Abstract
Reverse transcription quantitative polymerase chain reaction (RT-qPCR) has been recognized as the most accurate method for quantifying mRNA transcripts, but normalization of samples is a prerequisite for correct data interpretation. So, this study aimed to evaluate the most stable reference gene for RT-qPCR in human normal thyroid and goiter tissues. Beta-actin (ACTB); glyceraldehyde-3-phosphate dehydrogenase (GAPDH); succinate dehydrogenase, subunit A, flavoprotein (Fp) (SDHA); hypoxanthine phosphoribosyltransferase I (HPRTI); tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide (YWHAZ); and beta-2-microglobulin (B2M) were evaluated in 14 thyroid tissue samples (7 normal and 7 goiter tissues) by RT-qPCR. The mean Cq and the maximum fold change (MFC) and NormFinder software were used to assess the stability of the genes. As a result, ACTB gene was more stable than GAPDH, SDHA, HPRTI, YWHAZ, and B2M. In conclusion, ACTB could be used to normalize RT-qPCR data in normal thyroid and goiter tissues.
1. Introduction
Reverse transcription quantitative polymerase chain reaction (RT-qPCR) has been recognized as the most accurate, sensitive, and easy method for quantifying mRNA transcripts in biological samples [1, 2]. Nevertheless, normalization of samples is a prerequisite for correct data interpretation, since its accuracy is significantly affected by sample quality, reagent, and operator technique [3].
Cell number, quantification of rRNA or total RNA, and reference genes are some standardization methods that have been proposed so far [2, 4, 5]. Normalization with another gene, which is assumed to have stable expression, is the method of choice because this reference gene is exposed to the same preparation steps as the gene of interest [6]. Therefore, a full procedure control is obtained during the whole assay.
A good reference gene needs to present low variation in its expression in the different samples which would be evaluated and demonstrated to be minimally regulated during the individual experiment or pathological condition. Therefore, there is a need to validate genes to allow the accuracy of RNA transcription analysis [3, 6]. Nowadays, there is emerging evidence that reference genes could vary their expression in different cellular development stages and in other conditions [5, 7, 8].
The thyroid normal gland is a fairly homogenous structure, but in goiter a genetic heterogeneity is described in follicular cells; some nodules have a polyclonal origin and others a monoclonal origin. These changes may be related to mutations in oncogenes which do not produce malignancy per se but would alter growth and function [14, 15]. These alterations could modify the expression of many thyroid genes, including the ones usually described as reference genes, so the evaluation of candidates to normalize RT-qPCR is necessary.
Little is known about the expression stability of reference genes in thyroid cells or tissue, as shown in Table 1.
Table 1.
Stability of genes candidates to normalize RT-qPCR for thyroid derived biological material.
| Study | Material | Reference gene stability | Method |
|---|---|---|---|
| Santin et al., 2013 [9] | Human normal cells | ACTB>TBP>GAPDH>B2M | NormFinder |
| Chantawibul et al., 2012 [10] | Human tissues* | GAPDH>ACTB>B2M>RPLIA>HPRT1 | NormFinder |
| Lecchi et al., 2012 [11] | Bovine tissues, including thyroid | SF3A1=HBMS>ACTB | geNorm |
| HMBS>ACTB>GAPDH | NormFinder | ||
| Li et al., 2011 [12] | Porcine thyroid tissue | 18S rRNA>Ubiquitin>Histone H3>ACTB | geNorm |
| 18S rRNA>Ubiquitin>Histone H3>ACTB>GAPDH | NormFinder | ||
| Lisowski et al., 2008 [13] | Bovine thyroid tissue | TBP=HPRT1>YWHAZ>ACTB | geNorm |
*13 goiters, 6 adenomas, 4 carcinomas (2 follicular + 2 papillary), and 2 lymphocytic thyroiditis.
Considering the species, there are many differences in reference genes stability evaluated in these studies. Although in human tissues the same software was used to analyze data, differences in RT-qPCR and cells status could have contributed to these findings. In addition, a stable expression of a reference gene in one tumor type does not predict a stable expression in another tumor type [7]. In this way, there is no universal reference gene being necessary to validate potential reference genes for any experimental condition. So, this study aimed to evaluate the most stable reference gene for RT-qPCR in human normal thyroid and goiter tissues.
2. Material and Methods
2.1. Tissue Acquisition
Normal thyroid and goiter tissues were obtained from patients who underwent total thyroidectomy as part of treatment for differentiated thyroid cancer in the Hospital de Clínicas de Porto Alegre (HCPA). After the evaluation of macroscopic and frozen sections of surgical specimens by two pathologists, samples of normal thyroid or goiter tissue were frozen in liquid nitrogen and stored at −80°C until further processing. The pattern used to confirm the presence of goiter was a well-defined fibrous capsule with a mixture of macrofollicles and microfollicles and, in some cases, some degenerative changes such as fibrosis and hemorrhage [16]. This study was approved by the Ethics Committee of HCPA (GPPG: 12-0272).
2.2. Nucleic Acid Extraction and Reverse Transcription
Normal thyroid and goiter tissue were homogenized mechanically with Omnimix for 30 seconds in Trizol (Invitrogen, Life Technologies, Carlsbad, CA, USA), and total RNA was extracted with the same commercial kit, according to the manufacturer's protocol. The RNA concentration and purity were assessed with the NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). Samples were then stored at −80°C. 1 μg total RNA was reverse-transcribed to produce cDNA using random and oligo-dT primers and Superscript II reverse transcriptase (Invitrogen Life Technologies) in total reaction volumes of 20 μL. All cDNA samples were diluted 10-fold with diethyl pyrocarbonate- (DEPC-) treated water and stored at −20°C.
2.3. Gene Selection and Quantitative PCR
Six genes commonly used as reference in RT-qPCR gene expression experiments were selected (Table 1): beta-actin (ACTB, related to cell structure); glyceraldehyde-3-phosphate dehydrogenase (GAPDH, related to carbohydrate metabolism); succinate dehydrogenase, subunit A, flavoprotein (Fp) (SDHA, related to energy metabolism); hypoxanthine phosphoribosyltransferase I (HPRTI, related to nucleotide metabolism); tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide (YWHAZ, related to cell growth and death); and beta-2-microglobulin (B2M, related to major histocompatibility complex). Primers sequence shown in Table 2 was described previously [17]. Quantification of amplified samples was performed based on a standard curve with five successive tenfold dilution points of a pool of cDNA samples.
Table 2.
Primers sequence, product length, and NCBI reference sequence for reference genes by RT-qPCR.
| Gene symbol | Name | Forward and reverse primers | Product length (bp) | NCBI reference sequence |
|---|---|---|---|---|
| ACTB | Beta-actin | 5′-CTGGAACGGTGAAGGTGACA-3′ | 140 | NM_001101.3 |
| 5′-AAGGGACTTCCTGTAACAATGCA-3′ | ||||
|
| ||||
| B2M | Beta-2-microglobulin | 5′-CTATCCAGCGTACTCCAAAG-3′ | 165 | NM_004048.2 |
| 5′-ACAAGTCTGAATGCTCCACT-3′ | ||||
|
| ||||
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 5′-CTTTGTCAAGCTCATTTCCTGG-3′ | 133 | NM_002046.3 |
| 5′-TCTTCCTCTTGTGCTCTTGC-3′ | ||||
|
| ||||
| HPRT1 | Hypoxanthine phosphoribosyltransferase 1 | 5′-AGATGGTCAAGGTCGCAAG-3′ | 128 | NM_000194.2 |
| 5′-GTATTCATTATAGTCAAGGGCATATCC-3′ | ||||
|
| ||||
| SDHA | Succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 5′-TGGTTGTCTTTGGTCGGG-3′ | 85 | NM_004168.2 |
| 5′-GCGTTTGGTTTAATTGGAGGG-3′ | ||||
|
| ||||
| YWHAZ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | 5′-CAACACATCCTATCAGACTGGG-3′ 5′-AATGTATCAAGTTCAGCAATGGC-3′ |
133 | NM_001135699.1 |
bp: base pair. Modified from Souza et al., 2012 [17].
RT-qPCR was performed using Applied Biosystems StepOne Real-Time PCR System using Kit Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen Life Technologies, Carlsbad, CA, USA). The RT-qPCR was performed with an initial denaturation (10 min at 95°C), followed by 40 cycles of denaturation (15 s at 95°C), annealing, and extension (45 s at 60°C). Melting curve was acquired to ensure that a single product was amplified in each reaction.
2.4. Statistics
To evaluate the stability of the candidate reference genes in thyroid normal and goiter tissues, the NormFinder algorithm was used [18]. In addition, the analysis of raw quantification cycle (Cq) values of each gene was used to evaluate their stability. Mean Cq values, standard deviation (SD), coefficient of variation (CV), and maximum fold change (MFC, the ratio of the maximum and minimum values observed within the dataset) were calculated.
3. Results
Six reference genes were amplified in 14 thyroid samples: 7 normal and 7 goiter. All RT-qPCR assays produced a single peak in the melting curve. The mean Cq cycle values, SD, CV, and MFC obtained for each gene in goiter and normal thyroid are described in Table 3. Analyzing these data, none of the genes had a constant expression. Moreover, B2M showed higher variability in Cq values. Thereby, we proceeded the analysis of quantification data using the NormFinder algorithm. The values of stability of the candidate genes obtained from the NormFinder analysis are shown in Table 4. The most stable genes were ACTB for goiter and GAPDH for normal thyroid tissue. When all samples were analyzed together, the most stable genes were SDHA and ACTB. Stability values and intra/intergroup variations are shown, respectively, in Table 5 and Figure 1.
Table 3.
Dispersion data of raw Cq values for candidate reference genes in goiter and normal thyroid tissue.
| Tissue | Symbol gene | Mean Cq | SD | CV (%) | Minimum | Maximum | MFC |
|---|---|---|---|---|---|---|---|
| Goiter | ACTB | 21.88 | 1.78 | 8.1 | 19.78 | 25.00 | 1.26 |
| B2M | 20.83 | 2.59 | 9.3 | 18.10 | 26.11 | 1.44 | |
| GAPDH | 21.18 | 1.95 | 9.2 | 19.54 | 25.60 | 1.31 | |
| HPRT1 | 30.10 | 3.10 | 10.3 | 25.48 | 35.24 | 1.38 | |
| SDHA | 31.17 | 2.21 | 7.1 | 29.51 | 34.56 | 1.17 | |
| YWHAY | 27.48 | 2.47 | 9.0 | 22.71 | 30.93 | 1.36 | |
|
| |||||||
| Normal | ACTB | 23.56 | 1.92 | 8.1 | 19.99 | 25.69 | 1.29 |
| B2M | 24.63 | 5.08 | 20.6 | 18.53 | 32.65 | 1.76 | |
| GAPDH | 22.84 | 1.78 | 7.8 | 20.54 | 26.20 | 1.28 | |
| HPRT1 | 32.12 | 3.06 | 9.5 | 25.65 | 35.66 | 1.39 | |
| SDHA | 33.94 | 3.20 | 9.4 | 29.54 | 39.39 | 1.33 | |
| YWHAY | 26.29 | 2.35 | 8.9 | 22.83 | 29.25 | 1.28 | |
SD: standard deviation; CV: coefficient of variation; and MFC: maximum fold change (the ratio of the maximum and minimum values).
Table 4.
Stability values of reference genes candidates calculated by the NormFinder software for each tissue.
| Tissue | Symbol gene | Stability value |
|---|---|---|
| Goiter | ACTB | 0.228 |
| SDHA | 0.298 | |
| HPRT1 | 0.567 | |
| GAPDH | 0.685 | |
| YWHAZ | 0.688 | |
| B2M | 1.711 | |
|
| ||
| Normal | GAPDH | 0.114 |
| SDHA | 0.114 | |
| ACTB | 0.221 | |
| HPRT1 | 0.643 | |
| YWHAZ | 1.026 | |
| B2M | 2.867 | |
Table 5.
Stability values of reference candidate genes calculated by the NormFinder software for all tissues.
| Symbol gene | Stability value |
|---|---|
| SDHA | 0.076 |
| ACTB | 0.082 |
| GAPDH | 0.150 |
| HPRT1 | 0.221 |
| YWHAZ | 0.311 |
| B2M | 0.830 |
Figure 1.
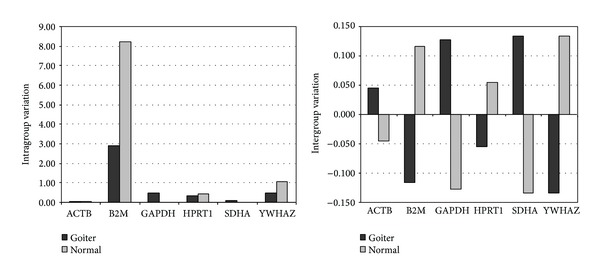
Intra- and intergroup variation of six candidate reference genes in goiter and normal thyroid tissues samples.
4. Discussion
In this study, we evaluated 6 reference genes in 14 thyroid specimens (7 normal thyroid and 7 goiter) using RT-qPCR. The genes with the lowest variations in expression for normal samples, goiter, and all samples were, respectively, GAPDH, ACTB, SDHA and ACTB, when evaluated by the NormFinder.
According to de Jonge et al. [19], a good candidate gene should present a small coefficient of variation and a MFC < 2. Moreover, a mean expression level lower than the maximum expression level subtracted with 2 SD is a prerequisite for a candidate reference gene. By these criteria, only B2M and GAPDH genes could be used as internal control for goiter, and none of these genes could be used for normal thyroid, although all had MFC < 2.
The NormFinder algorithm was used to rank the candidate reference genes based on their stability values. The gene expression is more stable when this value is closer to zero. The stability value less than 0.15 is the cutoff for an acceptable reference gene [4]. Considering these criteria, none of the selected genes could be used to normalize RT-qPCR data for goiter tissues, and GAPDH and SDHA could be used for normal thyroid. When all data were analyzed together, SDHA and ACTB genes had the lowest stability values, respectively, 0.076 and 0.082. We suggest using ACTB as reference gene, when comparing gene expression in normal thyroid and goiter, because this gene had the lowest intra- and intergroup variations. Our results differ somewhat from those of Chantawibul et al. [10] who found the highest stability value for HPRT1, while the GAPDH had the lowest stability value. These differences may partly be explained by the sample selection, which included goiters, adenomas, carcinomas, and lymphocytic thyroiditis.
ACTB mRNA is expressed at moderately abundant levels in most cell types and its protein plays a key role in the cytoskeleton maintenance. Recently, some studies have suggested that this gene does not satisfy the basic requirements for application as internal control due to change in expression under various biomedical stimuli [20, 21]. Nevertheless, in normal thyroid cells, ACTB expression appeared to be stable in response to different experimental treatments [9]. Other studies have demonstrated that ACTB can be used as reference gene [22–24] for different cell types.
One limitation of this study is the small number of samples of goiter; nevertheless, all samples were evaluated by two pathologists, who used the same criteria to define goiter.
In conclusion, the results of the present study suggest that ACTB gene is more stable than SDHA, GAPDH, HPRTI, YWHAZ, and B2M when evaluating human normal thyroid and goiter together.
Acknowledgments
The authors gratefully acknowledge FIPE/HCPA, CAPES/PROF, and CNPq for financial support.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnology Letters. 2004;26(6):509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 2.Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. Journal of Molecular Endocrinology. 2002;29(1):23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- 3.Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 4.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3(7) doi: 10.1186/gb-2002-3-7-research0034.RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thellin O, Zorzi W, Lakaye B, et al. Housekeeping genes as internal standards: use and limits. Journal of Biotechnology. 1999;75(2-3):291–295. doi: 10.1016/s0168-1656(99)00163-7. [DOI] [PubMed] [Google Scholar]
- 6.Radonić A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochemical and Biophysical Research Communications. 2004;313(4):856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- 7.Haller F, Kulle B, Schwager S, et al. Equivalence test in quantitative reverse transcription polymerase chain reaction: confirmation of reference genes suitable for normalization. Analytical Biochemistry. 2004;335(1):1–9. doi: 10.1016/j.ab.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 8.Dundas J, Ling M. Reference genes for measuring mRNA expression. Theory in Biosciences. 2012;131(4):215–223. doi: 10.1007/s12064-012-0152-5. [DOI] [PubMed] [Google Scholar]
- 9.Santin AP, Souza AF, Brum LS, Furlanetto TW. Validation of reference genes for normalizing gene expression in real-time quantitative reverse transcription PCR in human thyroid cells in primary culture treated with progesterone and estradiol. Molecular Biotechnology. 2013;54(2):278–282. doi: 10.1007/s12033-012-9565-0. [DOI] [PubMed] [Google Scholar]
- 10.Chantawibul S, Anuwong A, Leelawat K. Validation of appropriate reference genes for gene expression studies in human thyroid gland using real-time RT-PCR. Journal of the Medical Association of Thailand. 2012;95(supplement 3):S36–S40. [PubMed] [Google Scholar]
- 11.Lecchi C, Dilda F, Sartorelli P, Ceciliani F. Widespread expression of SAA and Hp RNA in bovine tissues after evaluation of suitable reference genes. Veterinary Immunology and Immunopathology. 2012;145(1-2):556–562. doi: 10.1016/j.vetimm.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 12.Li Q, Domig KJ, Ettle T, Windisch W, Mair C, Schedle K. Evaluation of potential reference genes for relative quantification by RT-qPCR in different porcine tissues derived from feeding studies. International Journal of Molecular Sciences. 2011;12(3):1727–1734. doi: 10.3390/ijms12031727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lisowski P, Pierzchała M, Gościk J, Pareek CS, Zwierzchowski L. Evaluation of reference genes for studies of gene expression in the bovine liver, kidney, pituitary, and thyroid. Journal of Applied Genetics. 2008;49(4):367–372. doi: 10.1007/BF03195635. [DOI] [PubMed] [Google Scholar]
- 14.Studer H, Peter HJ, Gerber H. Natural heterogeneity of thyroid cells: the basis for understanding thyroid function and nodular goiter growth. Endocrine Reviews. 1989;10(2):125–135. doi: 10.1210/edrv-10-2-125. [DOI] [PubMed] [Google Scholar]
- 15.Krohn K, Führer D, Bayer Y, et al. Molecular pathogenesis of euthyroid and toxic multinodular goiter. Endocrine Reviews. 2005;26(4):504–524. doi: 10.1210/er.2004-0005. [DOI] [PubMed] [Google Scholar]
- 16.Baloch ZW, LiVolsi VA. Follicular-patterned lesions of the thyroid: the bane of the pathologist. American Journal of Clinical Pathology. 2002;117(1):143–150. doi: 10.1309/8VL9-ECXY-NVMX-2RQF. [DOI] [PubMed] [Google Scholar]
- 17.Souza AF, Brum IS, Neto BS, Berger M, Branchini G. Reference gene for primary culture of prostate cancer cells. Molecular Biology Reports. 2012;40(4):2955–2962. doi: 10.1007/s11033-012-2366-5. [DOI] [PubMed] [Google Scholar]
- 18.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research. 2004;64(15):5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 19.de Jonge HJ, Fehrmann RSN, de Bont ESJM, et al. Evidence based selection of housekeeping genes. PLoS ONE. 2007;2(9, article e898) doi: 10.1371/journal.pone.0000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Selvey S, Thompson EW, Matthaei K, Lea RA, Irving MG, Griffiths LR. β-actin—an unsuitable internal control for RT-PCR. Molecular and Cellular Probes. 2001;15(5):307–311. doi: 10.1006/mcpr.2001.0376. [DOI] [PubMed] [Google Scholar]
- 21.Ruan W, Lai M. Actin, a reliable marker of internal control? Clinica Chimica Acta. 2007;385(1-2):1–5. doi: 10.1016/j.cca.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 22.Mori R, Wang Q, Danenberg KD, Pinski JK, Danenberg PV. Both β-actin and GAPDH are useful reference genes for normalization of quantitative RT-PCR in human FFPE tissue samples of prostate cancer. Prostate. 2008;68(14):1555–1560. doi: 10.1002/pros.20815. [DOI] [PubMed] [Google Scholar]
- 23.Mehta R, Birerdinc A, Hossain N, et al. Validation of endogenous reference genes for qRT-PCR analysis of human visceral adipose samples. BMC Molecular Biology. 2010;11, article 39 doi: 10.1186/1471-2199-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayashi R, Hayashi S, Arai K, Chikuda M, Obara Y. Effects of antioxidant supplementation on mRNA expression of glucose-6-phosphate dehydrogenase, β-actin and 18S rRNA in the anterior capsule of the lens in cataract patients. Experimental Eye Research. 2012;96(1):48–54. doi: 10.1016/j.exer.2012.01.001. [DOI] [PubMed] [Google Scholar]


