Abstract
Alcohol abuse and alcoholism incur a heavy socioeconomic cost in many countries. Both genetic and environmental factors contribute to variation in the inebriating effects of alcohol and alcohol addiction among individuals within and across populations. From a genetics perspective, alcohol sensitivity is a quantitative trait determined by the cumulative effects of multiple segregating genes and their interactions with the environment. This review summarizes insights from model organisms as well as human populations that represent our current understanding of the genetic and genomic underpinnings that govern alcohol metabolism and the sedative and addictive effects of alcohol on the nervous system.
Electronic supplementary material
The online version of this article (doi:10.1007/s00438-013-0808-y) contains supplementary material, which is available to authorized users.
Keywords: Addiction, Behavioral genetics, Genome-wide association, Quantitative trait loci, Meta-analysis
“Alcohol is the anaesthesia by which we endure the operation of life”.
George Bernard Shaw
Introduction
Alcoholic beverages have been around since time immemorial and have served economic, social, medical and religious purposes. Alcohol is unique among substance abuse drugs, as it is a natural by-product of fermentation. Moderate drinking of alcohol may offer health benefits (Marugame et al. 2007), including reduction in cardiovascular disease (Baer et al. 2002), ischemic strokes (Zeng et al. 2012; Peng et al. 2013), stress levels, incidence of type II diabetes (Conigrave et al. 2001; Koppes et al. 2005) and gallstone disease (Leitzmann et al. 1999). Excessive alcohol consumption, however, is a common cause of preventable death in most countries, and imposes a major socioeconomic burden. Alcohol abuse and dependence disorders are associated with marital instability, violent crime, fatal accidents, and injuries (Chick 2011). Heavy drinking is associated with increased risk of different types of cancer (Nelson et al. 2013; Touvier et al. 2013), higher cardiovascular disease mortality (Graff-Iversen et al. 2013), birth defects (Feldman et al. 2012), liver diseases (McCullough et al. 2011), and neuropsychiatric disorders (Rivas et al. 2013).
Effects of alcohol range from sedation, characterized by decreased awareness and ability to function, to addiction. The term “alcoholic” was introduced by the physician Magnus Huss in 1849 to describe alcohol addiction and can be defined as persistence of excessive drinking over a long period of time despite adverse health effects and disruption of social relations. Not all individuals who consume alcohol become alcoholics. Vulnerability to develop addiction depends on genetic, physiological and environmental conditions. Sustained alcohol intake can lead to functional alcohol tolerance, which enables increased alcohol consumption with fewer signs of intoxication. Alcohol tolerance allows escalation of drinking and eventually development of addiction. Alcohol “dependence” generally refers to physiological addiction, when cessation of alcohol intake precipitates withdrawal reactions, which range from anxiety and shakiness to severe complications, such as seizures and delirium tremens. The term “alcohol preference” refers to selectively bred strains of laboratory rats and mice that either prefer or avoid alcohol consumption. Alcohol preferring rodents voluntarily consume greater amounts of alcohol than non-preferring animals and have been studied extensively as models for alcohol addiction in humans.
Sensitivity to alcohol exposure varies among individuals within and across populations. From a genetics perspective, susceptibility to the inebriating effects of alcohol and alcohol addiction can be viewed as quantitative traits that result from the cumulative effects of multiple segregating genes and their interactions with the environment. Thus, there is no single locus that predisposes to alcohol abuse and dependence disorders, but rather many variants and their interactions with each other and the environment underlie alcohol-related phenotypes (Ducci and Goldman 2008; Morozova et al. 2012; Edenberg 2013). Disentangling the genetic and environmental contributions that shape alcohol-related phenotypes is complicated, because in human populations neither the genetic background nor the environment can be controlled precisely, results of excessive alcohol consumption are diverse, ranging from sedation to addiction and often confounded by neuropsychiatric conditions, and different studies have utilized different measurements to document alcohol-related phenotypes, such as frequency of drinks, total amount of alcohol consumed per drinking session, age at first drink, and withdrawal symptoms. This review integrates insights obtained from different model systems as well as human population studies to provide a comprehensive overview of the genetic factors that mediate sensitivity to alcohol.
Alcohol metabolism
The rate at which alcohol is metabolized and the nature and fate of its degradation products are important factors that determine its physiological effects. The primary pathway of ethanol metabolism in liver, and to a lesser extent in the stomach and intestinal tract, involves conversion of ethanol to acetaldehyde by alcohol dehydrogenase (ADH; Fig. 1), with acetaldehyde playing a major role in mediating aversive and rewarding effects of ethanol. Acetaldehyde is oxidized further to acetate by aldehyde dehydrogenase (ALDH; Fig. 1). The human genome contains five ADH classes with a total of seven closely related genes located on chromosome 4q. Candidate gene approaches, as well as family-based linkage studies together with genome-wide association studies (GWAS), have implicated many variants in and around the seven ADH genes that contribute to alcohol dependence or alcohol-related traits, including alleles of ADH1A, ADH1B, ADH1C, ADH2 and ADH4. While there are 18 genes encoding members of the ALDH enzyme family, only ALDH2 plays a major role in oxidizing acetaldehyde in the liver (Edenberg 2013).
Fig. 1.
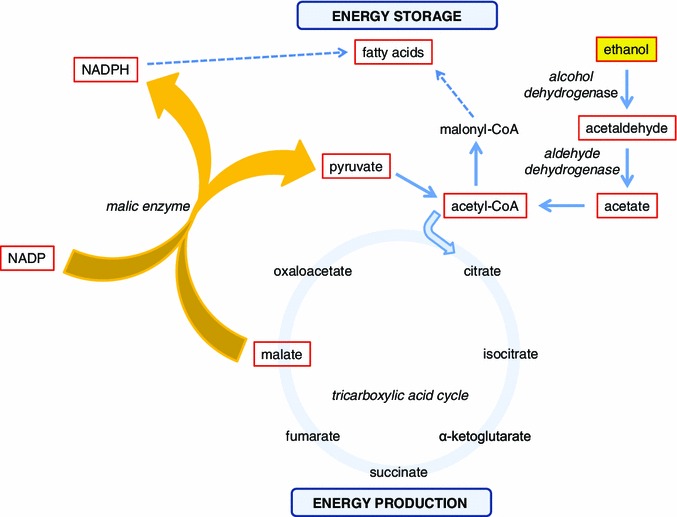
Schematic representation of the effects of ethanol on intermediary metabolism. The malic enzyme reaction is highlighted as a central mechanism that, in the presence of high ethanol concentrations, switches the flow of intermediary metabolism from energy production to energy storage by generating precursors for fatty acid biosynthesis. Central components in this pathway are illustrated in red bordered boxes
Alcoholism is less common in East Asian and Polynesian populations than in European populations, due to protective ADH and ALDH alleles. A variant of ADH1B (ADH1B*2; Arg47His), which occurs primarily in Asian and Polynesian populations, shows 100-fold higher enzymatic activity (Thomasson et al. 1995; Mulligan et al. 2003; Birley et al. 2009; Bierut et al. 2012; Hurley and Edenberg 2012), thereby attenuating the development of tolerance. Furthermore, the ALDH2*2 allele (Glu504Lys), which is also common in East Asia, is associated with aversive reactions to alcohol consumption, such as facial flushing, hypotension, headaches and nausea (Edenberg 2007). Polymorphisms in two other enzymes, ALDH1A1 (Lind et al. 2008) and ALDH1B1 (Linneberg et al. 2010), have also been associated with alcohol consumption in Finnish and Danish populations, respectively.
Studies on the Wistar-derived UChB line of rats, which were bred for high ethanol intake, replicated the protective ALDH2 phenotype observed in Asian populations following intravenous injection of an adenoviral vector with an Aldh2 antisense gene. This led to an 85 % decrease in ALDH2 activity in the liver and inhibited voluntary ethanol consumption up to 50 % (Ocaranza et al. 2008). Furthermore, studies with an adenoviral vector containing a multiple expression cassette showed that simultaneous increase of ADH and decrease of ALDH2 activities in the liver dramatically reduced voluntarily ethanol consumption of alcohol-dependent animals (Rivera-Meza et al. 2012).
Additional insights in the metabolism of alcohol come from studies on the fruit fly Drosophila melanogaster. Fruit flies encounter ethanol in their natural habitat, since larvae feed on fermented food sources, which provide substrates for lipid synthesis (Geer et al. 1985). They show preference for ethanol containing food over non ethanol containing food in laboratory experiments because of the caloric value of ethanol (Pohl et al. 2012). Both Adh and Aldh contribute to ethanol resistance in Drosophila (Fry and Saweikis 2006). There are two Adh alleles, designated Slow (AdhS) and Fast (AdhF) based on their electrophoretic mobility, that differ by a single amino acid (McDonald et al. 1980). Fast homozygotes have a higher level of enzymatic activity than Slow homozygotes and higher tolerance to alcohol in laboratory toxicity tests (McKenzie and McKechnie 1978). The physiological and behavioral aspects of ethanol in D. melanogaster have been reviewed (Kaun et al. 2012; Devineni and Heberlein 2013).
In addition to ADH, mutational analysis, artificial selection experiments, and analysis of transcript profiles following exposure to ethanol identified malic enzyme as a critical metabolic switch in response to alcohol exposure (Morozova et al. 2009). Malic enzyme converts malate, a Krebs cycle intermediate, to pyruvate, while generating NADPH. This provides the basic building blocks for the biosynthesis of fatty acids, thus switching the metabolism of energy-rich ethanol from combustion to lipid biosynthesis (Fig. 1). A similar pathway operates in people, where heavy drinking results in fatty liver disease, which can lead to inflammation and progress towards cirrhosis. Indeed an association study on the Framingham Offspring cohort revealed seven intronic SNPs in the cytoplasmic malic enzyme 1 gene that were associated with variation in cocktail drinking (Morozova et al. 2009).
In contrast to the liver, pathways for ethanol metabolism in the brain have been difficult to elucidate (Tabakoff and Hoffman 2013). Studies on rat brain homogenates suggest that ethanol metabolism proceeds here via catalase and cytochrome P450 (CYP2E1), which inactivate about 60–70 % and about 20 % of ethanol, respectively, via oxidation (Zimatkin et al. 2006). Studies on the UChB rats in which an antisense construct against catalase was delivered via a lentiviral vector in the ventral tegmental area (VTA) led to decreased levels of ALDH activity and abolished voluntary ethanol consumption (Karahanian et al. 2011; Tampier et al. 2013). When ADH was delivered into the VTA via a lentiviral vector alcohol intake increased significantly (Karahanian et al. 2011). Insights derived from these findings may contribute to the development of new strategies for the treatment of alcohol dependence in people.
Physiological effects of alcohol
Ethanol readily crosses the blood brain barrier and excessive alcohol consumption exerts both short-term and long-term effects on the nervous system. Acute effects are characterized by agitation and sedation, whereas long-term effects result in induction of tolerance and addiction. Different animal models can be utilized to optimally study these two aspects of alcohol consumption. Whereas alcohol-induced agitation and sedative effects of alcohol contribute in a major way to the socioeconomic cost of alcohol abuse (e.g., violence and drunk driving incidents, respectively), a vast proportion of human population studies has focused on genetic susceptibility to addiction. We will first demonstrate how D. melanogaster can serve as a powerful model to investigate the genetic basis of alcohol-induced sedation and how rodent models can be used to study alcohol addiction. We will then integrate studies from these model organisms with results from genetic studies on human populations.
A Drosophila model for alcohol sensitivity
Drosophila provides a powerful genetic model for studies on alcohol sensitivity, because large numbers of genetically identical individuals can be grown rapidly under controlled environmental conditions, and a wealth of community resources for genetic studies is available. Furthermore, flies exposed to ethanol undergo physiological and behavioral changes that resemble human alcohol intoxication. Assays have been developed to precisely quantify sensitivity to alcohol by measuring alcohol-induced knock-down time (Weber 1988). Low concentrations of ethanol stimulate locomotor activity, whereas high concentrations induce an intoxicated phenotype that shows marked similarities to human alcohol intoxication, characterized by locomotor impairments, loss of postural control, sedation and immobility (Singh and Heberlein 2000; Wolf et al. 2002). Ethanol provides an ecologically relevant chemical signal for flies to locate food and oviposition sites. Flies are attracted to low concentrations of ethanol via the olfactory system, but are repelled by high concentrations and this avoidance is mediated via gustatory perception (Devineni and Heberlein 2009). Conditions under which flies show preferential intake of ethanol have been reported and it has been proposed that such conditions could mimic aspects of addiction (Devineni and Heberlein 2009; Kaun et al. 2012; Peru y Colón de Portugal et al. 2013). Repeated exposure to ethanol induces tolerance in flies, similar to humans (Scholz et al. 2000).
Studies on flies have utilized two complementary strategies: single gene approaches aimed at the characterization of individual candidate genes identified through mutagenesis screens and systems genetics approaches to identify genetic networks associated with alcohol sensitivity. A P-element insertional mutagenesis screen for alcohol sensitivity revealed that almost 30 % of the P-element insertions tested affected the trait, indicating that a large fraction of the genome contributes to alcohol sensitivity and predicting extensive pleiotropy (Morozova et al. 2011; Fig. 2). Indeed, most of the mutations that affect alcohol sensitivity in Drosophila have pleiotropic effects on other complex traits (Harbison and Sehgal 2008; Edwards et al. 2009; Magwire et al. 2010).
Fig. 2.
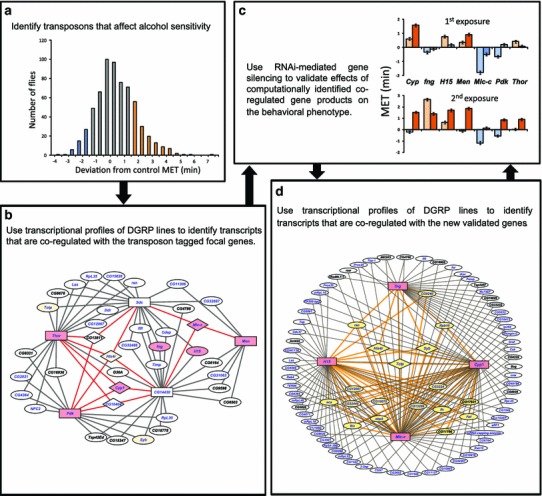
A strategy for the construction of genetic networks that underlie alcohol sensitivity in Drosophila melanogaster. The diagrams illustrate an iterative approach that combines P-element insertional mutagenesis (a), computational analysis of transcriptional profiles of DGRP lines (b, d) and RNAi-mediated target gene silencing (c) to generate a comprehensive network for alcohol sensitivity. a P-element insertional mutagenesis screen for alcohol sensitivity. Blue and orange colored bars designate lines with higher and lower alcohol sensitivity than control. b A network of co-regulated candidate genes associated with alcohol sensitivity. Rectangles indicate the most highly connected focal genes; genes targeted for validation studies are shown on pink and yellow backgrounds, and genes with annotated human orthologs are indicated in blue font. c Differences in mean elution time (MET) among RNAi lines after a first and a second exposure to ethanol. Blue bars indicate sensitive lines with MET lower than the control (P < 0.05); orange bars indicate resistant lines with MET higher than the control (P < 0.05); gray bars indicate no significant difference in MET. d An expanded network for alcohol sensitivity derived from highly interconnected genes from panel b is shown in pink boxes. Genes connected to two focal genes are shown at the periphery of the circle in white ovals and connected by gray lines. Genes interconnected by three and more networks are indicated on a yellow background and connected by orange lines. Diamond shapes indicate genes connected to all four focal genes. Genes with annotated human orthologs are indicated in blue. Modified from Morozova et al. (2011)
The effects of several mutants associated with alcohol sensitivity have been characterized in detail. From these studies cyclic AMP, PI3K/Akt and epidermal growth factor signaling pathways emerge as evolutionarily conserved signaling pathways that affect ethanol sensitivity in both flies and mammals (Eddison et al. 2011; Park et al. 2000; Neasta et al. 2011). Mutants associated with cyclic AMP signaling and alcohol sensitivity include cheapdate, an allele of amnesiac (Feany and Quinn 1995; Moore et al. 1998), the calcium/calmodulin-dependent adenylate cyclase rutabaga (Moore et al. 1998) and cyclic AMP-dependent protein kinase (Park et al. 2000). Arouser encodes a predicted adaptor protein homologous to the mammalian Epidermal Growth Factor Receptor Substrate 8 (Eps8) family (Eddison et al. 2011) and happy hour encodes a member of the Ste20 family of kinases that negatively regulate epidermal growth factor/extracellular signaling-related kinases (Corl et al. 2009).
Several synaptic neurotransmitter pathways have also been implicated in alcohol sensitivity, including signaling through dopamine (Bainton et al. 2000; Kong et al. 2010), the GABABR1 receptor (Dzitoyeva et al. 2003), neuropeptide F (Wen et al. 2005) and octopamine (Scholz et al. 2000; Rothenfluh and Heberlein 2002; Guarnieri and Heberlein 2003). In addition, the postsynaptic density protein Homer plays a role in regulation of synaptic plasticity and neuronal development (Foa and Gasperini 2009) and alcohol sensitivity, in both flies and mammals (Urizar et al. 2007; Cozzoli et al. 2009). Other mutants include slowpoke, which encodes a large-conductance calcium-activated potassium channel (Cowmeadow et al. 2005, 2006), and the transcription factors hangover (Scholz et al. 2005) and dLmo/Beadex. The mouse ortholog of dLmo/Beadex, Lmo3, also affects alcohol sensitivity (Lasek et al. 2011).
While “one-gene-at-a-time” studies can provide important information on the contributions of individual genes and evolutionarily conserved pathways to alcohol sensitivity, it is becoming increasingly clear that complex phenotypes are emergent features of dynamic networks of interacting genes (Ayroles et al. 2009; Morozova et al. 2012). Systems genetics approaches can generate insights in the relationship between DNA sequence variants, variation in transcript abundance and variation in organismal phenotype, gene-gene interactions and the effects of genetic or environmental perturbations on the composition of genetic networks (Mackay 2014).
Combining expression microarray analysis with artificial selection from a diverse base population identified candidate genes associated with alcohol sensitivity (Morozova et al. 2007). In addition, transcript profiles of isogenic Canton-S flies also identified differentially expressed genes following repeated alcohol exposures (Morozova et al. 2006). Acute exposure to concentrated ethanol vapor resulted in rapid down-regulation of genes affecting olfaction and up-regulation of genes encoding biotransformation enzymes. Subsequent development of tolerance was accompanied by changes in expression of transcriptional regulators, proteases and metabolic enzymes, including enzymes associated with fatty acid biosynthesis (Morozova et al. 2006).
Further insights in the complex underlying genetic architecture of alcohol sensitivity were obtained by capitalizing on whole genome transcriptional profiles of a subset of 40 lines from the D. melanogaster Genetic Reference Panel (Ayroles et al. 2009), a wild-derived population of inbred lines with fully sequenced genomes (Mackay et al. 2012). Variation in alcohol sensitivity and induction of tolerance was associated with variation in transcript abundance levels and significantly associated transcripts could be grouped in modules of genetically co-regulated transcripts (Morozova et al. 2009). Modules associated with acute alcohol sensitivity were distinct from those associated with induction of tolerance, indicating that the genetic architectures that underlie the immediate response to ethanol exposure and induction of tolerance are distinct. Notably, genes implicated in nervous system function were associated with natural variation in tolerance development but not with acute ethanol exposure. Connectivity of focal genes within these modules could be validated through P-element mediated mutagenesis, followed by measurements of transcript abundances of connected genes within the module, illustrating the power of complementary single gene and systems approaches (Morozova et al. 2009).
A reverse approach is to identify P-element mutations that affect sensitivity or resistance to alcohol exposure as focal genes and derive computational networks of covariant transcripts centered on these focal genes. The contributions of computationally recruited genes to alcohol sensitivity can then be confirmed by RNAi-mediated inhibition of their expression. Furthermore, these genes can then, in turn, serve as focal genes to expand the computationally derived networks by iteration, allowing a gradual buildup of the network with simultaneous functional validation (Morozova et al. 2011; Fig. 2). At least 60 % of Drosophila genes have human orthologs and these orthologs can be superimposed on the genetic networks from Drosophila. This can provide a stepping stone for the identification of candidate genes associated with alcohol-related phenotypes in human populations (Morozova et al. 2009).
Rodent models for alcohol dependence
Several animal systems have been used to model different aspects of human alcohol-related phenotypes, including preference for alcohol consumption and withdrawal (Bennett et al. 2006). Rodents are suitable models for studies on genetic susceptibility for alcohol dependence, since the organization of regions of the nervous system that mediate addiction is conserved among mice, rats and humans, including the projection from the VTA to the nucleus accumbens and the mesolimbic dopaminergic projection to the forebrain. Furthermore, neurotransmitter systems that regulate this reward pathway are similar. Studies on rodents show that different genes are associated with alcohol consumption and withdrawal effects, similar to the different genetic architectures for inebriation and induction of tolerance seen in the Drosophila model.
Single gene studies in mice have implicated more than 70 candidate genes in alcohol-related phenotypes (Crabbe et al. 2006). These include genes encoding calcium-stimulated adenylate cyclase and protein kinase A (Maas et al. 2005), calcium- and voltage-gated potassium channels (Blednov et al. 2003a; Martin et al. 2008), protein kinase C (Wand et al. 2001; Bowers et al. 2006), neuropeptide Y (Thiele et al. 1998; Thorsell 2007), and proteins involved in GABA neurotransmission (Blednov et al. 2003b; Boehm et al. 2004), dopamine and serotonin signaling (Fadda et al. 1991; Hall et al. 2003; Kelai et al. 2003; Crabbe et al. 2006; Bilbao 2013).
A large number of studies aimed at identifying genes that contribute to variation in alcohol-related phenotypes have relied on gene mapping strategies. At least 24 quantitative trait loci (QTL) have been identified in the mouse genome (Crabbe et al. 1999) and four genomic regions were found in rat (Saba et al. 2011). Meta-analysis of QTL mapping across eight different studies on murine alcohol consumption provided strong support for four QTL regions located on mouse chromosomes 2, 3, 4 and 9 (Belknap and Atkins 2001). Candidate genes have been identified within these QTL regions, including genes encoding a multiple PDZ domain protein (Mpdz) on mouse chromosome 4 (Fehr et al. 2002) and syntaxin binding protein 1 on mouse chromosome 2 (Stxbp1; Fehr et al. 2005) and genes for neuropeptide Y, α-synuclein and corticotrophin-releasing factor receptor 2 in rats (Spence et al. 2009). However, evidence that links candidate genes within QTL regions causally to the phenotype remains difficult to obtain.
Studies that integrate QTL mapping and gene expression analyses have been used both in mice and rats to facilitate the identification of candidate genes that contribute to linkage signals. Previously, two independent studies identified alcohol dependence QTLs on human chromosome 1q (Dick et al. 2002; Hill et al. 2004), and additional studies also provided support for association of markers on chromosome 1q with alcoholism (Aragaki et al. 1999; Turecki et al. 1999; Guerrini et al. 2005). QTL mapping studies in mice revealed 15 differentially expressed genes associated with alcohol withdrawal on chromosome 1, syntenic with human chromosome 1q. Among these genes Kcnj9 appeared the most promising candidate gene. Kcnj9 encodes GIRK3, which is a subunit of inward rectifying K+ channels that mediate inhibitory effects of Gi/o coupled receptors (Kozell et al. 2009). In addition, two independent gene profiling experiments identified candidate genes for alcohol consumption QTL within the rat chromosome 4 region (Carr et al. 2007; Liang et al. 2010), including Akr1b1, Copg2, Dgki, Grid2, Npy, Plxna4, Ppm1k, Qdpr, Scap2, Snca, Snx10 and Spr, which are involved in dopamine and serotonin signaling, protein trafficking and signal transduction. Furthermore, a transcriptome meta-analysis identified 20 cis-regulated candidates for alcohol preference QTL on mouse chromosome 9, including Carm1, Cryab, Pknox2 and Scn4b (Mulligan et al. 2006).
A combination of QTL mapping and microarray analyses in rats identified candidate genes associated with alcohol consumption and among them genes that were differentially expressed in brains as a result of alcohol intake. These genes implicated pathways associated with GABA release, activation of dopamine neurons and postsynaptic GABA receptor trafficking (Tabakoff et al. 2009). A similar strategy applied to high and low alcohol-consuming mice identified Gnb1 localized within a QTL region on chromosome 4 as a candidate gene, since it was also differentially expressed in brains (Saba et al. 2011). Gnb1 codes for the β1 subunit of guanine nucleotide binding proteins. A subsequent comparative genomics approach to study relationships among differentially expressed genes that contribute to alcohol drinking in rats, mice and humans revealed a neural signaling pathway that encompasses both presynaptic and postsynaptic elements of GABA signaling and includes Gβ1. These findings highlight cross-species similarities in genes and pathways that underlie alcohol consumption in animal models and humans and show that neural signaling pathways feature prominently in determining susceptibility to alcohol intake.
Mulligan et al. (2011) used a systems network approach to identify in different brain regions modules of co-regulated genes that showed changes in expression associated with acute drinking. Gene ontology enrichment analyses showed that the modules represented different physiological processes, functional groups, and cell types. Most genes with altered expression were specific for a brain region, suggesting that the response to acute alcohol exposure is cell type specific. Nevertheless, meta-analysis across all brain regions identified a subset of 42 alcohol responsive genes that were shared across multiple brain regions. Several of these genes, including Hba-a1, Hbb-b1, Fth1, Gstm1, Mt2, Nfkbia, Park7, Pltp, Prkcz, Qdpr and Scn4b, were associated with responses to ethanol in different species or were identified in multiple independent studies.
Combining transcript analyses with network and pathway analyses can provide functional context for candidate genes and help to prioritize them for functional validation. This strategy was successfully implemented by Blednov et al. (2012), who selected null mutant mice for six previously identified candidate genes related to peripheral immune and inflammatory signaling, encoding beta-2-microglobulin (B2m), cathepsin S (Ctss), cathepsin F (Ctsf), interleukin 1 receptor antagonist (Il6), CD14 molecule (Cd14) and interleukin 6 (Il6), and showed that all six mutants displayed changes in alcohol consumption. In contrast to previous studies that identified genes involved in neural signaling, none of these genes had previously been associated with alcohol-related phenotypes.
A transcriptional profiling study, which examined changes in transcript abundances in liver and prefrontal cortex in mice that voluntarily consume alcohol, identified a network of genes in prefrontal cortex associated with dopaminergic signaling as well as immune function. Orthologs of these genes include Drd1, Drd2, Fos, Fosb and Ppp1r1b, implicated previously in ethanol and drug addiction (Osterndorff-Kahanek et al. 2013). In liver, transcriptional changes were more numerous and comprised a genetic network associated with drug metabolism and glutathione depletion (Osterndorff-Kahanek et al. 2013). These studies indicate a link among ethanol intake, dopamine signaling and immune responses. The connection with the immune system is of interest, since bacterial lipopolysaccharide can leak from the gut as a result of chronic alcohol abuse and activate the immune system (Mandrekar and Szabo 2009). This bacterial endotoxin also increases alcohol consumption in mice (Blednov et al. 2011).
Following chronic alcohol exposure, removal of alcohol produces a range of withdrawal symptoms, which increase motivation to seek and consume alcohol. Withdrawal symptoms are a hallmark of physiological dependence and include convulsions, motor abnormalities, anxiety and irritability. Symptoms are qualitatively similar across species, but range in severity across individuals and last up to 48 h. Severity of alcohol withdrawal and alcohol preference drinking are genetically negatively correlated; genotypes that drink a lot of ethanol are genetically predisposed to have low withdrawal severity, and vice versa (Metten et al. 1998). Seminal studies by Buck and colleagues (Fehr et al. 2002; Shirley et al. 2004) identified the Mpdz gene within a QTL region on mouse chromosome 4 as a causal gene with large effects on alcohol and barbiturate withdrawal. MPDZ interacts physically with GABAB receptors (Balasubramanian et al. 2007), 5-HT2C receptors (Becamel et al. 2001) and postsynaptic density-associated GTPase-activating protein (SynGAP), which binds to the NMDA receptor 2B subunit/Ca2+-calmodulin kinase (MPDZ/NR2B/CaMKII) complex (Krapivinsky et al. 2004; Kim et al. 2005). Thus, MPDZ could modulate ethanol withdrawal symptoms by regulating GABAB and/or 5-HT2C receptor-mediated neurotransmission (Chen et al. 2011a). Using expression of the immediate early gene c-Fos as a marker for neuronal activation, Chen et al. (2008) found that animals congenic for the chromosome 4 QTL containing the Mpdz gene exhibited significantly less ethanol withdrawal-associated neuronal activation within the basal ganglia than background controls. This effect was particularly evident in the caudolateral region of the substantia nigra pars reticulata, where bilateral electrolytic lesions resulted in attenuation of the severity of ethanol withdrawal symptoms (Chen et al. 2011b). The mouse chromosome 4 QTL is syntenic with a region on human chromosome 9p. Indeed, human MPDZ has also been implicated in alcohol dependence (Karpyak et al. 2009, 2012; Buck et al. 2012).
Human genetic studies of alcohol-related phenotypes
Human genetic studies have identified polymorphisms associated with alcohol dependence in genes that comprise various neurotransmitter signaling pathways, including dopaminergic (e.g., MAOA, COMT, and DRD2, ANKK1, TTC12 and NCAM1; Kohnke et al. 2005; Yang et al. 2008; Tikkanen et al. 2009; Hendershot et al. 2011); serotonergic (e.g., 5-HTT, SLC6A4 and HTR3A, HTR3B, HTR1B; van der Zwaluw et al. 2010; Cao et al. 2013; Seneviratne et al. 2013); GABAergic (e.g., GABRA1, GABRA2 and GABRG1; Agrawal et al. 2006; Dick and Bierut 2006; Enoch 2008), glutamatergic (GRM8; Chen et al. 2009), and cholinergic systems (e.g., CHRM2 and CHRNA5, CHRNB2; Luo et al. 2005; Ehringer et al. 2007; Wang et al. 2009); opioid receptors (e.g., prodynorphin; PDYN; Flory et al. 2011, OPRM1, OPRD1 and OPRK1; Ray and Hutchison 2004; Zhang et al. 2008; Ashenhurst et al. 2012), and tachykinin receptor 3 (TACR3; Foroud et al. 2008).
In recent years, transcriptional profiling and GWAS also reported candidate genes associated with risk for alcohol dependence (for recent reviews, see Spanagel et al. 2010; Morozova et al. 2012; Rietschel and Treutlein 2013). However, with the exception of the large effects contributed by variation at ADH1B and ALDH2 in Asian populations (Edenberg 2007; Hurley and Edenberg 2012), there is little consistency across studies. Nevertheless, one gene encoding cadherin 13 (CDH13) was replicated in four independent studies among all SNPs that were significant at a nominal P value (Johnson et al. 2006; Liu et al. 2006b; Treutlein et al. 2009; Lind et al. 2010). In addition, several risk loci for alcohol dependence and consumption have been detected with samples from large datasets, such as the Collaborative Studies on the Genetics of Alcoholism (COGA), the Study of Addiction: Genetics and Environment (SAGE) and the Australian Twin-family study of alcohol use disorder (OZ-ALC). These include Rho GTPase-activating protein 28 (ARHGAP28), CUB and Suchi multiple domain 1 and 2 (CSMD1 and CSMD2), Catenin delta 2 (CTNND2), Kv channel interacting protein 1 (KCNIP1), Neuronal PAS domain protein 3 (NPAS3), Protein tyrosine phosphatase, receptor type D (PTPRD) and Usher syndrome 2A (USH2A; reviewed by Morozova et al. 2012). The lack of replication across GWAS could be explained by heterogeneity of study populations with different allele frequencies and epistatic interactions as well as different measurements of alcohol consumption. For example, in a GWAS study on individuals from the COGA dataset, Dick et al. (2008) found a gene associated with alcohol dependence on chromosome 7 encoding ACN9 homolog (ACN9), which is involved in gluconeogenesis (Dennis and McCammon 1999). However, a recent analysis of 14 genes involved in alcohol metabolism and oxidation revealed no association between polymorphisms in the ACN9 gene when blood and breath alcohol metabolites were used as phenotypic measurements. Instead, they found significant associations with a SNP in the promoter of CYP2E1 and SNPs in catalase (CAT), beta-enolase (ENO3) and glutamic-oxaloacetic transaminase 1 (GOT1; Lind et al. 2012).
To increase statistical power and ability to detect novel risk loci several groups have conducted meta-analyses using data from the available COGA, SAGE and OZ-ALC GWAS for alcohol dependence (Zuo et al. 2011, 2013a, b; Wang et al. 2011a, b). Interestingly, meta-analyses using similar datasets identified different risk alleles. One study which reanalyzed the COGA and SAGE datasets identified thrombospondin type I domain containing 7B (THSD7B), KIAA0040, nardilysin (N-arginine dibasic convertase, NRD1), PBX/knotted 1 homeobox 2 (PKNOX2), anaplastic lymphoma receptor tyrosine kinase (ALK), cancer susceptibility candidate 4 (CASC4) and semaphorin 5A (SEMA5A). Three of these genes, THSD7B, KIAA0040 and NRD1, were further replicated in OZ-ALC samples (Wang et al. 2011b). PKNOX2 and KIAA0040 genes have been also associated with alcohol dependence in other studies (Bierut et al. 2010; Chen et al. 2011c; Zuo et al. 2012).
A family-based association analysis for alcohol dependence that utilized both COGA and the OZ-ALC samples conducted by the same group (Wang et al. 2011a) found several additional genes associated with alcohol dependence, including endothelin receptor type B (EDNRB), Down syndrome cell adhesion molecule like 1 (DSCAML1), TCDD-inducible poly(ADP-ribose) polymerase (TIPARP), monoamine oxidase A (MAOA), Na+/K+ transporting ATPase interacting 2 (NKAIN2) and Usher syndrome 2A (USH2A), among which USH2A and MAOA genes have been previously associated with alcohol dependence (Johnson et al. 2006; Tikkanen et al. 2009; Heath et al. 2011).
A different set of studies, using GWAS and eQTL analyses, also reanalyzed data from the COGA, SAGE, and the OZ-ALC GWAS for alcohol dependence and identified several replicable risk regions for alcohol dependence (Zuo et al. 2011, 2013a, b). Candidate genes within these regions included Plant HomeoDomain finger protein 3-protein tyrosine phosphatase type IVA, member 1 (PHF3-PTP4A1; Zuo et al. 2011), Na+/K+ transporting ATPase interacting 1-serine incorporator 2 (NKAIN1-SERINC2; Zuo et al. 2013a); and importin 11-5-hydroxytryptamine (serotonin) receptor 1A (IPO11-HTR1A), associated with both alcohol and nicotine codependence (Zuo et al. 2013b). In addition, expression of the PHF3, PTP4A1, NKAIN1, and SERINC2 transcripts was significantly correlated with expression of numerous genes implicated in addiction, including those in dopaminergic (DRD1, DRD2, NCAM1, PPP1R1B) serotonergic (HTR1B, HTR2A), cholinergic (CHRNA3, CHRNB2), GABAergic (GABRA1, GABRA2, GABRG2), glutamatergic (GAD1, GRIK3, GRIN2C), neuropeptide Y (NPY1R, NPY5R) and opioid systems (OPRD1, OPRM1, POMC) (Zuo et al. 2011, 2013a) as well as genes associated with alcohol metabolism (ADH4, ADH5, ADH6). Moreover, injection of ethanol into the brains of mice resulted in altered expression of Ipo11 and Ptp4a1 (Kerns et al. 2005) and Nkain1, Nrd1 and Phf3 were differentially expressed in brains of alcohol-drinking mice (Mulligan et al. 2006).
Two recent studies on the same large datasets used the maximum number of alcoholic drinks consumed in a 24-h period (MaxDrinks) as phenotypic measurement (Kapoor et al. 2013; Pan et al. 2013). Both studies found different genes from those identified by previous meta-analyses and the results from these two studies showed little, if any, overlap.
One study which used the MaxDrinks criterion based on samples from the COGA and SAGE datasets found only two associated genes with genome-wide significance, LIM domain only 1 (LMO1) and phospholipase C-like 1 (PLCL1) (Kapoor et al. 2013). As mentioned earlier, orthologs of the LMO gene family regulate behavioral responses to ethanol in D. melanogaster and mice (Lasek et al. 2011). In addition, nominally significant SNPs were identified in autism susceptibility candidate 2 (AUTS2), inaD-like (INADL), chromosome 15 open reading frame 32 (C15orf32), and huntingtin interacting protein (HIP1) genes. These genes were also implicated in alcohol consumption previously (Heath et al. 2011). The Drosophila AUTS2 ortholog also contributes to alcohol sensitivity (Schumann et al. 2011).
A separate study on the COGA, SAGE and OZ-ALC datasets which also used MaxDrinks as the phenotype identified different candidate genes, including shugoshin-like 1 (SGOL1), nuclear receptor subfamily 4, group A, member 2 (NR4A2) gene and DTW domain containing 2 (DTWD2), N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 (NDST4), potassium voltage-gated channel, Shab-related subfamily, member 2 (KCNB2) and DOPA decarboxylase (DDC; Pan et al. 2013).The NR4A2 gene has been previously implicated in alcohol dependence in Mexican Americans (Wei et al. 2012), DDC has been associated with alcohol consumption in women (Agrawal et al. 2011) and SGOL1 has been implicated in alcohol dependence (Edenberg et al. 2010).
Several of the candidate risk genes for alcohol dependence identified in these studies contribute to alcohol-related behaviors in animal models. Orthologs of PKNOX2, SEMA5A, USH2A, DSCAML1, NR4A2, DDC, SGOL1, DTWD2 and NDST4 show differential expression in brains from alcohol-drinking mice compared to controls; (Mulligan et al. 2006, 2011; Wolstenholme et al. 2011). MAOA and DSCAML1 orthologs were also differentially expressed in alcohol preferring rats (Rimondini et al. 2002; Rodd et al. 2008) and a DSCAML1 ortholog was also found in flies selected for alcohol sensitivity (Morozova et al. 2007).
Zhao et al. (2012) performed a cross-species meta-analysis by ranking genes differentially expressed in mouse brains in response to ethanol (Kerns et al. 2005; Mulligan et al. 2006) based on orthologs implicated in alcohol-related phenotypes across multiple species, including humans (Reich et al. 1998; Mayfield et al. 2002; Kuo et al. 2006; Prescott et al. 2006; Hodgkinson et al. 2008); C. elegans (Kwon et al. 2004) and D. melanogaster (Morozova et al. 2006, 2007) and identified BDNF, GABRA2, GABRB1, MPDZ, NPY and NPY2R among the top ranked genes.
Using a similar approach we compiled findings from several transcriptional profiling studies that have identified differentially expressed genes from alcohol-related studies on D. melanogaster (Morozova et al. 2006, 2007, 2009, 2011; Urizar et al. 2007; Awofala 2010; Kong et al. 2010); from alcohol-related expression studies done on mice (Xu et al. 2001; Daniels and Buck 2002; Tabakoff et al. 2003; Hitzemann et al. 2004; Saito et al. 2004; Treadwell and Singh 2004; Kerns et al. 2005; MacLaren et al. 2006; Mulligan et al. 2006, 2011; Saba et al. 2006; Wang et al. 2007; Denmark and Buck 2008; Wolstenholme et al. 2011), and transcriptional profiling data on rats (Rimondini et al. 2002; Edenberg et al. 2005; Worst et al. 2005; Carr et al. 2007; Kimpel et al. 2007; Rodd et al. 2008), and identified human orthologs. In addition, we analyzed six published transcriptional profiling data sets performed on different areas of postmortem human brains and also included candidate genes for alcohol-related phenotypes from the HuGE Navigator database (Lewohl et al. 2000; Mayfield et al. 2002; Sokolov et al. 2003; Iwamoto et al. 2004; Flatscher-Bader et al. 2005; Liu et al. 2006a; Guo et al. 2009). Furthermore, we integrated information from GWAS, considering all candidate genes that were nominally significant (Johnson et al. 2006; Liu et al. 2006b; Dick et al. 2008; Treutlein et al. 2009; Bierut et al. 2010; Edenberg et al. 2010; Lind et al. 2010; Baik et al. 2011; Heath et al. 2011; Schumann et al. 2011; Wang et al. 2011a, b; Zuo et al. 2011, 2012, 2013a, b; Kapoor et al. 2013; Pan et al. 2013) and ranked these genes based on how many times within and between species they were replicated (supplementary Table 1). We found only seven genes that were replicated across all four species, ARIH1, COPB2, DLG2, IGF2R, IMPA2, MAX and SHC3, and 139 genes were replicated among any two model organisms and humans, including ALDH1A1, ADD1, APOD, AUTS2, CAT, CAST, CRYAB, GABBR1, NFKB1, NRD1, PDIA3, PRKCA and TACR3 (supplementary Table 1). We constructed interacting ensembles between candidate genes (http://www.BioProfiling.de; Antonov 2011) and could cluster 58 genes in networks that were significantly enriched (P < 0.005) for alcohol metabolism and biotransformation, inositol triphosphate metabolism, neurotransmitter biosynthesis and signaling, growth factor signaling and GTPase-dependent signal transduction (Fig. 3). Among the most highly interconnected genes in this integrated network are GRIA1 and GRIA4, which encode ionotropic AMPA 1 and 4 glutamate receptors, respectively, and GRIN1, GRIN2B and GRIN2C, which encode NMDA ionotropic glutamate receptor subunits 1, 2B and 2C, pointing at a central role for glutamatergic neurotransmission. Thus, by integrating data on alcohol-related phenotypes from GWAS and transcriptional profiling studies on both humans and model organisms it is possible to construct biologically meaningful networks of genes that contribute to alcohol dependence and identify “hub” genes as potential candidate genes for future follow-up studies.
Fig. 3.

A comprehensive genetic network for alcohol-related phenotypes. The network is derived from a meta-analysis that incorporates candidate genes from GWAS and differentially expressed genes from transcriptional profiling studies on humans and model organisms (supplementary Table 1). Ovals indicate metabolites that interconnect gene products. The network was constructed using the R-Spider algorithm in the bioprofiling.de web portal (Antonov 2011). This analysis tool incorporates data for ∼4,000 human genes and combines signaling and metabolic pathways from Reactome and KEGG databases to determine whether interactions between the input genes are greater than expected by chance. The network is built by connecting genes with known interactions in the two databases. The significance of the network is tested by Monte Carlo simulations, in which the same number of randomly selected genes is used to form the null distribution of the size of the network. The network presented here consists of 58 interconnected candidate genes (P < 0.005). When one missing gene is allowed (i.e., a gene that does not contain polymorphisms associated with phenotypic variation), the network can be expanded to encompass 258 candidate genes (P < 0.03)
Alcohol-induced epigenetic gene regulation: the next frontier
Epigenetic modifications are becoming increasingly appreciated as important contributors to the effects of alcohol on regulation of gene expression. Epigenetic modifications have been implicated especially in studies on fetal alcohol spectrum disorders (Perkins et al. 2013; Resendiz et al. 2013). Epigenetic alterations include DNA methylation and histone modifications, both of which remodel chromatin structure and, thereby, influence gene expression. Recent studies established that alcohol consumption induces epigenetic alterations in various organs, including brain (Ponomarev et al. 2012; Ponomarev 2013), the gastrointestinal tract (Shukla and Lim 2013) and liver (Mandrekar 2011; Shukla and Lim 2013). In addition, excessive alcohol consumption leads to hypomethylation and promotes histone acetylation in humans and rodents (Lu et al. 2000; Wolstenholme et al. 2011; Ponomarev et al. 2012; Warnault et al. 2013; Zhang et al. 2013). Furthermore, reduction in DNA methylation by administration of a DNA methyltransferase inhibitor as well as inhibition of histone deacetylase leads to reduction in alcohol consumption in mice (Warnault et al. 2013). These observations may open the door to develop chromatin modifying agents to treat alcoholism more effectively than currently available drugs, such as disulfiram, naltrexone and acamprosate (for comprehensive reviews see Heilig and Egli 2006; Johnson 2008; Franck and Jayaram-Lindstrom 2013).
Summary
Several GWAS and meta-analyses studies have reported a vast number of risk alleles for alcohol dependence with little overlap among studies. This is attributable to different phenotypic assessments, use of different species and different preparations, including different tissues or brain regions analyzed, and genome-by-environment interactions. The link between genotype and phenotype is likely also confounded by multidimensional gene-gene interactions, the magnitudes of which depend on allele frequencies (Mackay 2014). In addition, the genetic architectures that underlie different phenotypic manifestations of alcohol drinking behavior appear to be distinct. Nevertheless, different studies reveal different aspects of the genetic underpinnings of the physiological and behavioral effects of ethanol, while underscoring the underlying genomic complexity of the genotype-phenotype relationship. Combining and integrating information from experimentally tractable model systems with human genetic studies provides a powerful strategy to disentangle the genomic elements that contribute to alcohol-related phenotypes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
Work in the authors’ laboratories is supported by grant AA016560 from the National Institutes of Health.
References
- Agrawal A, Edenberg HJ, Foroud T, Bierut LJ, Dunne G, Hinrichs AL, Nurnberger JI, Crowe R, Kuperman S, Schuckit MA, et al. Association of GABRA2 with drug dependence in the collaborative study of the genetics of alcoholism sample. Behav Genet. 2006;36(5):640–650. doi: 10.1007/s10519-006-9069-4. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Lynskey MT, Todorov AA, Schrage AJ, Littlefield AK, Grant JD, Zhu Q, Nelson EC, Madden PA, Bucholz KK, et al. A candidate gene association study of alcohol consumption in young women. Alcohol Clin Exp Res. 2011;35(3):550–558. doi: 10.1111/j.1530-0277.2010.01372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonov AV. BioProfiling.de: analytical web portal for high-throughput cell biology. Nucleic Acids Res. 2011;39:W323–W327. doi: 10.1093/nar/gkr372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragaki C, Quiaoit F, Hsu L, Zhao LP. Mapping alcoholism genes using linkage/linkage disequilibrium analysis. Genet Epidemiol. 1999;17(suppl 1):S43–S48. doi: 10.1002/gepi.1370170708. [DOI] [PubMed] [Google Scholar]
- Ashenhurst JR, Bujarski S, Ray LA. Delta and kappa opioid receptor polymorphisms influence the effects of naltrexone on subjective responses to alcohol. Pharmacol Biochem Behav. 2012;103(2):253–259. doi: 10.1016/j.pbb.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awofala AA. Genetic approaches to alcohol addiction: gene expression studies and recent candidates from Drosophila. Invert Neurosci. 2010;11(1):1–7. doi: 10.1007/s10158-010-0113-y. [DOI] [PubMed] [Google Scholar]
- Ayroles JF, Carbone M, Stone EA, Jordan KW, Lyman RF, Magwire MM, Rollmann SR, Laura HD, Lawrence F, Anholt RRH, et al. Systems genetics of complex traits in Drosophila melanogaster. Nat Genet. 2009;41(3):299–307. doi: 10.1038/ng.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer DJ, Judd JT, Clevidence BA, Muesing RA, Campbell WS, Brown ED, Taylor PR. Moderate alcohol consumption lowers risk factors for cardiovascular disease in postmenopausal women fed a controlled diet. Am J Clin Nutr. 2002;75(3):593–599. doi: 10.1093/ajcn/75.3.593. [DOI] [PubMed] [Google Scholar]
- Baik I, Cho NH, Kim SH, Han BG, Shin C. Genome-wide association studies identify genetic loci related to alcohol consumption in Korean men. Am J Clin Nutr. 2011;93(4):809–816. doi: 10.3945/ajcn.110.001776. [DOI] [PubMed] [Google Scholar]
- Bainton RJ, Tsai LTY, Singh CM, Moore MS, Neckameyer WS, Heberlein U. Dopamine modulates acute responses to cocaine, nicotine and ethanol in Drosophila. Curr Biol. 2000;10(4):187–194. doi: 10.1016/S0960-9822(00)00336-5. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Fam SR, Hall RA. GABAB receptor association with the PDZ scaffold Mupp1 alters receptor stability and function. J Biol Chem. 2007;282(6):4162–4171. doi: 10.1074/jbc.M607695200. [DOI] [PubMed] [Google Scholar]
- Becamel C, Figge A, Poliak S, Dumuis A, Peles E, Bockaert J, Lubbert H, Ullmer C. Interaction of serotonin 5-hydroxytryptamine type 2C receptors with PDZ10 of the multi-PDZ domain protein MUPP1. J Biol Chem. 2001;276(16):12974–12982. doi: 10.1074/jbc.M008089200. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Atkins AL. The replicability of QTLs for murine alcohol preference drinking behavior across eight independent studies. Mamm Genome. 2001;12(12):893–899. doi: 10.1007/s00335-001-2074-2. [DOI] [PubMed] [Google Scholar]
- Bennett B, Downing C, Parker C, Johnson TE. Mouse genetic models in alcohol research. Trends Genet. 2006;22(7):367–374. doi: 10.1016/j.tig.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, et al. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci USA. 2010;107(11):5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Goate AM, Breslau N, Johnson EO, Bertelsen S, Fox L, Agrawal A, Bucholz KK, Grucza R, Hesselbrock V, et al. ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Mol Psychiatry. 2012;17(4):445–450. doi: 10.1038/mp.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilbao A. Advanced transgenic approaches to understand alcohol-related phenotypes in animals. Curr Top Behav Neurosci. 2013;13:271–311. doi: 10.1007/978-3-642-28720-6_204. [DOI] [PubMed] [Google Scholar]
- Birley AJ, James MR, Dickson PA, Montgomery GW, Heath AC, Martin NG, Whitfield JB. Adh snp associations with alcohol metabolism in vivo. Hum Mol Genet. 2009;18(8):1533–1542. doi: 10.1093/hmg/ddp060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Stoffel M, Alva H, Harris RA. A pervasive mechanism for analgesia: activation of GIRK2 channels. Proc Natl Acad Sci USA. 2003;100(1):277–282. doi: 10.1073/pnas.012682399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Walker D, Alva H, Creech K, Findlay G, Harris RA. GABAA receptor alpha 1 and beta 2 subunit null mutant mice: behavioral responses to ethanol. J Pharmacol Exp Ther. 2003;305(3):854–863. doi: 10.1124/jpet.103.049478. [DOI] [PubMed] [Google Scholar]
- Blednov YA, Benavidez JM, Geil C, Perra S, Morikawa H, Harris RA. Activation of inflammatory signaling by lipopolysaccharide produces a prolonged increase of voluntary alcohol intake in mice. Brain Behav Immun. 2011;25(Suppl 1):S92–S105. doi: 10.1016/j.bbi.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Ponomarev I, Geil C, Bergeson S, Koob GF, Harris RA. Neuroimmune regulation of alcohol consumption: behavioral validation of genes obtained from genomic studies. Addict Biol. 2012;17(1):108–120. doi: 10.1111/j.1369-1600.2010.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm SL, 2nd, Ponomarev I, Jennings AW, Whiting PJ, Rosahl TW, Garrett EM, Blednov YA, Harris RA. Gamma-aminobutyric acid A receptor subunit mutant mice: new perspectives on alcohol actions. Biochem Pharmacol. 2004;68(8):1581–1602. doi: 10.1016/j.bcp.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Bowers BJ, Miyamoto-Ditmon J, Wehner JM. Regulation of 5-HT2A/C receptors and DOI-induced behaviors by protein kinase C gamma. Pharmacol Biochem Behav. 2006;85(2):441–447. doi: 10.1016/j.pbb.2006.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck KJ, Milner LC, Denmark DL, Grant SG, Kozell LB. Discovering genes involved in alcohol dependence and other alcohol responses: role of animal models. Alcohol Res. 2012;34(3):367–374. doi: 10.35946/arcr.v34.3.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, LaRocque E, Li D. Associations of the 5-hydroxytryptamine (serotonin) receptor 1B gene (HTR1B) with alcohol, cocaine, and heroin abuse. Am J Med Genet B Neuropsychiatr Genet. 2013;162B(2):169–176. doi: 10.1002/ajmg.b.32128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr LG, Kimpel MW, Liang T, McClintick JN, McCall K, Morse M, Edenberg HJ. Identification of candidate genes for alcohol preference by expression profiling of congenic rat strains. Alcohol Clin Exp Res. 2007;31(7):1089–1098. doi: 10.1111/j.1530-0277.2007.00397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Kozell LB, Hitzemann R, Buck KJ. Involvement of the limbic basal ganglia in ethanol withdrawal convulsivity in mice is influenced by a chromosome 4 locus. J Neurosci. 2008;28(39):9840–9849. doi: 10.1523/JNEUROSCI.1713-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AC, Tang Y, Rangaswamy M, Wang JC, Almasy L, Foroud T, Edenberg HJ, Hesselbrock V, Nurnberger J, Jr, Kuperman S, et al. Association of single nucleotide polymorphisms in a glutamate receptor gene (GRM8) with theta power of event-related oscillations and alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(3):359–368. doi: 10.1002/ajmg.b.30818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Cuzon Carlson VC, Wang J, Beck A, Heinz A, Ron D, Lovinger DM, Buck KJ. Striatal involvement in human alcoholism and alcohol consumption, and withdrawal in animal models. Alcohol Clin Exp Res. 2011;35(10):1739–1748. doi: 10.1111/j.1530-0277.2011.01520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Kozell LB, Buck KJ. Substantia nigra pars reticulata is crucially involved in barbiturate and ethanol withdrawal in mice. Behav Brain Res. 2011;218(1):152–157. doi: 10.1016/j.bbr.2010.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Cho K, Singer BH, Zhang H. The nuclear transcription factor PKNOX2 is a candidate gene for substance dependence in European-origin women. PLoS One. 2011;6(1):e16002. doi: 10.1371/journal.pone.0016002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chick J. The WHO global strategy to reduce the harmful use of alcohol. Alcohol Alcohol. 2011;46(3):223. doi: 10.1093/alcalc/agr035. [DOI] [PubMed] [Google Scholar]
- Conigrave KM, Hu BF, Camargo CA, Jr, Stampfer MJ, Willett WC, Rimm EB. A prospective study of drinking patterns in relation to risk of type 2 diabetes among men. Diabetes. 2001;50(10):2390–2395. doi: 10.2337/diabetes.50.10.2390. [DOI] [PubMed] [Google Scholar]
- Corl AB, Berger KH, Ophir-Shohat G, Gesch J, Simms JA, Bartlett SE, Heberlein U. Happyhour, a Ste20 family kinase, implicates EGFR signaling in ethanol-induced behaviors. Cell. 2009;137(5):949–960. doi: 10.1016/j.cell.2009.03.020. [DOI] [PubMed] [Google Scholar]
- Cowmeadow RB, Krishnan HR, Atkinson NS. The slowpoke gene is necessary for rapid ethanol tolerance in Drosophila. Alcohol Clin Exp Res. 2005;29:1777–1786. doi: 10.1097/01.alc.0000183232.56788.62. [DOI] [PubMed] [Google Scholar]
- Cowmeadow RB, Krishnan HR, Ghezzi A, Al’Hasan YM, Wang YZ, Atkinson NS. Ethanol tolerance caused by slowpoke induction in Drosophila. Alcohol Clin Exp Res. 2006;30:745–753. doi: 10.1111/j.1530-0277.2006.00087.x. [DOI] [PubMed] [Google Scholar]
- Cozzoli DK, Goulding SP, Zhang PW, Xiao B, Hu JH, Ary AW, Obara I, Rahn A, Abou-Ziab H, Tyrrel B, et al. Binge drinking upregulates accumbens mGluR5-Homer2-PI3K signaling: functional implications for alcoholism. J Neurosci. 2009;29(27):8655–8668. doi: 10.1523/JNEUROSCI.5900-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Buck KJ, Cunningham CL, Belknap JK. Identifying genes for alcohol and drug sensitivity: recent progress and future directions. Trends Neurosci. 1999;22:173–179. doi: 10.1016/S0166-2236(99)01393-4. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Harris RA, Arends MA, Koob GF. Alcohol-related genes: contributions from studies with genetically engineered mice. Addict Biol. 2006;11(3–4):195–269. doi: 10.1111/j.1369-1600.2006.00038.x. [DOI] [PubMed] [Google Scholar]
- Daniels GM, Buck KJ. Expression profiling identifies strain-specific changes associated with ethanol withdrawal in mice. Genes Brain Behav. 2002;1(1):35–45. doi: 10.1046/j.1601-1848.2001.00008.x. [DOI] [PubMed] [Google Scholar]
- Denmark DL, Buck KJ. Molecular analyses and identification of promising candidate genes for loci on mouse chromosome 1 affecting alcohol physical dependence and associated withdrawal. Genes Brain Behav. 2008;7(5):599–608. doi: 10.1111/j.1601-183X.2008.00396.x. [DOI] [PubMed] [Google Scholar]
- Dennis RA, McCammon MT. Acn9 is a novel protein of gluconeogenesis that is located in the mitochondrial intermembrane space. Eur J Biochem. 1999;261(1):236–243. doi: 10.1046/j.1432-1327.1999.00267.x. [DOI] [PubMed] [Google Scholar]
- Devineni AV, Heberlein U. Preferential ethanol consumption in Drosophila models features of addiction. Curr Biol. 2009;19(24):2126–2132. doi: 10.1016/j.cub.2009.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devineni AV, Heberlein U. The evolution of Drosophila melanogaster as a model for alcohol research. Annu Rev Neurosci. 2013;36:121–138. doi: 10.1146/annurev-neuro-062012-170256. [DOI] [PubMed] [Google Scholar]
- Dick DM, Bierut LJ. The genetics of alcohol dependence. Curr Psychiatry Rep. 2006;8(2):151–157. doi: 10.1007/s11920-006-0015-1. [DOI] [PubMed] [Google Scholar]
- Dick DM, Nurnberger J, Jr, Edenberg HJ, Goate A, Crowe R, Rice J, Bucholz KK, Kramer J, Schuckit MA, Smith TL, et al. Suggestive linkage on chromosome 1 for a quantitative alcohol-related phenotype. Alcohol Clin Exp Res. 2002;26(10):1453–1460. doi: 10.1111/j.1530-0277.2002.tb02443.x. [DOI] [PubMed] [Google Scholar]
- Dick DM, Aliev F, Wang JC, Saccone S, Hinrichs A, Bertelsen S, Budde J, Saccone N, Foroud T, Nurnberger J, Jr, et al. A systematic single nucleotide polymorphism screen to fine-map alcohol dependence genes on chromosome 7 identifies association with a novel susceptibility gene ACN9. Biol Psychiatry. 2008;63(11):1047–1053. doi: 10.1016/j.biopsych.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducci F, Goldman D. Genetic approaches to addiction: genes and alcohol. Addiction. 2008;103(9):1414–1428. doi: 10.1111/j.1360-0443.2008.02203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzitoyeva S, Dimitrijevic N, Manev H. Gamma-aminobutyric acid B receptor 1 mediates behavior-impairing actions of alcohol in Drosophila: adult RNA interference and pharmacological evidence. Proc Natl Acad Sci USA. 2003;100:5485–5490. doi: 10.1073/pnas.0830111100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddison M, Guarnieri DJ, Cheng L, Liu CH, Moffat KG, Davis G, Heberlein U. Arouser reveals a role for synapse number in the regulation of ethanol sensitivity. Neuron. 2011;70(5):979–990. doi: 10.1016/j.neuron.2011.03.030. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health. 2007;30(1):5–13. [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ. Genetics of alcohol use disorders. In: Miller PM, editor. Biological research on addiction. London: Elsevier Science & Technology Books/Academic Press; 2013. pp. 500–508. [Google Scholar]
- Edenberg HJ, Strother WN, McClintick JN, Tian H, Stephens M, Jerome RE, Lumeng L, Li TK, McBride WJ. Gene expression in the hippocampus of inbred alcohol-preferring and -nonpreferring rats. Genes Brain Behav. 2005;4(1):20–30. doi: 10.1111/j.1601-183X.2004.00091.x. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut LJ, Bucholz KK, Goate A, Aliev F, et al. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res. 2010;34(5):840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AC, Zwarts L, Yamamoto A, Callaerts P, Mackay TFC. Mutations in many genes affect aggressive behavior in Drosophila melanogaster. BMC Biol. 2009;7:29. doi: 10.1186/1741-7007-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehringer MA, Clegg HV, Collins AC, Corley RP, Crowley T, Hewitt JK, Hopfer CJ, Krauter K, Lessem J, Rhee SH, et al. Association of the neuronal nicotinic receptor beta2 subunit gene (CHRNB2) with subjective responses to alcohol and nicotine. Am J Med Genet B Neuropsychiatr Genet. 2007;144B(5):596–604. doi: 10.1002/ajmg.b.30464. [DOI] [PubMed] [Google Scholar]
- Enoch MA. The role of GABA(A) receptors in the development of alcoholism. Pharmacol Biochem Behav. 2008;90(1):95–104. doi: 10.1016/j.pbb.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadda F, Garau B, Marchei F, Colombo G, Gessa GL. MDL 72222, a selective 5-HT3 receptor antagonist, suppresses voluntary ethanol consumption in alcohol-preferring rats. Alcohol Alcohol. 1991;26(2):107–110. doi: 10.1093/oxfordjournals.alcalc.a045088. [DOI] [PubMed] [Google Scholar]
- Feany MB, Quinn WG. A neuropeptide gene defined by the Drosophila memory mutant amnesiac. Science. 1995;268:869–873. doi: 10.1126/science.7754370. [DOI] [PubMed] [Google Scholar]
- Fehr C, Shirley RL, Belknap JK, Crabbe JC, Buck KJ. Congenic mapping of alcohol and pentobarbital withdrawal liability loci to a <1 centimorgan interval of murine chromosome 4: identification of Mpdz as a candidate gene. J Neurosci. 2002;22:3730–3738. doi: 10.1523/JNEUROSCI.22-09-03730.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr C, Shirley RL, Crabbe JC, Belknap JK, Buck KJ, Phillips TJ. The syntaxin binding protein 1 gene (Stxbp1) is a candidate for an ethanol preference drinking locus on mouse chromosome 2. Alcohol Clin Exp Res. 2005;29(5):708–720. doi: 10.1097/01.ALC.0000164366.18376.EF. [DOI] [PubMed] [Google Scholar]
- Feldman HS, Jones KL, Lindsay S, Slymen D, Klonoff-Cohen H, Kao K, Rao S, Chambers C. Prenatal alcohol exposure patterns and alcohol-related birth defects and growth deficiencies: a prospective study. Alcohol Clin Exp Res. 2012;36(4):670–676. doi: 10.1111/j.1530-0277.2011.01664.x. [DOI] [PubMed] [Google Scholar]
- Flatscher-Bader T, van der Brug M, Hwang JW, Gochee PA, Matsumoto I, Niwa S, Wilce PA. Alcohol-responsive genes in the frontal cortex and nucleus accumbens of human alcoholics. J Neurochem. 2005;93(2):359–370. doi: 10.1111/j.1471-4159.2004.03021.x. [DOI] [PubMed] [Google Scholar]
- Flory JD, Pytte CL, Hurd Y, Ferrell RE, Manuck SB. Alcohol dependence, disinhibited behavior and variation in the prodynorphin gene. Biol Psychol. 2011;88(1):51–56. doi: 10.1016/j.biopsycho.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foa L, Gasperini R. Developmental roles for Homer: more than just a pretty scaffold. J Neurochem. 2009;108(1):1–10. doi: 10.1111/j.1471-4159.2008.05726.x. [DOI] [PubMed] [Google Scholar]
- Foroud T, Wetherill LF, Kramer J, Tischfield JA, Nurnberger JI, Jr, Schuckit MA, Xuei X, Edenberg HJ. The tachykinin receptor 3 is associated with alcohol and cocaine dependence. Alcohol Clin Exp Res. 2008;32(6):1023–1030. doi: 10.1111/j.1530-0277.2008.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck J, Jayaram-Lindstrom N. Pharmacotherapy for alcohol dependence: status of current treatments. Curr Opin Neurobiol. 2013;23(4):692–699. doi: 10.1016/j.conb.2013.05.005. [DOI] [PubMed] [Google Scholar]
- Fry JD, Saweikis M. Aldehyde dehydrogenase is essential for both adult and larval ethanol resistance in Drosophila melanogaster. Genet Res. 2006;87(2):87–92. doi: 10.1017/S0016672306008032. [DOI] [PubMed] [Google Scholar]
- Geer BW, Langevin ML, McKechnie SW. Dietary ethanol and lipid synthesis in Drosophila melanogaster. Biochem Genet. 1985;23(7–8):607–622. doi: 10.1007/BF00504295. [DOI] [PubMed] [Google Scholar]
- Graff-Iversen S, Jansen MD, Hoff DA, Hoiseth G, Knudsen GP, Magnus P, Morland J, Normann PT, Naess OE, Tambs K. Divergent associations of drinking frequency and binge consumption of alcohol with mortality within the same cohort. J Epidemiol Community Health. 2013;67(4):350–357. doi: 10.1136/jech-2012-201564. [DOI] [PubMed] [Google Scholar]
- Guarnieri DJ, Heberlein U. Drosophila melanogaster, a genetic model system for alcohol research. Int Rev Neurobiol. 2003;54:199–228. doi: 10.1016/S0074-7742(03)54006-5. [DOI] [PubMed] [Google Scholar]
- Guerrini I, Cook CC, Kest W, Devitgh A, McQuillin A, Curtis D, Gurling HM. Genetic linkage analysis supports the presence of two susceptibility loci for alcoholism and heavy drinking on chromosome 1p22.1-11.2 and 1q21.3-24.2. BMC Genet. 2005;6:11. doi: 10.1186/1471-2156-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo AY, Webb BT, Miles MF, Zimmerman MP, Kendler KS, Zhao Z. ERGR: an ethanol-related gene resource. Nucleic Acids Res. 2009;37(database issue):D840–D845. doi: 10.1093/nar/gkn816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall FS, Sora I, Uhl GR. Sex-dependent modulation of ethanol consumption in vesicular monoamine transporter 2 (VMAT2) and dopamine transporter (DAT) knockout mice. Neuropsychopharmacology. 2003;28(4):620–628. doi: 10.1038/sj.npp.1300070. [DOI] [PubMed] [Google Scholar]
- Harbison ST, Sehgal A. Quantitative genetic analysis of sleep in Drosophila melanogaster. Genetics. 2008;178(4):2341–2360. doi: 10.1534/genetics.107.081232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM, Lind PA, McEvoy BP, Schrage AJ, Grant JD, Chou YL, et al. A quantitative-trait genome-wide association study of alcoholism risk in the community: findings and implications. Biol Psychiatry. 2011;70(6):513–518. doi: 10.1016/j.biopsych.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilig M, Egli M. Pharmacological treatment of alcohol dependence: target symptoms and target mechanisms. Pharmacol Ther. 2006;111(3):855–876. doi: 10.1016/j.pharmthera.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Hendershot CS, Lindgren KP, Liang T, Hutchison KE. COMT and ALDH2 polymorphisms moderate associations of implicit drinking motives with alcohol use. Addict Biol. 2011;17(1):192–201. doi: 10.1111/j.1369-1600.2010.00286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SY, Shen S, Zezza N, Hoffman EK, Perlin M, Allan W. A genome wide search for alcoholism susceptibility genes. Am J Med Genet B Neuropsychiatr Genet. 2004;128B(1):102–113. doi: 10.1002/ajmg.b.30013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitzemann R, Reed C, Malmanger B, Lawler M, Hitzemann B, Cunningham B, McWeeney S, Belknap J, Harrington C, Buck K, et al. On the integration of alcohol-related quantitative trait loci and gene expression analyses. Alcohol Clin Exp Res. 2004;28(10):1437–1448. doi: 10.1097/01.ALC.0000139827.86749.DA. [DOI] [PubMed] [Google Scholar]
- Hodgkinson CA, Yuan Q, Xu K, Shen PH, Heinz E, Lobos EA, Binder EB, Cubells J, Ehlers CL, Gelernter J, et al. Addictions biology: haplotype-based analysis for 130 candidate genes on a single array. Alcohol Alcohol. 2008;43(5):505–515. doi: 10.1093/alcalc/agn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley TD, Edenberg HJ. Genes encoding enzymes involved in ethanol metabolism. Alcohol Res. 2012;34(3):339–344. doi: 10.35946/arcr.v34.3.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto K, Bundo M, Yamamoto M, Ozawa H, Saito T, Kato T. Decreased expression of NEFH and PCP4/PEP19 in the prefrontal cortex of alcoholics. Neurosci Res. 2004;49(4):379–385. doi: 10.1016/j.neures.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Johnson BA. Update on neuropharmacological treatments for alcoholism: scientific basis and clinical findings. Biochem Pharmacol. 2008;75(1):34–56. doi: 10.1016/j.bcp.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Drgon T, Liu QR, Walther D, Edenberg H, Rice J, Foroud T, Uhl GR. Pooled association genome scanning for alcohol dependence using 104,268 SNPs: validation and use to identify alcoholism vulnerability loci in unrelated individuals from the collaborative study on the genetics of alcoholism. Am J Med Genet B Neuropsychiatr Genet. 2006;141B(8):844–853. doi: 10.1002/ajmg.b.30346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M, Wang JC, Wetherill L, Le N, Bertelsen S, Hinrichs AL, Budde J, Agrawal A, Bucholz K, Dick D, et al. A meta-analysis of two genome-wide association studies to identify novel loci for maximum number of alcoholic drinks. Hum Genet. 2013;132(10):1141–1151. doi: 10.1007/s00439-013-1318-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karahanian E, Quintanilla ME, Tampier L, Rivera-Meza M, Bustamante D, Gonzalez-Lira V, Morales P, Herrera-Marschitz M, Israel Y. Ethanol as a prodrug: brain metabolism of ethanol mediates its reinforcing effects. Alcohol Clin Exp Res. 2011;35(4):606–612. doi: 10.1111/j.1530-0277.2011.01439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpyak VM, Kim JH, Biernacka JM, Wieben ED, Mrazek DA, Black JL, Choi DS. Sequence variations of the human MPDZ gene and association with alcoholism in subjects with European ancestry. Alcohol Clin Exp Res. 2009;33(4):712–721. doi: 10.1111/j.1530-0277.2008.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpyak VM, Geske JR, Colby CL, Mrazek DA, Biernacka JM. Genetic variability in the NMDA-dependent AMPA trafficking cascade is associated with alcohol dependence. Addict Biol. 2012;17(4):798–806. doi: 10.1111/j.1369-1600.2011.00338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaun KR, Devineni AV, Heberlein U. Drosophila melanogaster as a model to study drug addiction. Hum Genet. 2012;131(6):959–975. doi: 10.1007/s00439-012-1146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelai S, Aissi F, Lesch KP, Cohen-Salmon C, Hamon M, Lanfumey L. Alcohol intake after serotonin transporter inactivation in mice. Alcohol Alcohol. 2003;38(4):386–389. doi: 10.1093/alcalc/agg095. [DOI] [PubMed] [Google Scholar]
- Kerns RT, Ravindranathan A, Hassan S, Cage MP, York T, Sikela JM, Williams RW, Miles MF. Ethanol-responsive brain region expression networks: implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. J Neurosci. 2005;25(9):2255–2266. doi: 10.1523/JNEUROSCI.4372-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Dunah AW, Wang YT, Sheng M. Differential roles of NR2A- and NR2B-containing NMDA receptors in Ras-ERK signaling and AMPA receptor trafficking. Neuron. 2005;46(5):745–760. doi: 10.1016/j.neuron.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Kimpel MW, Strother WN, McClintick JN, Carr LG, Liang T, Edenberg HJ, McBride WJ. Functional gene expression differences between inbred alcohol-preferring and -non-preferring rats in five brain regions. Alcohol. 2007;41(2):95–132. doi: 10.1016/j.alcohol.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohnke MD, Batra A, Kolb W, Kohnke AM, Lutz U, Schick S, Gaertner I. Association of the dopamine transporter gene with alcoholism. Alcohol Alcohol. 2005;40(5):339–342. doi: 10.1093/alcalc/agh179. [DOI] [PubMed] [Google Scholar]
- Kong EC, Allouche L, Chapot PA, Vranizan K, Moore MS, Heberlein U, Wolf FW. Ethanol-regulated genes that contribute to ethanol sensitivity and rapid tolerance in Drosophila. Alcohol Clin Exp Res. 2010;34(2):302–316. doi: 10.1111/j.1530-0277.2009.01093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppes LL, Dekker JM, Hendriks HF, Bouter LM, Heine RJ. Moderate alcohol consumption lowers the risk of type 2 diabetes: a meta-analysis of prospective observational studies. Diabetes Care. 2005;28(3):719–725. doi: 10.2337/diacare.28.3.719. [DOI] [PubMed] [Google Scholar]
- Kozell LB, Walter NA, Milner LC, Wickman K, Buck KJ. Mapping a barbiturate withdrawal locus to a 0.44 Mb interval and analysis of a novel null mutant identify a role for Kcnj9 (GIRK3) in withdrawal from pentobarbital, zolpidem, and ethanol. J Neurosci. 2009;29(37):11662–11673. doi: 10.1523/JNEUROSCI.1413-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapivinsky G, Medina I, Krapivinsky L, Gapon S, Clapham DE. SynGAP–MUPP1–CaMKII synaptic complexes regulate p38 MAP kinase activity and NMDA receptor-dependent synaptic AMPA receptor potentiation. Neuron. 2004;43(4):563–574. doi: 10.1016/j.neuron.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Kuo PH, Neale MC, Riley BP, Webb BT, Sullivan PF, Vittum J, Patterson DG, Thiselton DL, van den Oord EJ, Walsh D, et al. Identification of susceptibility loci for alcohol-related traits in the Irish affected sib pair study of alcohol dependence. Alcohol Clin Exp Res. 2006;30(11):1807–1816. doi: 10.1111/j.1530-0277.2006.00217.x. [DOI] [PubMed] [Google Scholar]
- Kwon JY, Hong M, Choi MS, Kang S, Duke K, Kim S, Lee S, Lee J. Ethanol-response genes and their regulation analyzed by a microarray and comparative genomic approach in the nematode Caenorhabditis elegans. Genomics. 2004;83(4):600–614. doi: 10.1016/j.ygeno.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Lasek AW, Giorgetti F, Berger KH, Tayor S, Heberlein U. Lmo genes regulate behavioral responses to ethanol in Drosophila melanogaster and the mouse. Alcohol Clin Exp Res. 2011;35(9):1600–1606. doi: 10.1111/j.1530-0277.2011.01506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitzmann MF, Giovannucci EL, Stampfer MJ, Spiegelman D, Colditz GA, Willett WC, Rimm EB. Prospective study of alcohol consumption patterns in relation to symptomatic gallstone disease in men. Alcohol Clin Exp Res. 1999;23(5):835–841. doi: 10.1111/j.1530-0277.1999.tb04191.x. [DOI] [PubMed] [Google Scholar]
- Lewohl JM, Wang L, Miles MF, Zhang L, Dodd PR, Harris RA. Gene expression in human alcoholism: microarray analysis of frontal cortex. Alcohol Clin Exp Res. 2000;24(12):1873–1882. doi: 10.1111/j.1530-0277.2000.tb01993.x. [DOI] [PubMed] [Google Scholar]
- Liang T, Kimpel MW, McClintick JN, Skillman AR, McCall K, Edenberg HJ, Carr LG. Candidate genes for alcohol preference identified by expression profiling in alcohol-preferring and -nonpreferring reciprocal congenic rats. Genome Biol. 2010;11(2):R11. doi: 10.1186/gb-2010-11-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, Eriksson CJ, Wilhelmsen KC. The role of aldehyde dehydrogenase-1 (ALDH1A1) polymorphisms in harmful alcohol consumption in a Finnish population. Hum Genomics. 2008;3(1):24–35. doi: 10.1186/1479-7364-3-1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, Macgregor S, Vink JM, Pergadia ML, Hansell NK, de Moor MH, Smit AB, Hottenga JJ, Richter MM, Heath AC, et al. A genomewide association study of nicotine and alcohol dependence in Australian and Dutch populations. Twin Res Hum Genet. 2010;13(1):10–29. doi: 10.1375/twin.13.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, Macgregor S, Heath AC, Madden PA, Montgomery GW, Martin NG, Whitfield JB. Association between in vivo alcohol metabolism and genetic variation in pathways that metabolize the carbon skeleton of ethanol and NADH reoxidation in the alcohol challenge twin study. Alcohol Clin Exp Res. 2012;36(12):2074–2085. doi: 10.1111/j.1530-0277.2012.01829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linneberg A, Gonzalez-Quintela A, Vidal C, Jorgensen T, Fenger M, Hansen T, Pedersen O, Husemoen LL. Genetic determinants of both ethanol and acetaldehyde metabolism influence alcohol hypersensitivity and drinking behaviour among Scandinavians. Clin Exp Allergy. 2010;40(1):123–130. doi: 10.1111/j.1365-2222.2009.03398.x. [DOI] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Iyer VR, Dodd PR, Randall PK, Mayfield RD. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology. 2006;31(7):1574–1582. doi: 10.1038/sj.npp.1300947. [DOI] [PubMed] [Google Scholar]
- Liu QR, Drgon T, Johnson C, Walther D, Hess J, Uhl GR. Addiction molecular genetics: 639,401 SNP whole genome association identifies many “cell adhesion” genes. Am J Med Genet B Neuropsychiatr Genet. 2006;141B(8):918–925. doi: 10.1002/ajmg.b.30436. [DOI] [PubMed] [Google Scholar]
- Lu SC, Huang ZZ, Yang H, Mato JM, Avila MA, Tsukamoto H. Changes in methionine adenosyltransferase and S-adenosylmethionine homeostasis in alcoholic rat liver. Am J Physiol Gastrointest Liver Physiol. 2000;279(1):G178–G185. doi: 10.1152/ajpgi.2000.279.1.G178. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Wang S, Blumberg HP, Gelernter J. CHRM2 gene predisposes to alcohol dependence, drug dependence and affective disorders: results from an extended case-control structured association study. Hum Mol Genet. 2005;14(16):2421–2434. doi: 10.1093/hmg/ddi244. [DOI] [PubMed] [Google Scholar]
- Maas JW, Jr, Vogt SK, Chan GC, Pineda VV, Storm DR, Muglia LJ. Calcium-stimulated adenylyl cyclases are critical modulators of neuronal ethanol sensitivity. J Neurosci. 2005;25(16):4118–4126. doi: 10.1523/JNEUROSCI.4273-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC (2014) Epistasis and quantitative traits: using model organisms to study gene–gene interactions. Nat Rev Genet 15(1):22–33 [DOI] [PMC free article] [PubMed]
- Mackay TFC, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, et al. The Drosophila melanogaster genetic reference panel. Nature. 2012;482(7384):173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLaren EJ, Bennett B, Johnson TE, Sikela JM. Expression profiling identifies novel candidate genes for ethanol sensitivity QTLs. Mamm Genome. 2006;17(2):147–156. doi: 10.1007/s00335-005-0065-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magwire MM, Yamamoto A, Carbone MA, Roshina NV, Pasyukova EG, Morozova TV, Mackay TFC. Quantitative and molecular genetic analyses of mutations increasing Drosophila life span. PLoS Genet. 2010;6(7):e1001037. doi: 10.1371/journal.pgen.1001037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandrekar P. Epigenetic regulation in alcoholic liver disease. World J Gastroenterol. 2011;17(20):2456–2464. doi: 10.3748/wjg.v17.i20.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandrekar P, Szabo G. Signalling pathways in alcohol-induced liver inflammation. J Hepatol. 2009;50(6):1258–1266. doi: 10.1016/j.jhep.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GE, Hendrickson LM, Penta KL, Friesen RM, Pietrzykowski AZ, Tapper AR, Treistman SN. Identification of a BK channel auxiliary protein controlling molecular and behavioral tolerance to alcohol. Proc Natl Acad Sci USA. 2008;105(45):17543–17548. doi: 10.1073/pnas.0801068105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marugame T, Yamamoto S, Yoshimi I, Sobue T, Inoue M, Tsugane S. Patterns of alcohol drinking and all-cause mortality: results from a large-scale population-based cohort study in Japan. Am J Epidemiol. 2007;165(9):1039–1046. doi: 10.1093/aje/kwk112. [DOI] [PubMed] [Google Scholar]
- Mayfield RD, Lewohl JM, Dodd PR, Herlihy A, Liu J, Harris RA. Patterns of gene expression are altered in the frontal and motor cortices of human alcoholics. J Neurochem. 2002;81(4):802–813. doi: 10.1046/j.1471-4159.2002.00860.x. [DOI] [PubMed] [Google Scholar]
- McCullough AJ, O’Shea RS, Dasarathy S. Diagnosis and management of alcoholic liver disease. J Dig Dis. 2011;12(4):257–262. doi: 10.1111/j.1751-2980.2010.00470.x. [DOI] [PubMed] [Google Scholar]
- McDonald JF, Anderson SM, Santos M. Biochemical differences between products of the Adh locus in Drosophila. Genetics. 1980;95(4):1013–1022. doi: 10.1093/genetics/95.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie JA, McKechnie SW. Ethanol tolerance and the Adh polymorphism in a natural population of Drosophila melanogaster. Nature. 1978;272(5648):75–76. doi: 10.1038/272075a0. [DOI] [PubMed] [Google Scholar]
- Metten P, Phillips TJ, Crabbe JC, Tarantino LM, McClearn GE, Plomin R, Erwin VG, Belknap JK. High genetic susceptibility to ethanol withdrawal predicts low ethanol consumption. Mamm Genome. 1998;9(12):983–990. doi: 10.1007/s003359900911. [DOI] [PubMed] [Google Scholar]
- Moore MS, DeZazzo J, Luk AY, Tully T, Singh CM, Heberlein U. Ethanol intoxication in Drosophila: genetic and pharmacological evidence for regulation by the cAMP signaling pathway. Cell. 1998;93:997–1007. doi: 10.1016/S0092-8674(00)81205-2. [DOI] [PubMed] [Google Scholar]
- Morozova TV, Anholt RRH, Mackay TFC. Transcriptional response to alcohol exposure in Drosophila melanogaster. Genome Biol. 2006;7(10):R95. doi: 10.1186/gb-2006-7-10-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova TV, Anholt RRH, Mackay TFC. Phenotypic and transcriptional response to selection for alcohol sensitivity in Drosophila melanogaster. Genome Biol. 2007;8(10):R231. doi: 10.1186/gb-2007-8-10-r231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova TV, Ayroles JF, Jordan KW, Duncan LH, Carbone MA, Lyman RF, Stone EA, Govindaraju DR, Ellison RC, Mackay TFC, et al. Alcohol sensitivity in Drosophila: translational potential of systems genetics. Genetics. 2009;183(2):733–745. doi: 10.1534/genetics.109.107490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova TV, Mackay TFC, Anholt RRH. Transcriptional networks for alcohol sensitivity in Drosophila melanogaster. Genetics. 2011;187(4):1193–1205. doi: 10.1534/genetics.110.125229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova TV, Goldman D, Mackay TFC, Anholt RRH. The genetic basis of alcoholism: multiple phenotypes, many genes, complex networks. Genome Biol. 2012;13(2):239. doi: 10.1186/gb-2012-13-2-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan CJ, Robin RW, Osier MV, Sambuughin N, Goldfarb LG, Kittles RA, Hesselbrock D, Goldman D, Long JC. Allelic variation at alcohol metabolism genes (ADH1B, ADH1C, ALDH2) and alcohol dependence in an American Indian population. Hum Genet. 2003;113(4):325–336. doi: 10.1007/s00439-003-0971-z. [DOI] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, Crabbe JC, Blednov YA, Grahame NJ, Phillips TJ, et al. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci USA. 2006;103:6368–6373. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Rhodes JS, Crabbe JC, Mayfield RD, Adron Harris R, Ponomarev I. Molecular profiles of drinking alcohol to intoxication in C57BL/6J mice. Alcohol Clin Exp Res. 2011;35(4):659–670. doi: 10.1111/j.1530-0277.2010.01384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neasta J, Ben Hamida S, Yowell QV, Carnicella S, Ron D. AKT signaling pathway in the nucleus accumbens mediates excessive alcohol drinking behaviors. Biol Psychiatry. 2011;70(6):575–582. doi: 10.1016/j.biopsych.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Jarman DW, Rehm J, Greenfield TK, Rey G, Kerr WC, Miller P, Shield KD, Ye Y, Naimi TS. Alcohol-attributable cancer deaths and years of potential life lost in the United States. Am J Public Health. 2013;103(4):641–648. doi: 10.2105/AJPH.2012.301199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocaranza P, Quintanilla ME, Tampier L, Karahanian E, Sapag A, Israel Y. Gene therapy reduces ethanol intake in an animal model of alcohol dependence. Alcohol Clin Exp Res. 2008;32(1):52–57. doi: 10.1111/j.1530-0277.2007.00553.x. [DOI] [PubMed] [Google Scholar]
- Osterndorff-Kahanek E, Ponomarev I, Blednov YA, Harris RA. Gene expression in brain and liver produced by three different regimens of alcohol consumption in mice: comparison with immune activation. PLoS One. 2013;8(3):e59870. doi: 10.1371/journal.pone.0059870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Luo X, Liu X, Wu LY, Zhang Q, Wang L, Wang W, Zuo L, Wang KS. Genome-wide association studies of maximum number of drinks. J Psychiatr Res. 2013;47(11):1717–1724. doi: 10.1016/j.jpsychires.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SK, Sedore SA, Cronmiller C, Hirsh J. Type II cAMP-dependent protein kinase-deficient Drosophila are viable but show developmental, circadian, and drug response phenotypes. J Biol Chem. 2000;275:20588–20596. doi: 10.1074/jbc.M002460200. [DOI] [PubMed] [Google Scholar]
- Peng C, Li WA, Fu P, Chakraborty T, Hussain M, Guthikonda M, Rafols JA, Ding Y. At low doses ethanol maintains blood-brain barrier (BBB) integrity after hypoxia and reoxygenation: a brain slice study. Neurol Res. 2013;35(8):790–797. doi: 10.1179/1743132813Y.0000000198. [DOI] [PubMed] [Google Scholar]
- Perkins A, Lehmann C, Lawrence RC, Kelly SJ. Alcohol exposure during development: impact on the epigenome. Int J Dev Neurosci. 2013;31(6):391–397. doi: 10.1016/j.ijdevneu.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peru y Colón de Portugal RL, Ojelade SA, Penninti PS, Dove RJ, Nye MJ, Acevedo SF, Lopez A, Rodan AR, Rothenfluh A (2013) Long-lasting, experience-dependent alcohol preference in Drosophila. Addict Biol doi:10.1111/adb.12105 [DOI] [PMC free article] [PubMed]
- Pohl JB, Baldwin BA, Dinh BL, Rahman P, Smerek D, Prado FJ, 3rd, Sherazee N, Atkinson NS. Ethanol preference in Drosophila melanogaster is driven by its caloric value. Alcohol Clin Exp Res. 2012;36(11):1903–1912. doi: 10.1111/j.1530-0277.2012.01817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I. Epigenetic control of gene expression in the alcoholic brain. Alcohol Res. 2013;35(1):69–76. doi: 10.35946/arcr.v35.1.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 2012;32(5):1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott CA, Sullivan PF, Kuo PH, Webb BT, Vittum J, Patterson DG, Thiselton DL, Myers JM, Devitt M, Halberstadt LJ, et al. Genomewide linkage study in the Irish affected sib pair study of alcohol dependence: evidence for a susceptibility region for symptoms of alcohol dependence on chromosome 4. Mol Psychiatry. 2006;11(6):603–611. doi: 10.1038/sj.mp.4001811. [DOI] [PubMed] [Google Scholar]
- Ray LA, Hutchison KE. A polymorphism of the mu-opioid receptor gene (OPRM1) and sensitivity to the effects of alcohol in humans. Alcohol Clin Exp Res. 2004;28(12):1789–1795. doi: 10.1097/01.ALC.0000148114.34000.B9. [DOI] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, et al. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81(3):207–215. doi: 10.1002/(SICI)1096-8628(19980508)81:3<207::AID-AJMG1>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Resendiz M, Chen Y, Oztürk NC, Zhou FC. Epigenetic medicine and fetal alcohol spectrum disorders. Epigenomics. 2013;5(1):73–86. doi: 10.2217/epi.12.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietschel M, Treutlein J. The genetics of alcohol dependence. Ann N Y Acad Sci. 2013;1282:39–70. doi: 10.1111/j.1749-6632.2012.06794.x. [DOI] [PubMed] [Google Scholar]
- Rimondini R, Arlinde C, Sommer W, Heilig M. Long-lasting increase in voluntary ethanol consumption and transcriptional regulation in the rat brain after intermittent exposure to alcohol. Faseb J. 2002;16(1):27–35. doi: 10.1096/fj.01-0593com. [DOI] [PubMed] [Google Scholar]
- Rivas I, Sanvisens A, Bolao F, Fuster D, Tor J, Pujol R, Torrens M, Rey-Joly C, Muga R. Impact of medical comorbidity and risk of death in 680 patients with alcohol use disorders. Alcohol Clin Exp Res. 2013;37(suppl 1):E221–E227. doi: 10.1111/j.1530-0277.2012.01861.x. [DOI] [PubMed] [Google Scholar]
- Rivera-Meza M, Quintanilla ME, Tampier L. Reduction of ethanol consumption in alcohol-preferring rats by dual expression gene transfer. Alcohol Alcohol. 2012;47(2):102–108. doi: 10.1093/alcalc/agr161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodd ZA, Kimpel MW, Edenberg HJ, Bell RL, Strother WN, McClintick JN, Carr LG, Liang T, McBride WJ. Differential gene expression in the nucleus accumbens with ethanol selfadministration in inbred alcohol-preferring rats. Pharmacol Biochem Behav. 2008;89:481–498. doi: 10.1016/j.pbb.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenfluh A, Heberlein U. Drugs, flies, and videotape: the effects of ethanol and cocaine on Drosophila locomotion. Curr Opin Neurobiol. 2002;12(6):639–645. doi: 10.1016/S0959-4388(02)00380-X. [DOI] [PubMed] [Google Scholar]
- Saba L, Bhave SV, Grahame N, Bice P, Lapadat R, Belknap J, Hoffman PL, Tabakoff B. Candidate genes and their regulatory elements: alcohol preference and tolerance. Mamm Genome. 2006;17(6):669–688. doi: 10.1007/s00335-005-0190-0. [DOI] [PubMed] [Google Scholar]
- Saba LM, Bennett B, Hoffman PL, Barcomb K, Ishii T, Kechris K, Tabakoff B. A systems genetic analysis of alcohol drinking by mice, rats and men: influence of brain GABAergic transmission. Neuropharmacology. 2011;60(7–8):1269–1280. doi: 10.1016/j.neuropharm.2010.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Szakall I, Toth R, Kovacs KM, Oros M, Prasad VV, Blumenberg M, Vadasz C. Mouse striatal transcriptome analysis: effects of oral self-administration of alcohol. Alcohol. 2004;32(3):223–241. doi: 10.1016/j.alcohol.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Scholz H, Ramond J, Singh CM, Heberlein U. Functional ethanol tolerance in Drosophila. Neuron. 2000;28:261–271. doi: 10.1016/S0896-6273(00)00101-X. [DOI] [PubMed] [Google Scholar]
- Scholz H, Franz M, Heberlein U. The hangover gene defines a stress pathway required for ethanol tolerance development. Nature. 2005;436:845–847. doi: 10.1038/nature03864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D, Desrivieres S, Aliev FA, Khan AA, Amin N, et al. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc Natl Acad Sci USA. 2011;108(17):7119–7124. doi: 10.1073/pnas.1017288108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seneviratne C, Franklin J, Beckett K, Ma JZ, Ait-Daoud N, Payne TJ, Johnson BA, Li MD. Association, interaction, and replication analysis of genes encoding serotonin transporter and 5-HT3 receptor subunits A and B in alcohol dependence. Hum Genet. 2013;132(10):1165–1176. doi: 10.1007/s00439-013-1319-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley RL, Walter NA, Reilly MT, Fehr C, Buck KJ. Mpdz is a quantitative trait gene for drug withdrawal seizures. Nat Neurosci. 2004;7(7):699–700. doi: 10.1038/nn1271. [DOI] [PubMed] [Google Scholar]
- Shukla SD, Lim RW. Epigenetic effects of ethanol on the liver and gastrointestinal system. Alcohol Res. 2013;35(1):47–55. doi: 10.35946/arcr.v35.1.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh CM, Heberlein U. Genetic control of acute ethanol-induced behaviors in Drosophila. Alcohol Clin Exp Res. 2000;24:1127–1136. doi: 10.1111/j.1530-0277.2000.tb02075.x. [DOI] [PubMed] [Google Scholar]
- Sokolov BP, Jiang L, Trivedi NS, Aston C. Transcription profiling reveals mitochondrial, ubiquitin and signaling systems abnormalities in postmortem brains from subjects with a history of alcohol abuse or dependence. J Neurosci Res. 2003;72(6):756–767. doi: 10.1002/jnr.10631. [DOI] [PubMed] [Google Scholar]
- Spanagel R, Bartsch D, Brors B, Dahmen N, Deussing J, Eils R, Ende G, Gallinat J, Gebicke-Haerter P, Heinz A, et al. An integrated genome research network for studying the genetics of alcohol addiction. Addict Biol. 2010;15(4):369–379. doi: 10.1111/j.1369-1600.2010.00276.x. [DOI] [PubMed] [Google Scholar]
- Spence JP, Liang T, Liu L, Johnson PL, Foroud T, Carr LG, Shekhar A. From QTL to candidate gene: a genetic approach to alcoholism research. Curr Drug Abuse Rev. 2009;2(2):127–134. doi: 10.2174/1874473710902020127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabakoff B, Hoffman PL. The neurobiology of alcohol consumption and alcoholism: an integrative history. Pharmacol Biochem Behav. 2013;113:20–37. doi: 10.1016/j.pbb.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabakoff B, Bhave SV, Hoffman PL. Selective breeding, quantitative trait locus analysis, and gene arrays identify candidate genes for complex drug-related behaviors. J Neurosci. 2003;23(11):4491–4498. doi: 10.1523/JNEUROSCI.23-11-04491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabakoff B, Saba L, Printz M, Flodman P, Hodgkinson C, Goldman D, Koob G, Richardson HN, Kechris K, Bell RL, et al. Genetical genomic determinants of alcohol consumption in rats and humans. BMC Biol. 2009;7:70. doi: 10.1186/1741-7007-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tampier L, Quintanilla ME, Karahanian E, Rivera-Meza M, Herrera-Marschitz M, Israel Y. The alcohol deprivation effect: marked inhibition by anticatalase gene administration into the ventral tegmental area in rats. Alcohol Clin Exp Res. 2013;37(8):1278–1285. doi: 10.1111/acer.12101. [DOI] [PubMed] [Google Scholar]
- Thiele TE, Marsh DJ, Ste Marie L, Bernstein IL, Palmiter RD. Ethanol consumption and resistance are inversely related to neuropeptide Y levels. Nature. 1998;396(6709):366–369. doi: 10.1038/24614. [DOI] [PubMed] [Google Scholar]
- Thomasson HR, Beard JD, Li TK. ADH2 gene polymorphisms are determinants of alcohol pharmacokinetics. Alcohol Clin Exp Res. 1995;19(6):1494–1499. doi: 10.1111/j.1530-0277.1995.tb01013.x. [DOI] [PubMed] [Google Scholar]
- Thorsell A. Neuropeptide Y (NPY) in alcohol intake and dependence. Peptides. 2007;28(2):480–483. doi: 10.1016/j.peptides.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Tikkanen R, Sjoberg RL, Ducci F, Goldman D, Holi M, Tiihonen J, Virkkunen M. Effects of MAOA-genotype, alcohol consumption, and aging on violent behavior. Alcohol Clin Exp Res. 2009;33(3):428–434. doi: 10.1111/j.1530-0277.2008.00853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touvier M, Druesne-Pecollo N, Kesse-Guyot E, Andreeva VA, Galan P, Hercberg S, Latino-Martel P. Demographic, socioeconomic, disease history, dietary and lifestyle cancer risk factors associated with alcohol consumption. Int J Cancer. 2013 doi: 10.1002/ijc.28365. [DOI] [PubMed] [Google Scholar]
- Treadwell JA, Singh SM. Microarray analysis of mouse brain gene expression following acute ethanol treatment. Neurochem Res. 2004;29(2):357–369. doi: 10.1023/B:NERE.0000013738.06437.a6. [DOI] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, et al. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry. 2009;66(7):773–784. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turecki G, Rouleau GA, Alda M. Family density of alcoholism and linkage information in the analysis of the COGA data. Genet Epidemiol. 1999;17(Suppl 1):S361–S366. doi: 10.1002/gepi.1370170761. [DOI] [PubMed] [Google Scholar]
- Urizar NL, Yang Z, Edenberg HJ, Davis RL. Drosophila homer is required in a small set of neurons including the ellipsoid body for normal ethanol sensitivity and tolerance. J Neurosci. 2007;27(17):4541–4551. doi: 10.1523/JNEUROSCI.0305-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zwaluw CS, Engels RC, Vermulst AA, Rose RJ, Verkes RJ, Buitelaar J, Franke B, Scholte RH. A serotonin transporter polymorphism (5-HTTLPR) predicts the development of adolescent alcohol use. Drug Alcohol Depend. 2010;112(1–2):134–139. doi: 10.1016/j.drugalcdep.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Wand G, Levine M, Zweifel L, Schwindinger W, Abel T. The cAMP-protein kinase A signal transduction pathway modulates ethanol consumption and sedative effects of ethanol. J Neurosci. 2001;21(14):5297–5303. doi: 10.1523/JNEUROSCI.21-14-05297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gutala R, Sun D, Ma JZ, Sheela RC, Ticku MK, Li MD. Regulation of platelet-derived growth factor signaling pathway by ethanol, nicotine, or both in mouse cortical neurons. Alcohol Clin Exp Res. 2007;31(3):357–375. doi: 10.1111/j.1530-0277.2006.00331.x. [DOI] [PubMed] [Google Scholar]
- Wang JC, Grucza R, Cruchaga C, Hinrichs AL, Bertelsen S, Budde JP, Fox L, Goldstein E, Reyes O, Saccone N, et al. Genetic variation in the CHRNA5 gene affects mRNA levels and is associated with risk for alcohol dependence. Mol Psychiatry. 2009;14(5):501–510. doi: 10.1038/mp.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KS, Liu X, Aragam N, Jian X, Mullersman JE, Liu Y, Pan Y. Family-based association analysis of alcohol dependence in the COGA sample and replication in the Australian twin-family study. J Neural Transm. 2011;118(9):1293–1299. doi: 10.1007/s00702-011-0628-3. [DOI] [PubMed] [Google Scholar]
- Wang KS, Liu X, Zhang Q, Pan Y, Aragam N, Zeng M. A meta-analysis of two genome-wide association studies identifies 3 new loci for alcohol dependence. J Psychiatr Res. 2011;45(11):1419–1425. doi: 10.1016/j.jpsychires.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Warnault V, Darcq E, Levine A, Barak S, Ron D. Chromatin remodeling—a novel strategy to control excessive alcohol drinking. Transl Psychiatry. 2013;3:e231. doi: 10.1038/tp.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber KE. An apparatus for measurement of resistance to gas-phase reagents. Drosophila Inform Serv. 1988;67:90–92. [Google Scholar]
- Wei YM, Du YL, Nie YQ, Li YY, Wan YJ. Nur-related receptor 1 gene polymorphisms and alcohol dependence in Mexican Americans. World J Gastroenterol. 2012;18(37):5276–5282. doi: 10.3748/wjg.v18.i37.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen T, Parrish CA, Xu D, Wu Q, Shen P. Drosophila neuropeptide F and its receptor, NPFR1, define a signaling pathway that acutely modulates alcohol sensitivity. Proc Natl Acad Sci USA. 2005;102:2141–2146. doi: 10.1073/pnas.0406814102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FW, Rodan AR, Tsai LT, Heberlein U. High-resolution analysis of ethanol-induced locomotor stimulation in Drosophila. J Neurosci. 2002;22:11035–11044. doi: 10.1523/JNEUROSCI.22-24-11035.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolstenholme JT, Warner JA, Capparuccini MI, Archer KJ, Shelton KL, Miles MF. Genomic analysis of individual differences in ethanol drinking: evidence for non-genetic factors in C57BL/6 mice. PLoS One. 2011;6(6):e21100. doi: 10.1371/journal.pone.0021100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worst TJ, Tan JC, Robertson DJ, Freeman WM, Hyytia P, Kiianmaa K, Vrana KE. Transcriptome analysis of frontal cortex in alcohol-preferring and nonpreferring rats. J Neurosci Res. 2005;80(4):529–538. doi: 10.1002/jnr.20496. [DOI] [PubMed] [Google Scholar]
- Xu Y, Ehringer M, Yang F, Sikela JM. Comparison of global brain gene expression profiles between inbred long-sleep and inbred short-sleep mice by high-density gene array hybridization. Alcohol Clin Exp Res. 2001;25(6):810–818. doi: 10.1111/j.1530-0277.2001.tb02284.x. [DOI] [PubMed] [Google Scholar]
- Yang BZ, Kranzler HR, Zhao H, Gruen JR, Luo X, Gelernter J. Haplotypic variants in DRD2, ANKK1, TTC12, and NCAM1 are associated with comorbid alcohol and drug dependence. Alcohol Clin Exp Res. 2008;32(12):2117–2127. doi: 10.1111/j.1530-0277.2008.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Asmaro K, Ren C, Gao M, Peng C, Ding JY, Fredrickson V, Ji X, Ding Y. Acute ethanol treatment reduces blood-brain barrier dysfunction following ischemia/reperfusion injury. Brain Res. 2012;1437:127–133. doi: 10.1016/j.brainres.2011.12.012. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kranzler HR, Yang BZ, Luo X, Gelernter J. The OPRD1 and OPRK1 loci in alcohol or drug dependence: OPRD1 variation modulates substance dependence risk. Mol Psychiatry. 2008;13(5):531–543. doi: 10.1038/sj.mp.4002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Miao Q, Wang C, Zhao R, Li W, Haile CN, Hao W, Zhang XY. Genome-wide DNA methylation analysis in alcohol dependence. Addict Biol. 2013;18(2):392–403. doi: 10.1111/adb.12037. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Guo AY, van den Oord EJ, Aliev F, Jia P, Edenberg HJ, Riley BP, Dick DM, Bettinger JC, Davies AG, et al. Multi-species data integration and gene ranking enrich significant results in an alcoholism genome-wide association study. BMC Genom. 2012;13(suppl 8):S16. doi: 10.1186/1471-2164-13-S8-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimatkin SM, Pronko SP, Vasiliou V, Gonzalez FJ, Deitrich RA. Enzymatic mechanisms of ethanol oxidation in the brain. Alcohol Clin Exp Res. 2006;30(9):1500–1505. doi: 10.1111/j.1530-0277.2006.00181.x. [DOI] [PubMed] [Google Scholar]
- Zuo L, Zhang CK, Wang F, Li CS, Zhao H, Lu L, Zhang XY, Zhang H, Zhang F, Krystal JH, et al. A novel, functional and replicable risk gene region for alcohol dependence identified by genome-wide association study. PLoS One. 2011;6(11):e26726. doi: 10.1371/journal.pone.0026726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Gelernter J, Zhang CK, Zhao H, Lu L, Kranzler HR, Malison RT, Li CS, Wang F, Zhang XY, et al. Genome-wide association study of alcohol dependence implicates KIAA0040 on chromosome 1q. Neuropsychopharmacology. 2012;37(2):557–566. doi: 10.1038/npp.2011.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Wang K, Zhang XY, Krystal JH, Li CS, Zhang F, Zhang H, Luo X. NKAIN1-SERINC2 is a functional, replicable and genome-wide significant risk gene region specific for alcohol dependence in subjects of European descent. Drug Alcohol Depend. 2013;129(3):254–264. doi: 10.1016/j.drugalcdep.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Zhang XY, Wang F, Li CS, Lu L, Ye L, Zhang H, Krystal JH, Deng HW, Luo X. Genome-wide significant association signals in IPO11-HTR1A region specific for alcohol and nicotine codependence. Alcohol Clin Exp Res. 2013;37(5):730–739. doi: 10.1111/acer.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


