Rudofsky et al. associated the TLR4 SNPs rs4986790 and rs4986791 with a reduced prevalence of diabetic neuropathy in type 2 diabetes [29]. Our group associated TLR4 variants with impairment of wound healing in T2DM patients [10], and only one report associated SNPs rs4986790 and rs4986791 with DR in the Polish population [18]. Hence, our study is a step toward improving our understanding of the missing link between innate immunity, T2DM, and its complications, such as DR. Further studies with different ethnic groups will demonstrate whether the same results can be obtained in other areas of the Indian subcontinent.
Acknowledgments
This work was funded by the Department of Science and Technology, New Delhi, India (P-07-523). Financial assistance by the Department of Biotechnology, Ministry of Science and Technology, New Delhi, in the form of a Senior Research Fellowship to the first author is thankfully acknowledged.
References
- 1.Singh K, Agrawal NK, Gupta SK, Singh K. Association of Variant rs7903146 (C/T) Single nucleotide polymorphism of TCF7L2 gene with impairment in wound healing among north Indian Type 2 Diabetes population: A Case–Control Study. Int J Low Extrem Wounds. 2013;12:310–5. doi: 10.1177/1534734613504435. [DOI] [PubMed] [Google Scholar]
- 2.Singh K, Agrawal NK, Gupta SK, Singh K. A Functional SNP-1562C>T in the Matrix Metalloproteinases-9 Promoter is associated with type 2 diabetes and Diabetic Foot Ulcers. Int J Low Extrem Wounds. 2013;12:199–204. doi: 10.1177/1534734613493289. [DOI] [PubMed] [Google Scholar]
- 3.Kohener EM, Stratton IM, Aldinton SJ. Prevalence of diabetic retinopathy at diagnosis of NIDDM in the UKPDS. Invest Ophthalmol Vis Sci. 1993;34:713. [Google Scholar]
- 4.Navarro JF, Mora C. Role of inflammation in diabetic complications. Nephrol Dial Transplant. 2005;20:2601–4. doi: 10.1093/ndt/gfi155. [DOI] [PubMed] [Google Scholar]
- 5.Petrovic D. Candidate Genes for Proliferative Diabetic Retinopathy. BioMed Research International 2013;2013; 540416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abu El-Asrar AM, Struyf S, Opdenakker G, Damme JV, Geboes K. Expression of stem cell factor/c-kit signaling pathway components in diabetic fibrovascular epiretinal membranes. Mol Vis. 2010;16:1098–107. [PMC free article] [PubMed] [Google Scholar]
- 7.Kunicki TJ, Kritzik M, Annis DS, Nugent DJ. Hereditary variation in platelet integrin α2β1 density is associated with two silent polymorphisms in the α2 gene coding sequence. Blood. 1997;89:1939–43. [PubMed] [Google Scholar]
- 8.McLeod DS, Lefer DJ, Merges C, Lutty GA. Enhanced expression of intracellular adhesion molecule-1 and P-selectin in the diabetic human retina and choroid. Am J Pathol. 1995;147:642–53. [PMC free article] [PubMed] [Google Scholar]
- 9.Kanhaiya, Agrawal NK, Gupta SK, Singh K. Differential expression of Toll like Receptor 4 in Type 2 Diabetic patients with impaired wound healing. Journal of Diabetes and Metabolism. 2013;4:260. [Google Scholar]
- 10.Singh K, Singh VK, Agrawal NK, Gupta SK, Singh K. Association of Toll-like receptor 4 polymorphisms with diabetic foot ulcers and application of artificial neural network in DFU risk assessment in type 2 diabetes patients. Biomed Res Int. 2013;2013:318686. doi: 10.1155/2013/318686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 12.Armant MA, Fenton MJ. Toll-like receptors: a family of pattern-recognition receptors inmammals. Genome Biol. 2002;3:3011. doi: 10.1186/gb-2002-3-8-reviews3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zarember KA, Godowski PJ. Tissue expression of human Toll-like receptors and differential regulation of Toll-like receptors mRNAs in leukocytes in response to microbes, their products and cytokines. J Immunol. 2002;168:554–61. doi: 10.4049/jimmunol.168.2.554. [DOI] [PubMed] [Google Scholar]
- 14.Akira S, Takeda K, Kaisho T. Toll-like receptors: Critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–80. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 15.Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A, Mignot G, Maiuri MC, Ullrich E, Saulnier P, Yang H, Amigorena S, Ryffel B, Barrat FJ, Saftig P, Levi F, Lidereau R, Nogues C, Mira JP, Chompret A, Joulin V, Clavel-Chapelon F, Bourhis J, Andre F, Delaloge S, Tursz T, Kroemer G, Zitvogel L. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat Med. 2007;13:1050–9. doi: 10.1038/nm1622. [DOI] [PubMed] [Google Scholar]
- 16.Prohinar P, Rallabhandi P, Weiss JP, Gioannini TL. Expression of functional D299G.T399I polymorphic variant of TLR4 depends more on co-expression of MD-2 than does wild-type TLR4. J Immunol. 2010;184:4362–7. doi: 10.4049/jimmunol.0903142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song J, Kim DY, Kim CS, Kim HJ, Lee DH, Lee HM, Ko W, Lee G. The association between Toll-like receptor 4 (TLR4) polymorphisms and the risk of prostate cancer in Korean men. Cancer Genet Cytogenet. 2009;190:88–92. doi: 10.1016/j.cancergencyto.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 18.Buraczynska M, Baranowicz-Gaszczyk I, Tarach J, Ksiazek A. Toll-like receptor 4 gene polymorphism and early onset of diabetic retinopathy in patients with type 2 diabetes. Hum Immunol. 2009;70:121–4. doi: 10.1016/j.humimm.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Zareparsi S, Buraczynska M, Branham KE, Shah S, Eng D, Li M, Pawar H, Yashar BM, Moroi SE, Lichter PR, Petty HR, Richards JE, Abecasis GR, Elner VM, Swaroop A. Toll-like receptor 4 variant D299G is associated with susceptibility to age-related macular degeneration. Hum Mol Genet. 2005;14:1449–55. doi: 10.1093/hmg/ddi154. [DOI] [PubMed] [Google Scholar]
- 20.Early Treatment Diabetic Retinopathy Study Research Group Grading diabetic retinopathy from stereoscopic color fundus photographs—an extension of the modified Airlie House classification. ETDRS report number 10. Ophthalmology. 1991;98:806–96. [PubMed] [Google Scholar]
- 21.Oostenbrug LE, Drenth JP, de Jong DJ, Nolte IM, Oosterom E, van Dullemen HM, van der Linde K, te Meerman GJ, van der Steege G, Kleibeuker JH, Jansen PL. Association between Toll-like receptor 4 and inflammatory bowel disease. Inflamm Bowel Dis. 2005;11:567–75. doi: 10.1097/01.mib.0000161305.81198.0f. [DOI] [PubMed] [Google Scholar]
- 22.Leung TF, Tang NL, Wong GW, Fok TF. CD14 and Toll-like receptors: Potential contribution of genetic factors and mechanisms to inflammation and allergy. Curr Drug Targets Inflamm Allergy. 2005;4:169–75. doi: 10.2174/1568010053586336. [DOI] [PubMed] [Google Scholar]
- 23.Shibuya E, Meguro A, Ota M, Kashiwagi K, Mabuchi F, Iijima H, Kawase K, Yamamoto T, Nakamura M, Negi A, Sagara T, Nishida T, Inatani M, Tanihara H, Aihara M, Araie M, Fukuchi T, Abe H, Higashide T, Sugiyama K, Kanamoto T, Kiuchi Y, Iwase A, Ohno S, Inoko H, Mizuki N. Association of Toll-like Receptor 4 Gene Polymorphisms with Normal Tension Glaucoma. Invest Ophthalmol Vis Sci. 2008;49:4453–7. doi: 10.1167/iovs.07-1575. [DOI] [PubMed] [Google Scholar]
- 24.Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 25.Hold GL, Rabkin CS, Chow WH, Smith MG, Gammon MD, Risch HA, Vaughan TL, McColl KE, Lissowska J, Zatonski W, Schoenberg JB, Blot WJ, Mowat NA, Fraumeni JF, Jr, El-Omar EM. A functional polymorphism of Toll-like receptor 4 gene increases risk of gastric carcinoma and its precursors. Gastroenterology. 2007;132:905–12. doi: 10.1053/j.gastro.2006.12.026. [DOI] [PubMed] [Google Scholar]
- 26.Achyut BR, Ghoshal UC, Moorchung N, Mittal B. Association of Toll-like receptor-4 (Asp299Gly and Thr399Ile) gene polymorphism with gastritis and precancerous lesions. Hum Immunol. 2007;68:901–7. doi: 10.1016/j.humimm.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 27.Ferronato S, Gomez-Lira M, Menegazzi M, Diani E, Olivato S, Sartori M, Scuro A, Malerba G, Pignatti PF, Romanelli MG, Mazzucco S. Polymorphism −2604G>A variants in TLR4 promoter are associated with different gene expression level in peripheral blood of atherosclerotic patients. J Hum Genet. 2013;58:812–4. doi: 10.1038/jhg.2013.98. [DOI] [PubMed] [Google Scholar]
- 28.Suh W, Kim S. Ki Chang-Seok, Kee C. Toll-like Receptor 4 gene polymorphisms do not associate with normal tension glaucoma in a Korean population. Mol Vis. 2011;17:2343–8. [PMC free article] [PubMed] [Google Scholar]
- 29.Rudofsky G, Jr, Reismann P, Witte S, Humpert PM, Isermann B, Chavakis T, Tafel J, Nosikov VV, Hamann A, Nawroth P, Bierhaus A. Asp299Gly and Thr399Ile Genotypes of the TLR4 Gene Are Associated With a Reduced Prevalence of Diabetic Neuropathy in Patients with Type 2 Diabetes. Diabetes Care. 2004;27:179–83. doi: 10.2337/diacare.27.1.179. [DOI] [PubMed] [Google Scholar]