Abstract
We generated draft genome sequences for two cold-adapted Archaea, Methanogenium frigidum and Methanococcoides burtonii, to identify genotypic characteristics that distinguish them from Archaea with a higher optimal growth temperature (OGT). Comparative genomics revealed trends in amino acid and tRNA composition, and structural features of proteins. Proteins from the cold-adapted Archaea are characterized by a higher content of noncharged polar amino acids, particularly Gln and Thr and a lower content of hydrophobic amino acids, particularly Leu. Sequence data from nine methanogen genomes (OGT 15°–98°C) were used to generate 1111 modeled protein structures. Analysis of the models from the cold-adapted Archaea showed a strong tendency in the solvent-accessible area for more Gln, Thr, and hydrophobic residues and fewer charged residues. A cold shock domain (CSD) protein (CspA homolog) was identified in M. frigidum, two hypothetical proteins with CSD-folds in M. burtonii, and a unique winged helix DNA-binding domain protein in M. burtonii. This suggests that these types of nucleic acid binding proteins have a critical role in cold-adapted Archaea. Structural analysis of tRNA sequences from the Archaea indicated that GC content is the major factor influencing tRNA stability in hyperthermophiles, but not in the psychrophiles, mesophiles or moderate thermophiles. Below an OGT of 60°C, the GC content in tRNA was largely unchanged, indicating that any requirement for flexibility of tRNA in psychrophiles is mediated by other means. This is the first time that comparisons have been performed with genome data from Archaea spanning the growth temperature extremes from psychrophiles to hyperthermophiles.
The analysis of thermal adaptation across the full spectrum of known growth temperatures has been hampered by the lack of genome sequences for cold-adapted organisms. Of the 16 complete archaeal genomes (December 2002), 13 are for thermophiles or hyperthermophiles, and the remainder are for mesophiles. Even though Archaea contribute significantly to biomass in the predominantly cold biosphere (e.g., ∼1028 cells in the world's oceans; Karner et al. 2001), the only free living isolates that have been formally characterized are a handful of methanogens (Franzmann et al. 1992, 1997; Simankova et al. 2001; Chong et al. 2002; von Klein et al. 2002). Consequently, our understanding of cold adaptation in this domain of life is limited (Cavicchioli et al. 2000).
The methanogen with the lowest known optimum growth temperature (OGT) is Methanogenium frigidum (15°C), which is unable to grow above 18°C (Franzmann et al. 1997). It was isolated from Ace Lake in the Vestfold Hills region of Antarctica, where the bottom waters are saturated in methane and permanently 1°–2°C. Ace Lake was also the source of Methanococcoides burtonii (Franzmann et al. 1992). Its ability to grow on methyl-substrates and tolerate a broader range of growth temperatures (< 4° to 29°C) has led to its use for studies on protein adaptation (Thomas and Cavicchioli 1998, 2000, 2002; Thomas et al. 2001; Siddiqui et al. 2002) and gene regulation (Lim et al. 2000).
The methanogens stand out as the only group of organisms that have species capable of growth at 0°C (M. frigidum and M. burtonii) and 110°C (Methanopyrus kandleri). Moreover, there are complete genome sequences for M. kandleri (OGT 98°C; Slesarev et al. 2002), Methanocaldococcus jannaschii (85°C; Bult et al. 1996), Methanothermobacter thermautotrophicus (65°C; Smith et al. 1997), Methanosarcina acetivorans (37°C; Galagan et al. 2002), Methanosarcina mazei (37°C; Deppenmeier et al. 2002), and high-quality draft sequences for Methanococcus maripaludis (37°C) and Methanosarcina barkeri (35°C).
In this study we report the draft sequencing of M. frigidum and M. burtonii. These data provide the missing link for enabling a rigorous assessment of thermal adaptation at the genomic level. Using comparative genomics with the methanogen and/or all available archaeal genomes, we identified genome-wide characteristics of cold adaptation. In particular, we identified trends in amino acid and tRNA composition, and structural and compositional features of protein homology models, that distinguish the genomes of the cold-adapted Archaea from other Archaea. In addition, we highlight the value of careful processing of draft genomes for identifying targets for functional studies, by identifying unique or hypothetical genes that may be novel signatures of cold adaptation.
RESULTS
Draft Genome Statistics of M. frigidum and M. burtonii
Descriptive statistics from the current genome assemblies are presented in Table 1. Genome size has not been measured experimentally for these two organisms. However, based on the coverage and on the increase in assembly size as reads were added to the assembly, we estimate genome sizes of approximately 2–2.5 Mbp and 2.8–3 Mbp for M. frigidum and M. burtonii, respectively. These estimates fall within the known range of genome sizes for methanogens (1.6–5.7 Mbp). The number of candidate protein-coding genes in each case (M. frigidum 1815, M. burtonii 2676) agrees well with the estimated genome sizes and with the coding density of other procaryotic genomes. Due to the draft state of the genomes, we have defined three sets of gene data: predicted coding regions, putative open reading frames (ORFs), and stringent ORFs (see Methods).
Table 1.
Genome Assembly Statistics for M. frigidum and M. burtonii
| Organism | M. frigidum | M. burtonii |
|---|---|---|
| Total sequence reads | 9870 | 41,957 |
| Contigs | 1413 | 287 (114 scaffolds) |
| Total assembled bases | 1,597,795 | 2,668,325 |
| Coverage | ∼2× | ∼9.4× |
| Estimated genome size | ∼2-2.5 Mbp | ∼2.8 Mbp |
| Protein coding regions | 1815 | 2676 |
| Hits to nr database | 1200 | 2168 |
| Putative full-length ORFs | 1096 | 2557 |
In M. burtonii, 81% of the coding regions had a BLAST hit to the nonredundant protein database (nrdb). This reflects the high number of sequences with similarity to the genomes of M. acetivorans and M. mazei. The proportion of sequences from M. burtonii with at least one BLAST hit to a sequence from these genomes is 73% and 72%, respectively. In contrast, the M. frigidum sequence contains a far higher proportion of coding regions with no significant BLAST hit to the nrdb (∼34%). The proportion of M. frigidum sequences with BLAST hits to other methanogen genomes is also lower than for M. burtonii (53% for both M. acetivorans and M. mazei). This indicates a substantial level of novel coding potential in the M. frigidum genome.
Predicted Genes Unique to M. frigidum and M. burtonii
Five predicted genes were identified that were present in both M. frigidum and M. burtonii and unique to these two organisms (no BLAST hit to the nrdb at E ≤ e-10). Threading against the SCOP database of protein folds was used to assign putative functions. One of the pairs of genes (M. frigidum Contig474.292.11, M. burtonii Scaffold8.gene3587) was identified as a 3-helical bundle DNA/RNA-binding protein. The best threading template, with z-scores of 28.92 (M. frigidum) and 40.04 (M. burtonii), was the ArsR-like transcriptional regulator SmtB from Synechococcus and the top nine templates were all members of the `winged helix' DNA-binding domain superfamily.
Bacterial Cold Shock Protein (Csp) Homologs
Proteins homologous to E. coli CspA and Bacillus CspB are associated with cold adaptation in a broad range of bacteria, but are not widely dispersed in Archaea (Cavicchioli et al. 2000, 2002). Using bacterial Csp sequences as BLAST queries, we identified a Csp in M. frigidum but not in M. burtonii. We also searched the genome and nucleotide databases for other archaeal csp genes. Apart from those recently annotated in the Halobacterium genome (Kennedy et al. 2001) and an uncultured marine crenarchaeote (Beja et al. 2002), only one other was identified in Haloferax volcanii (GenBank accession AF442116). Residues known to be involved in RNA-binding in the bacterial Csp proteins are conserved in the archaeal sequences (Fig. 1).
Figure 1.
Multiple alignment (CLUSTALW) of archaeal Csp sequences. (*) Conserved residues in M. frigidum Csp involved with RNA binding.
Csp proteins contain the conserved cold shock domain (CSD; Sommerville 1999). The absence of a Csp homolog in M. burtonii prompted us to search for the CSD-fold in sequences which did not have primary sequence similarity to Csp proteins. The S1 RNA-binding domain of polynucleotide phosphorylase was identified as the best threading template for two sequences from M. burtonii (Scaffold3.gene1629 and Scaffold5.gene2863, z-scores 29.53 and 32.66, respectively). The top three threading templates in both cases belonged to the CSD-fold family.
Amino Acid Composition
The amino acid composition of predicted archaeal proteomes was analyzed using principal components analysis (PCA). PCA can be applied to a multivariate data set in order to reduce the complexity of the data and to determine whether there are underlying trends that explain the observed variation (for discussion of the method, see Kreil and Ouzounis 2001).
The first two principal components (PCs) accounted for 71% of the variance in amino acid composition. PCs 1 and 2 correlated strongly with genome GC content and OGT (correlation coefficients for PC scores vs. these variables were ∼0.95 and -0.95, respectively). The two PCs were plotted for each organism (Fig. 2A). Organisms with the highest scores were exclusively hyperthermophiles, with OGTs of 83°–103°C, and those with the lowest scores were the cold-adapted methanogens and Halobacterium. The presence of Halobacterium as an outlier can be attributed to the overrepresentation of acidic amino acids as an adaptation of this halophilic mesophile to growth in highly saline environments (Kennedy et al. 2001).
Figure 2.
PCA of amino acid composition in archaeal proteomes. (A) Component scores for organisms; (B) component loadings for amino acids. af, Archaeoglobus fulgidus; ap, Aeropyrum pernix; fa, Ferroplasma acidarmanus; halo, Halobacterium sp. NRC-1; ma, Methanosarcina acetivorans; mbar, Methanosarcina barkeri; mbur, Methanococcoides burtonii; mf, Methanogenium frigidum; mj, Methanocaldococcus jannaschii; mk, Methanopyrus kandleri; mm, Methanosarcina mazei; mmar, Methanococcus maripaludis; mt, Methanobacter thermautotrophicus; pa, Pyrobaculum aerophilum; pab, Pyrococcus abyssi; pf, Pyrococcus furiosus; ph, Pyrococcus horikoshii; ss, Sulfolobus solfataricus; st, Sulfolobus tokodaii; ta, Thermoplasma acidophilum; tv, Thermoplasma volcanium.
Loadings for PCs 1 and 2 were plotted for each amino acid (Fig. 2B). The influence of GC content on PC 1 was apparent, with clusters at the extremes of the axis that contain amino acids encoded by high (Gly, Ala, Arg, Val, Pro) and low (Tyr, Lys, Phe, Ile, Asn) GC codons. On the PC 2 axis, the strongest PC loadings were observed for Gln, Thr, and to a lesser extent Asp and His, whereas at the other extreme, Leu exhibited the strongest loading with weaker contributions from Trp and Glu. These amino acids were analyzed further using regression analysis. High regression coefficients for amino acid composition versus OGT were observed for Leu (0.77), Gln (-0.80), and Thr (-0.79), all with P-values significant at 0.01%. Regression coefficients for Trp, His, and Asp were 0.69, -0.63, and -0.58, respectively, all significant at 1%. Regression of Glu content versus OGT was less significant (coefficient, 0.36; 0.12% confidence level).
Comparative Homology Modeling of Proteins From Methanogens
Sequences of predicted proteins from the methanogen genomes were used to generate 1111 three-dimensional models. The proportion of sequences that could be modeled varied from 2.3% (M. frigidum) to 7.5% (M. maripaludis; Table 2). The complete set of models from each organism was analyzed for structural features previously associated with cold adaptation (Russell 2000; Zecchinon et al. 2001; Cavicchioli et al. 2002): (1) H-bonds/100 residues, (2) salt-bridges/100 residues, (3) fraction of atoms/residues in loops, (4) fraction of Pro in loops, (5) percentage of completely inaccessible residues, (6) fraction of accessible or inaccessible area composed of charged residues, and (7) fraction of accessible or inaccessible area composed of hydrophobic residues. The contributions of Gln, Thr, and Leu residues to the accessible or inaccessible area of the modeled proteins were also analyzed.
Table 2.
Summary of Comparative Homology Modeling for Protein Sequences from Nine Methanogens
| Organism | Putative ORFs | Top BLAST hits to PDB | Prospect templates | Prospect model files | Models generated |
|---|---|---|---|---|---|
| M. frigidum | 1096 | 30 | 29 | 28 | 25 |
| M. burtonii | 2676 | 146 | 141 | 139 | 116 |
| M. acetivorans | 4540 | 226 | 223 | 220 | 182 |
| M. barkeri | 3145 | 180 | 175 | 173 | 142 |
| M. maripaludis | 1729 | 168 | 163 | 160 | 130 |
| M. mazei | 3371 | 210 | 206 | 203 | 168 |
| M. thermautotrophicus | 1873 | 173 | 169 | 164 | 124 |
| M. jannaschii | 1729 | 165 | 159 | 157 | 123 |
| M. kandleri | 1687 | 129 | 124 | 121 | 101 |
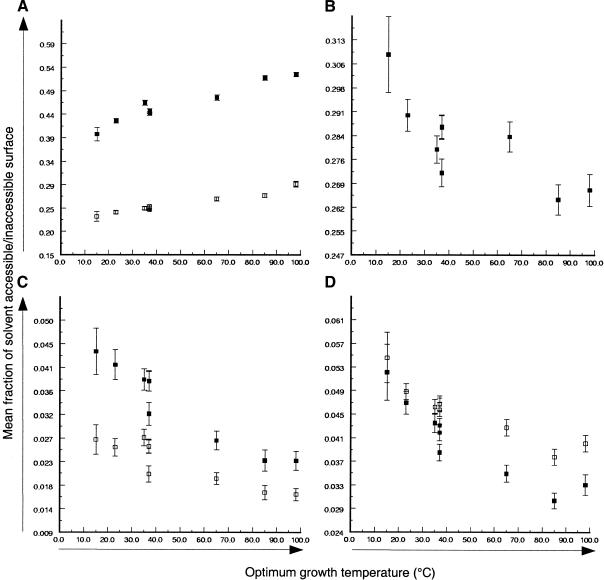
Significant trends were observed across the entire OGT range for the accessibility of charged and hydrophobic residues. The mean fraction of accessible area composed of charged residues increased with OGT (Fig. 3A). A similar trend was observed for the contribution of charged residues to the total inaccessible area (Fig. 3A), but the mean inaccessible fraction was ∼50% that of the accessible fraction. The contribution of hydrophobic residues to the accessible area decreased with increasing OGT (Fig. 3B), although there was no significant trend in the inaccessible area (data not shown). The summed amino acid composition of charged or hydrophobic residues did not correlate significantly with OGT, highlighting the importance of their structural context. The number of predicted salt-bridges decreased significantly at lower OGT for several models; however, this was not a consistent finding in all models (data not shown). No significant trends were identified in any of the models for the other variables tested.
Figure 3.
Contribution of amino acids to the surfaces of modeled proteins from methanogens. (A) Mean fraction of solvent-accessible (▪) and buried (□) surface that is charged residues. (B) Mean fraction of solvent-accessible surface that is hydrophobic residues. (C) Mean fraction of solvent-accessible (▪) and buried (□) surface that is Gln. (D) Mean fraction of solvent-accessible (▪) and buried (□) surface that is Thr. Error bars are standard error of the mean (s.e.m.).
The compositional decrease in Gln and Thr content with OGT seen in the PCA analysis (Fig. 2B) was also reflected by a decrease in their contribution to both accessible and inaccessible areas (Fig. 3C,D). Gln had a preference for accessible regions, whereas Thr distributed approximately equally between accessible and inaccessible areas. The accessibility of Leu residues showed no significant trend with OGT (data not shown), despite that observed for hydrophobic residues (Fig. 3B). Moreover, the proportion of hydrophobic residues in the solvent-accessible fraction increased at low OGT (Fig. 3B), whereas Leu (hydrophobic residue) strongly associated with hyperthermophiles in PCA (Fig. 2B). All of the trends described for the models were observed for both all-atoms and only-side-chain atoms.
Composition and Structure of tRNA in Archaeal Genomes
The genome sequence data from M. frigidum and M. burtonii contained 28 and 46 predicted tRNA sequences, respectively. The mean GC content was calculated for these and for predicted tRNA sequences from all available archaeal genome data, and mean GC content was plotted versus OGT for both stems and total tRNA (Fig. 4). There was a clear nonlinear relationship between OGT and tRNA GC content, with a significant elevation in GC content in the hyperthermophiles. However, GC content in tRNA from the cold-adapted Archaea was comparable to that of mesophiles and moderate thermophiles. These trends are mainly due to tRNA in the stem regions.
Figure 4.
Mean GC fraction of predicted tRNA sequences from archaeal genomes. Total GC (□), stem GC (○). Error bars: s.e.m.
DISCUSSION
Amino Acid Composition and Structural Context
Our aim in this study was to make maximum use of novel genome sequence data from the cold-adapted Archaea and all currently available sequence data from archaeal genomes to address the question of thermal adaptation, particularly in the low temperature range. The analysis of amino acid composition was achieved through PCA, a method which has previously been applied using different genome data sets (Kreil and Ouzounis 2001; Tekaia et al. 2002). Importantly, the inclusion of our genome data shows that the amino acid composition in proteins from cold-adapted Archaea is sufficiently different to distinguish them from both mesophilic and hyper/thermophilic organisms. Strong evidence for decreased Gln and increased Glu content (Kreil and Ouzounis 2001; Tekaia et al. 2002), and moderate evidence for a decrease in Thr, His, and Ser content at high OGTs was reported (Kreil and Ouzounis 2001). Our results show that at least in the Archaea, there is an almost linear trend in the content of Gln, Thr, and Leu over the complete range of OGTs from psychrophile to hyperthermophile. The trend for increased Leu content with OGT has not been previously described. This may be due to the inclusion of data sets for the cold-adapted Archaea and the availability of more archaeal genomes.
We performed large-scale comparative modeling using protein sequences from the methanogen genomes and generated the largest number of models (141) from cold-adapted organisms described to date. The use of proteins from a common phylogenetic and metabolic group (Slesarev et al. 2002) minimized problems associated with genetic diversity and physiology that have been faced by other studies. The main factor, which may affect amino acid composition, was the variation in genome GC content (31%–61%), but this would have more influence on asymmetrical substitution patterns and was accounted for in the PCA analysis. The models were analyzed for structural features that have been proposed as important for protein function at low temperature (Russell 2000; Zecchinon et al. 2001; Cavicchioli et al. 2002). Rather than focus on the details of specific subsets of proteins, we calculated mean values for the set of models from each organism in order to determine whether general trends were apparent.
The most significant findings relate to the solvent accessibility of charged and hydrophobic residues. The contribution of charged amino acids to both accessible and inaccessible area is lowest in proteins from the cold-adapted Archaea and increases with increasing OGT. This reflects a general increase in the proportion of Lys+Arg+Glu in hyperthermophiles. However, these amino acids contribute approximately twofold greater area to the solvent-accessible area than to the inaccessible area, and the increase in contribution to accessibility with OGT is also more pronounced for the accessible area. In contrast, the exposure of hydrophobic residues in the accessible area was greatest for proteins from the cold-adapted Archaea and decreased with increasing OGT, indicating that exposure of hydrophobic residues to the solvent is linked to thermal adaptation. The increased exposure of hydrophobic residues and decreased charge is likely to destabilize the surface of proteins from cold-adapted Archaea. Surface energy and consequent thermal expansion have previously been correlated to overall protein stability (Palma and Curmi 1999; Rees 2001; Rees and Robertson 2001). This is consistent with the findings from studies of individual cold-active proteins (Russell 2000; Zecchinon et al. 2001; Cavicchioli et al. 2002) and supports the concept that increased flexibility may reduce the activation energy of the protein-substrate transition state and increase catalytic efficiency at low temperature (Zecchinon et al. 2001; Siddiqui et al. 2002). Moreover, the reduced fraction of charged surface may also reduce the effects of cold denaturation which will also act on the accessible surface of the protein (Graziano et al. 1997). The lack of a trend for hydrophobic residues in the inaccessible area suggests a more complex relationship between internal hydrophobic interactions and thermal characteristics.
We analyzed the positioning of Gln, Thr, and Leu in the models to determine whether their marked compositional bias had a structural basis. The increased proportion of Gln and Thr in the cold-adapted Archaea is reflected in an increase in their contribution to both solvent-accessible and -inaccessible area. However, Gln makes up a significantly higher proportion of the solvent-accessible area as OGT decreases (Fig. 3C), whereas Thr contributes more equally to both areas (Fig. 3D). No trends in solvent accessibility were observed for Leu despite its decreasing abundance with decreasing OGT. It was reported that fewer polar noncharged residues and more hydrophobic residues occur in proteins from thermophilic Methanococcus species (Haney et al. 1999). It was also reported that the proportion of solvent-accessible charge increases and that of polar noncharged residues decreases in proteins from hyperthermophiles (Cambillau and Claverie 2000). Clearly, these trends are not limited to a particular thermal class but occur from psychrophiles to hyperthermophiles and so reflect adaptations in amino acid composition at all growth temperatures. Moreover, we found that the polar noncharged amino acids which contributed the most to the protein accessible area were Gln and Thr. Gln residues are susceptible to deamidation, and Thr may catalyze peptide bond cleavage (Wright 1991). The increase in Gln and Thr may compensate for the general reduction in surface charged residues. This would minimize problems with aggregation that would otherwise occur if they were only replaced by hydrophobic residues. The increased abundance of Gln and Thr may also reflect a tolerance in cold-active proteins, in contrast to their thermal lability in proteins from hyper/thermophiles.
The largest structural study of psychrophilic enzymes prior to this study examined 21 proteins using both crystal structures and homology models (Gianese et al. 2001). A tendency was observed for the replacement of Arg, Glu, and Lys at exposed sites in the direction thermophile psychrophile. In a later comparative study, it was concluded that although specific structural features were important for function in individual protein families, characteristics generally conserved in the crystal structures of cold-adapted proteins included a decrease in the number of ion pairs, side chain contribution to exposed surface, and apolar fraction of the buried surface (Gianese et al. 2002). Our modeling study suggests that although specific structural features can be important adaptations in individual cases, the most important general trends in proteins across the entire range of OGTs relate to the interaction of the accessible surface area with the intracellular solvent.
Predicting Novel Genes for Cold-Adaptation
Five predicted genes were found to be present in both M. frigidum and M. burtonii that had no significant BLAST hit in the nrdb. One of these gene pairs very likely belongs to the superfamily of `winged helix' DNA-binding proteins (Gajiwala and Burley 2000). Proteins of this superfamily in all three domains of life are transcription factors which regulate a wide variety of cellular processes. The unique presence of this gene pair encoding winged-helix structures in the two cold-adapted Archaea is therefore the strongest indicator of its possible function.
The csp gene in M. frigidum was the first such gene identified in a cold-adapted archaeon. Aside from its presence in the mesophilic halophiles, the other Archaean in which it was found also inhabits cold environments (Beja et al. 2002). Moreover, the gene was absent from the mesophilic methanogens and hyper/thermophilic Archaea. The fact that the protein also contained all ten residues (conserved or conserved substitutions) that are involved in RNA binding (Sommerville 1999) implies it is likely to bind nucleic acid and have a cellular function in cold adaptation.
Interestingly, no csp gene was found in M. burtonii, but two hypothetical proteins were predicted to have a CSD-fold. Despite the lack of obvious primary sequence similarity between members of the CSD-fold family, they appear to have evolved common structural properties which may have been retained from an ancestral protein (Graumann and Marahiel 1998). Some members of the CSD-fold family also perform similar cellular functions; for example, the S1 domain can functionally complement an E. coli quadruple csp deletion mutant (Xia et al. 2001). In this context it is tempting to speculate that the novel CSD-fold proteins may have a role in cold adaptation, similar to that of the Csp protein in M. frigidum. All four of these putative nucleic acid binding proteins are clearly attractive targets for experimental study.
tRNA Flexibility
Previous studies (Galtier and Lobry 1997) have shown, and our analyses confirm, that although there is no relationship between genome GC content and OGT, the GC content of structural RNAs is elevated in hyper/thermophiles. However, without the inclusion of extensive data from psychrophiles, it had not been possible to predict whether the GC content would continue to decrease with low OGT. Our analysis shows that lowered GC content is not an adaptation in the cold-adapted Archaea. The tRNAs from these organisms are comparable in GC content to those from mesophiles and moderate thermophiles, suggesting a threshold temperature of around 60°C, above which GC-mediated structural integrity increases in importance. Based on genome sequences, our data are assembled from complete (or nearly complete) sets of tRNA, which would avoid biases introduced from sampling specific tRNA species. Moreover, although our data are exclusively for Archaea, the inclusion of tRNA from the complete genomes of psychrophilic, mesophilic, and hyperthermophilic bacteria did not change the trends we observed (data not shown).
It is likely that a minimum GC content is required for the structural integrity of tRNA, even at low OGT. However, studies of psychrophilic bacteria indicate that the maintenance of flexibility at low temperature is important for tRNA function, and that this is largely achieved through posttranscriptional incorporation of dihydrouridine (Dalluge et al. 1997), in contrast to the incorporation of different modified nucleosides in archaeal hyperthermophiles (McCloskey et al. 2001). The recent identification of the gene encoding dihydrouridine synthase (Bishop et al. 2002) enabled us to search our genome sequences, and candidate genes were identified in both organisms (data not shown). To examine this further, nucleoside modifications in tRNA from M. burtonii were examined by liquid chromatography mass spectrometry (LC-MS), and a significant amount of dihydrouridine was detected, a component which had not previously been identified in other methanogens (McCloskey et al. 2001; K. Noon, R. Guymon, P. Crain, J. McCloskey, M. Thomm, J. Lim, R. Cavicchioli, in prep.). This further highlights the specific adaptation strategies which have evolved in cold-adapted Archaea.
METHODS
Genome Sequencing and Assembly
M.burtonii was grown at 23°C (Franzmann et al. 1992) and M. frigidum at 15°C (Franzmann et al. 1997). Small-insert (2–4 and 4–7 kb) libraries of M. frigidum DNA, prepared by Sau3AI partial digestion of genomic DNA cloned in pUC18, were sequenced at the Australian Genome Research Facility and Amersham Biosciences. M. burtonii small- and large-insert libraries were prepared and sequenced at the Joint Genome Institute (JGI; http://www.jgi.doe.gov/Internal/prots_index.html). Sequencing was carried out by using dye terminator reactions and ABI 377, ABI 3700, and MegaBACE 4000 sequencers. Base calling and shotgun assembly were performed using Phred (Ewing et al. 1998) and Phrap (P. Green, unpubl.) and the JGI JAZZ assembler for M. burtonii. The Staden package (Dear and Staden 1991) was used where inspection and editing of the assembly were required.
Prediction and Analysis of ORFs
Critica (Badger and Olsen 1999), Glimmer (Delcher et al. 1999), and Generation were used to identify putative coding regions, and contigs were searched using tBLASTN to identify genes that may have been missed. Because our genome sequences are at a draft stage, we defined three data sets. Coding regions were sequences derived from raw output of gene finding. These sequences can contain nonstandard amino acid letters (due to, e.g., frameshifts), but were used for initial BLAST searches. Putative ORFs were coding regions that contained in-frame sequence between start and stop codons. These may be truncated due to errors in sequencing or ORF prediction, but can be used for, for example, threading. Stringent ORFs were defined as ORFs that had a BLAST hit (< 1e-10) in the COG database (Tatusov et al. 2001) where the sequences differ in length by < 10%. These were considered genuine ORFs and used where full-length, error-free sequences were required. The naming scheme for M. burtonii ORFs was ScaffoldXX.geneYY. M. frigidum ORFs were named ContigXX.YY.ZZ, where XX is the contig number and YY.ZZ represents the start and end positions of the ORF (bp) within the contig. Scripts for data analysis were written using the BioPerl modules (Stajich et al. 2002). The M. burtonii and M. frigidum data are available at the JGI (http://www.jgi.doe.gov/JGI_microbial/html/index.html) or at our Web site (http://psychro.bioinformatics.unsw.edu.au/), and sequences are to be submitted to the public databases.
Analysis of Amino Acid Composition
Predicted protein sequences in Fasta format from complete archaeal genomes were downloaded from the NCBI. Sequence data were obtained from the JGI (M. barkeri and Ferroplasma acidarmanus) and from the University of Washington Genome Center (M. maripaludis, http://www.genome.washington.edu/UWGC/methanococus/). In the latter case, Critica was used to predict coding regions, as only contig sequence data were available. Values for OGT and genome GC content were found at the TIGR CMR (http://www.tigr.org/tigr-scripts/CMR2/CMRHomePage.spl) and from a previous study (Galtier and Lobry 1997). For draft genomes, stringent ORF data sets were used and translated to protein sequence. Amino acid composition for each sequence set was calculated using a Perl script. Very similar compositions were obtained regardless of whether the stringent ORF or larger putative ORF data sets were used. Principal components analysis (PCA) was performed using the packages prcomp and princomp, part of the R statistical analysis package (Ihaka and Gentleman 1996). The input data were a matrix of 20 columns (percentage composition of each amino acid in the sequence set) with 21 rows (organisms).
Threading
PROSPECT version 2.0 beta (Xu and Xu 2000) was used for threading. Folds were assigned to novel proteins by threading sequences against templates from the SCOP database (Murzin et al. 1995) using global-local alignment. To identify potential cold shock domain (CSD-like) folds in proteins, all putative ORFs from M. frigidum and M. burtonii were threaded against a nonredundant set of CSDs from SCOP using global alignment. Sequences annotated as hypothetical or conserved hypothetical and with a z-score of >6 were then threaded against the complete set of SCOP domains using global-local alignment. Threading output was ranked by z-score and examined for significant scores against CSDs.
Comparative Homology Modeling and Analysis
Putative ORFs from the five complete and four incomplete methanogen genomes were used to search the current release of the PDB using BLAST (cutoff e-10). A parser was written in Perl to extract query sequences with >40% identity and length +/-30% or 30 residues of the BLAST hit, plus the PDB code of the top hit. A nonredundant list of PDB codes was compiled, and the corresponding structures downloaded from the PDB. The PROSPECT program make_template was used to generate threading templates not already present in the supplied PROSPECT database. PROSPECT was used to align query sequences to their best PDB template, taking into account sequence similarity (PSI-BLAST matrix) and predicted secondary structure (PHD prediction). Output from the threading alignment was processed using modellerProspect, to generate input files for MODELLER version 6v2 (Marti-Renom et al. 2000). Secondary structure in the protein models was analyzed using DSSP (Kabsch and Sander 1983). Salt bridges were calculated using the WWW interface to WHATIF (Rodriguez et al. 1998). The program naccess (Hubbard and Thornton 1993) was used for area accessibility calculations using the defaults for probe size (1.40Å) and van der Waals radii. The total solvent-accessible and -inaccessible areas were calculated in each case. The contribution of a residue type to either the solvent-accessible or -inaccessible area was defined as the summed accessible or inaccessible area of that residue type divided by the total accessible or inaccessible area, respectively.
Prediction and Analysis of tRNA
tRNA sequences in archaeal genomes were predicted using tRNAScan-SE (Lowe and Eddy 1997). RNAfold, part of the Vienna RNA package (Hofacker et al. 1994) was used to predict tRNA secondary structure.
Acknowledgments
Thanks to Jim McCloskey for data on M. burtonii tRNA, Greg Tyrelle, Steve Harrop, Mark Tanaka, and Tassia Kolesnikow for helpful discussions. This work was supported by the Australian Research Council and the United States Department of Energy.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1180903.
Footnotes
Article published online before print in June 2003.
References
- Badger, J.H. and Olsen, G.J. 1999. CRITICA: Coding region identification tool invoking comparative analysis. Mol. Biol. Evol. 16: 512-524. [DOI] [PubMed] [Google Scholar]
- Beja, O., Koonin, E.V., Aravind, L., Taylor, L.T., Seitz, H., Stein, J.L., Bensen, D.C., Feldman, R.A., Swanson, R.V., and DeLong, E.F. 2002. Comparative genomic analysis of archaeal genotypic variants in a single population and in two different oceanic provinces. Appl. Environ. Microbiol. 68: 335-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, A.C., Xu, J., Johnson, R.C., Schimmel, P., and de Crecy-Lagard, V. 2002. Identification of the tRNA-dihydrouridine synthase family. J. Biol. Chem. 277: 25090-25095. [DOI] [PubMed] [Google Scholar]
- Bult, C.J., White, O., Olsen, G.J., Zhou, L., Fleischmann, R.D., Sutton, G.G., Blake, J.A., FitzGerald, L.M., Clayton, R.A., Gocayne, J.D., et al. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273: 1058-1073. [DOI] [PubMed] [Google Scholar]
- Cambillau, C. and Claverie, J.-M. 2000. Structural and genomic correlates of hyperthermostability. J. Biol. Chem. 275: 32383-32386. [DOI] [PubMed] [Google Scholar]
- Cavicchioli, R., Thomas, T., and Curmi, P.M.G. 2000. Cold stress response in Archaea. Extremophiles 4: 321-331. [DOI] [PubMed] [Google Scholar]
- Cavicchioli, R., Saunders, N., and Thomas, T. 2002. In Encyclopedia of life support systems (EOLSS), Eolss Publishers, Oxford, UK (http://www.eolss.net).
- Chong, S.C., Liu, Y., Cummins, M., Valentine, D.L., and Boone, D.R. 2002. Methanogenium marinum sp. nov., a H2-using methanogen from Skan Bay, Alaska, and kinetics of H2 utilization. Antonie van Leeuwenhoek 81: 263-270. [DOI] [PubMed] [Google Scholar]
- Dalluge, J.J., Hamamoto, T., Horikoshi, K., Morita, R.Y., Stetter, K.O., and McCloskey, J.A. 1997. Posttranscriptional modification of tRNA in psychrophilic bacteria. J. Bacteriol. 179: 1918-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dear, S. and Staden, R. 1991. A sequence assembly and editing program for efficient management of large projects. Nucleic Acids Res. 19: 3907-3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher, A.L., Harmon, D., Kasif, S., White, O., and Salzberg, S.L. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27: 4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deppenmeier, U., Johann, A., Hartsch, T., Merkl, R., Schmitz, R.A., Martinez-Arias, R., Henne, A., Wiezer, A., Baumer, S., Jacobi, C., et al. 2002. The genome of Methanosarcina mazei: Evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 4: 453-461. [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175-185. [DOI] [PubMed] [Google Scholar]
- Franzmann, P.D., Springer, N., Ludwig, W., Conway de Macario, E., and Rohde, M. 1992. A methanogenic archaeon from Ace Lake, Antarctica: Methanococcoides burtonii sp.nov. Syst. Appl. Microbiol. 15: 573-581. [Google Scholar]
- Franzmann, P.D., Liu, Y., Balkwill, D.L., Aldrich, H.C., Conway de Macario, E., and Boone D.R. 1997. Methanogenium frigidum sp. nov., a psychrophilic, H2-using methanogen from Ace Lake, Antarctica. Int. J. Syst. Bacteriol. 47: 1068-1072. [DOI] [PubMed] [Google Scholar]
- Gajiwala, K.S. and Burley, S.K. 2000. Winged helix proteins. Curr. Opin. Struct. Biol. 10: 110-116. [DOI] [PubMed] [Google Scholar]
- Galagan, J.E., Nusbaum, C., Roy, A., Endrizzi, M.G., Macdonald, P., FitzHugh, W., Calvo, S., Engels, R., Smirnov, S., Atnoor, D., et al. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12: 532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier, N. and Lobry, J.R. 1997. Relationships between genomic G+C content, RNA secondary structures, and optimal growth temperature in prokaryotes. J. Mol. Evol. 44: 632-636. [DOI] [PubMed] [Google Scholar]
- Gianese, G., Argos, P., and Pascarella, S. 2001. Structural adaptation of enzymes to low temperatures. Protein Eng. 14: 141-148. [DOI] [PubMed] [Google Scholar]
- Gianese, G., Bossa, F., and Pascarella, S. 2002. Comparative structural analysis of psychrophilic and meso- and thermophilic enzymes. Proteins 47: 236-249. [DOI] [PubMed] [Google Scholar]
- Graumann, P.L. and Marahiel, M.A. 1998. A superfamily of proteins that contain the cold-shock domain. Trends Biochem. Sci. 23: 286-290. [DOI] [PubMed] [Google Scholar]
- Graziano, G., Catanzano, F., Riccio, A., and Barone G. 1997. A reassessment of the molecular origin of cold denaturation. J. Biochem. 122: 395-401. [DOI] [PubMed] [Google Scholar]
- Haney, P.J., Badger, J.H., Buldak, G.L., Reich, C.I., Woese, C.R., and Olsen, G.J. 1999. Thermal adaptation analyzed by comparison of protein sequences from mesophilic and extremely thermophilic Methanococcus species. Proc. Natl. Acad. Sci. 96: 3578-3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker, I.L., Fontana, W., Stadler, P.F., Bonhoeffer, L.S., Tacker, M., and Schuster, P. 1994. Fast folding and comparison of RNA secondary structures. Monatsh. Chem. 125: 167-188. [Google Scholar]
- Hubbard, S.J. and Thornton, J.M. 1993. naccess computer program, Department of Biochemistry and Molecular Biology, University College London, UK.
- Ihaka, R. and Gentleman, R. 1996. R: A language for data analysis and graphics. J. Comp. Graph. Stat. 5: 299-314. [Google Scholar]
- Kabsch, W. and Sander, C. 1983. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22: 2577-2637. [DOI] [PubMed] [Google Scholar]
- Karner, M.B., DeLong, E.F., and Karl, D.M. 2001. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409: 507-510. [DOI] [PubMed] [Google Scholar]
- Kennedy, S.P., Ng, W.V., Salzberg, S.L., Hood, L., and DasSarma, S. 2001. Understanding the adaptation of Halobacterium species NRC-1 to its extreme environment through computational analysis of its genome sequence. Genome Res. 11: 1641-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreil, D.P. and Ouzounis, C.A. 2001. Identification of thermophilic species by the amino acid compositions deduced from their genomes. Nucleic Acids Res. 29: 1608-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, J., Thomas, T., and Cavicchioli, R. 2000. Low temperature regulated DEAD-box RNA helicase from the Antarctic archaeon, Methanococcoides burtonii. J. Mol. Biol. 297: 553-567. [DOI] [PubMed] [Google Scholar]
- Lowe, T.M. and Eddy, S.R. 1997. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti-Renom, M.A., Stuart, A., Fiser, A., Sánchez, R., Melo, F., and Sali, A. 2000. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29: 291-325. [DOI] [PubMed] [Google Scholar]
- McCloskey, J.A., Graham, D.E., Zhou, S., Crain, P.F., Ibba, M., Konisky, J., Soll, D., and Olsen, G.J. 2001. Posttranscriptional modification in archaeal tRNAs: Identities and phylogenetic relations of nucleotides from mesophilic and hyperthermophilic Methanococcales. Nucleic Acids Res. 29: 4699-4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin, A.G., Brenner, S.E., Hubbard, T., and Chothia, C. 1995. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247: 536-540. [DOI] [PubMed] [Google Scholar]
- Palma, R. and Curmi, P.M.G. 1999. Computational studies on mutant protein stability: The correlation between surface thermal expansion and protein stability. Protein Sci. 8: 913-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees, D.C. 2001. Crystallographic analyses of hyperthermophilic proteins. Methods Enzymol. 334: 423-437. [DOI] [PubMed] [Google Scholar]
- Rees, D.C. and Robertson, A.D. 2001. Some thermodynamic implications for the thermostability of proteins. Protein Sci. 10: 1187-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, R., Chinea, G., Lopez, N., Pons, T., and Vriend, G. 1998. Homology modeling, model and software evaluation: Three related resources. Comput. Appl. Biosci. 14: 523-528. [DOI] [PubMed] [Google Scholar]
- Russell, N.J. 2000. Toward a molecular understanding of cold activity of enzymes from psychrophiles. Extremophiles 4: 83-90. [DOI] [PubMed] [Google Scholar]
- Siddiqui, K.S., Cavicchioli, R., and Thomas, T. 2002. Thermodynamic activation properties of elongation factor 2 (EF-2) proteins from psychrotolerant and thermophilic Archaea. Extremophiles 6: 143-150. [DOI] [PubMed] [Google Scholar]
- Simankova, M.V., Parshina, S.N., Tourova, T.P., Kolganova, T.V., Zehnder, A.J.B., and Nozhevnikova, A.N. 2001. Methanosarcina lacustris sp. nov., a new psychrotolerant methanogenic archaeon from anoxic lake sediments. Syst. Appl. Microbiol. 24: 362-367. [DOI] [PubMed] [Google Scholar]
- Slesarev, A.I., Mezhevaya, K.V., Makarova, K.S., Polushin, N.N., Shcherbinina, O.V., Shakhova, V.V., Belova, G.I., Aravind, L., Natale, D.A., Rogozin, I.B., et al. 2002. The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc. Natl. Acad. Sci. 99: 4644-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D.R., Doucette-Stamm, L.A., Deloughery, C., Lee, H., Dubois, J., Aldredge, T., Bashirzadeh, R., Blakely, D., Cook, R., Gilbert, K., et al. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum δH: Functional analysis and comparative genomics. J. Bacteriol. 179: 7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommerville, J. 1999. Activities of cold-shock domain proteins in translation control. Bioessays 21: 319-325. [DOI] [PubMed] [Google Scholar]
- Stajich, J.E., Block, D., Boule, K., Brenner, S.E., Chervitz, S.A., Dagdigian, C., Fuellen, G., Gilbert, J.G., Korf, I., Lapp, H., et al. 2002. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 12: 1611-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov, R.L., Natale, D.A., Garkavtsev, I.V., Tatusova, T.A., Shankavaram, U.T., Rao, B.S., Kiryutin, B., Galperin, M.Y., Fedorova, N.D., and Koonin, E.V. 2001. The COG database: New developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29: 22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekaia, F, Yeramian, E., and Dujon, B. 2002. Amino acid composition of genomes, lifestyles of organisms, and evolutionary trends: A global picture with correspondence analysis. Gene 297: 51-60. [DOI] [PubMed] [Google Scholar]
- Thomas, T. and Cavicchioli, R. 1998. Archaeal cold-adapted proteins: Structural and evolutionary analysis of the elongation factor 2 proteins from psychrophilic, mesophilic and thermophilic methanogens. FEBS Lett. 439: 281-286. [DOI] [PubMed] [Google Scholar]
- Thomas, T. and Cavicchioli, R. 2000. Effect of temperature on stability and activity of elongation factor 2 proteins from Antarctic and thermophilic methanogens. J. Bacteriol. 182: 1328-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, T. and Cavicchioli, R. 2002. Cold adaptation of archaeal elongation factor 2 (EF-2) proteins. Curr. Prot. Pep. Sci. 3: 223-230. [DOI] [PubMed] [Google Scholar]
- Thomas, T., Kumar, N., and Cavicchioli, R. 2001. Effects of ribosomes and intracellular solutes on activities and stabilities of elongation factor 2 proteins from psychrotolerant and thermophilic methanogens. J. Bacteriol. 183: 1974-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Klein, D., Arab, H., Volker, H., and Thomm, M. 2002. Methanosarcina baltica, sp. nov., a novel methanogen isolated from the Gotland Deep of the Baltic Sea. Extremophiles 6: 103-110. [DOI] [PubMed] [Google Scholar]
- Wright, H.T. 1991. Nonenzymatic deamidation of asparaginyl and glutaminyl residues in proteins. Crit. Rev. Biochem. Mol. Biol. 26: 1-52. [DOI] [PubMed] [Google Scholar]
- Xia, B., Ke, H. and Inouye, M. 2001. Acquirement of cold sensitivity by quadruple deletion of the cspA family and its suppression by PNPase S1 domain in Escherichia coli. Mol. Microbiol. 40: 179-188. [DOI] [PubMed] [Google Scholar]
- Xu, Y. and Xu, D. 2000. Protein threading using PROSPECT: Design and evaluation. Prot. 40: 343-354. [PubMed] [Google Scholar]
- Zecchinon, L., Claverie, P., Collins, T., D'Amico, S., Delille, D., Feller, G., Georlette, D., Gratia, E., Hoyoux, A., Meuwis, M.-A., et al. 2001. Did psychrophilic enzymes really win the challenge? Extremophiles 5: 313-321. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.jgi.doe.gov/Internal/prots_index.html; JGI sequencing protocols.
- http://www.jgi.doe.gov/JGI_microbial/html/index.html; JGI microbial genomes.
- http://psychro.bioinformatics.unsw.edu.au/; Cavicchioli lab bioinformatics site.
- http://www.genome.washington.edu/UWGC/methanococus/; UWGC Methanococcus site.
- http://www.tigr.org/tigr-scripts/CMR2/CMRHomePage.spl; TIGR CMR database.