Abstract
The transcriptional network of Escherichia coli may well be the most complete experimentally characterized network of a single cell. A rule-based approach was built to assess the degree of consistency between whole-genome microarray experiments in different experimental conditions and the accumulated knowledge in the literature compiled in RegulonDB, a data base of transcriptional regulation and operon organization in E. coli. We observed a high and statistical significant level of consistency, ranging from 70%-87%. When effector metabolites of regulatory proteins are not considered in the prediction of the active or inactive state of the regulators, consistency falls by up to 40%. Similarly, consistency decreases when rules for multiple regulatory interactions are altered or when “on” and “off” entries were assigned randomly. We modified the initial state of regulators and evaluated the propagation of errors in the network that do not correlate linearly with the connectivity of regulators. We interpret this deviation mainly as a result of the existence of redundant regulatory interactions. Consistency evaluation opens a new space of dialogue between theory and experiment, as the consequences of different assumptions can be evaluated and compared.
Regulatory gene networks in the cell play an essential role in controlling the expression of specific genes according to environmental changes. The resulting patterns of gene expression vary temporally and spatially, as the outcome of a set of decisions executed by the regulatory network (Oosawa and Savageau 2002).
Extensive molecular studies in Escherichia coli have determined details of regulatory mechanisms and have also revealed many global aspects of the gene regulatory network (Neidhardt and Savageau 1996; Oosawa and Savageau 2002). RegulonDB is a database that contains information about transcriptional regulation and operon organization of E. coli derived from the careful examination of pertinent literature (Salgado et al. 2001). Although the information is still far from complete, there is currently data for about 83 (of the total possible 314) regulatory proteins (Pérez-Rueda and Collado-Vides 2000) that regulate 600 genes. All of this knowledge together, comprising around 25% of the cellular network, provides us with the largest known regulatory network of a single cell.
The understanding of how structural properties of regulatory networks determine the dynamics of their regulated genes has been the subject of studies for four decades (Kauffman 1974; Savageau 1998; Thieffry et al. 1998; Thomas and D'Ari 1990). The comparison of whole-genome expression profiles with a known network as large as that of E. coli provides a great opportunity to test established hypotheses, to assess more recent theoretical models (Palsson 2001; Pilpel et al. 2001), and in general, to evaluate all aspects on our modeling of the knowledge of gene regulation, such as, for instance, general rules governing the expression by multiple regulators.
Given an initial condition specifying the state of regulatory proteins derived from an experiment, the network of regulatory interactions and conformations of regulators determines theoretical expression states of the regulated genes that can be compared with experimental data. This comparison, which generates a single number, the consistency between experiment and theory, opens a new space of dialogue between theory and experiment. The effects of different assumptions and their corresponding propagation of errors can be tested and compared. Contrary to most recent studies aimed at reconstructing the regulatory wiring from microarray experimental data alone (Eisen et al. 1998; Brown et al. 2000), we used the regulatory interactions described in RegulonDB to establish the network of causal relationships that allowed us to evaluate the congruence between literature and whole-genome expression profiles of E. coli in different conditions. The levels of consistency obtained with this approach range from 70%-87%, and fall by up to 40% when metabolites affecting the conformations of regulatory proteins are not considered. The degree of consistency depends both on the quality of the experiment and on the quality and quantity of knowledge about the regulatory elements governing gene expression.
RESULTS
We analyzed expression profiles of E. coli under four conditions, minimal medium (the common control condition), heat shock, stationary phase, and anaerobic growth. Three control, independently repeated experiments, showed a correlation coefficient varying between 79% and 87%. The filtering of noise, as explained in the Methods section, left a set of 2157 (49%) genes for all analyses presented here. Similarly, of 170 known regulatory proteins, only 83 (49%) satisfied these conditions and were used in our analyses. We used a relative expression scale to transform expression ratios into on and off states, as described in the Methods section. This discretization could be done because all experimental values are relative to a unique control condition, that of minimal medium. Minimal medium shows a larger fraction of on genes, contrary to anaerobic and stationary phase conditions, consistent with overall results obtained in other laboratories (Richmond et al. 1999; Tao et al. 1999; Oh and Liao 2000).
To get an initial estimate of the expected consistency, we performed a comparison of microarray expression values with well-defined known sets of genes of each stimulon. The results of literature comparison are detailed in Tables 1-4S in the Supplemental material, available online at www.genome.org. Table 1 shows that in all conditions except stationary phase, the expression of genes corresponds to that reported in the literature for 86%-88% of the cases. The lower consistency of 69% in stationary phase is not a surprise, as the experimental settings of deprivation and stresses to induce this condition are rather variable in the literature and do not correspond exactly with those used in the microarray experiment. We considered in stationary phase, the subset of genes induced by sigmaS, which precedes the most significant changes involved in the transcription of most of the genes when the cell enters to stationary phase (Ishihama 2000).
Table 1.
Comparison of Microarray Expression With Previous Reports in Literature
| Condition | Number of genes compared | Fraction of genes consistent with literature | |
|---|---|---|---|
| Heat | As reported in literature | 19 | 0.86 |
| Shock | Different from literature | 3 | 0.14 |
| Total | 22 | ||
| Stationary | As reported in literature | 51 | 0.69 |
| Phase | Different from literature | 23 | 0.31 |
| Total | 74 | ||
| Anaerobic growth | As reported in literature | 66 | 0.88 |
| Different from literature | 9 | 0.12 | |
| Total | 75 | ||
| Minimal | As reported in literature | 103 | 0.88 |
| Medium | Different from literature | 14 | 0.12 |
| Total | 117 |
Literature search of genes induced or repressed in each condition were compared with expression-discretized values. Consistency ranges from 88% to 69% in the four conditions tested. The complete set of compared genes can be found in Table 1-4S of Supplemental material.
Definitions of Complex, Simple, and Strict Regulons and Homogeneity of Their Expression
Consistency can be evaluated because of the rich structure of the transcriptional regulatory network, in which regulatory proteins work together in several combinations, governing the expression of several genes. We used the term regulon, as initially defined by Maas for the ArgR regulon in E. coli (Maas 1964), as the set of genes regulated by only one transcriptional regulator. We call these simple regulons. We define as complex regulons, a group of genes regulated by exactly the same set of several regulatory proteins.
Furthermore, complex and simple regulons are strict regulons if the role (activator or repressor) of each regulatory protein is the same for every gene in the regulon. For instance, the group of genes fruB, fruA, fruK, and pykF define the negative strict simple regulon of FruR. On the other hand, the genes edp and pgk conform the complex regulon regulated by FruR and CRP. Once we have discretized individual genes, we identify those simple and complex strict regulons that are homogeneously expressed in a given condition (as explained in the Methods section). Now, we can refer to the on or off state for each strict regulon. We performed this process in all cases, excluding the regulator from the regulon set, even if subject to autoregulation, to prevent noise in the homogeneity due to possible conflicts in the expression of the regulatory gene, such as oscillations or other complex behavior. In the four conditions tested, 77% of the regulons, on average, show a homogeneous expression as shown in Table 2. This is rather high, considering the experimental noise inherent in the methodology of microarrays, messenger stability, and the amount of plausible incomplete knowledge of gene regulation in the database, such as unknown additional regulators affecting transcription initiation, and alternative levels of regulation. In addition, we are not certain that the experiments as performed were done under steady-state conditions. This could be an additional source of error that may have a consequence in our estimates of consistency. The subsequent analysis of consistency is limited to regulons with a homogeneous expression profile. We have information for 77 simple regulons in RegulonDB, with an average of 4.71 genes per regulon. Of these, only on average, 18.25 were shown homogeneous in at least one condition. There are 171 strict complex regulons in the database, of which 39.5, on average, were homogeneously expressed and are analyzed. Table 2 also shows that, on average, 57 simple regulons in each condition are regulating just one gene. Of these, only 20, on average, were used in evaluations of consistency. These left us ∼78 simple and complex strict regulons to work with.
Table 2.
Homogeneity of Expression Levels in Strictly Coregulated Groups Coming From Transcriptome Data
| Conditions | Two or more genes per complex regulon | Simple regulons | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group state | ||||||||||||||
| on | off | Rejected | Fraction of homogeneous complex regulons | on | off | Total of regulons | ||||||||
| Heat shock | 21 | 30 | 20 | 0.72 | 27 | 31 | 129 | |||||||
| Stationary phase | 31 | 32 | 11 | 0.85 | 28 | 26 | 128 | |||||||
| Anaerobic growth | 30 | 26 | 16 | 0.78 | 32 | 26 | 130 | |||||||
| Minimal medium | 36 | 25 | 18 | 0.77 | 25 | 34 | 138 | |||||||
Homogeneity of expression within strict regulons was evaluated on the basis of a binomial probability using the Boolean values of expression in the four conditions tested.
Prediction of Conformation and State of Regulators
The initial conditions of the network in terms of regulatory proteins being present or absent, and active or inactive, are derived from their discretized expression values in the microarray experiment, as well as from the expression of their corresponding strict simple regulons. Presence or absence of regulators is based on their discretized on or off values. In the absence of the regulatory protein, if it is a repressor, the genes of its simple regulon should be on assuming that a strong promoter transcribes them. In the case of a promoter regulated by an activator, we assumed a weak promoter, therefore, in the absence of the activator, the gene should be off (Neidhardt and Savageau 1996). We refer to these as the promoter rules when determining consistency. We found 32% of the 83 regulators to be off in all conditions.
When a regulator is present, a more elaborated process, using our basic knowledge of gene regulation, is required. First, we assume that when a gene gives an on value on the microarray, its protein product is present too. But presence of a regulator does not mean it is in a conformation able to exert its positive (if an activator) or its negative (if a repressor) effect on regulated genes. We assume that regulators have two conformations—which is the case, except for very few exceptions—one in which the allosteric metabolite (i.e., cAMP for CRP, allolactose for LacI, arabinose for AraC, etc.) is bound to the protein and one in which it is unbound. In some cases, alternative conformations involve phosphorylation or other processes, which, in the model, are similarly treated. Conformations are defined as active (for present regulators) or inactive (for present regulators—depending on the presence or absence of its effector—and for absent regulators), and are deduced from the expression of the strict simple regulon for each regulator. Conformations, as defined, can in fact be assigned also for regulators whose effector metabolites have not been identified.
Briefly, we can say that a simple regulon is expressed either when its transcriptional activator is present and active, or when its transcriptional repressor is absent or inactive. We call this the simple rule, as it can be applied only to genes whose promoters are regulated by only one transcription factor, that is to say, to strict simple regulons. Table 3 summarizes the rules used to determine all possible active and inactive states of regulators determining simple regulons to be on or off. The first half of the table, when the regulatory gene is absent, is based on the promoter rules, whereas the second half, when regulatory proteins are present, uses the simple rule. We were able to predict in all of the conditions, on average, the conformation state as active or inactive for 64% of regulators from the initial set of 83. We found that 51% of the regulatory proteins have an inactive state (on average, around 25 are activators and 17 are repressors), whereas 13% are active (around five are activators and six are repressors). For 36% of the regulators (around 30), we were unable to predict whether the proteins were in an active or inactive state, either because the simple regulon did not behave homogeneously, or because there is no known strict simple regulon. With these data at hand, we are now in a position to exploit the structure of the network and evaluate consistency between simple and complex regulons.
Table 3.
Simple Regulatory Rules
| Regulator presence | Regulator function | DNA binding conformation | Effector presence (i) | Discrete expression level | |
|---|---|---|---|---|---|
| P | Absent | → | Off | ||
| Positive | Present | → | Off | ||
| P-i | Absent | → | Off | ||
| Absence | Present | → | Off | ||
| P | Absent | → | On | ||
| Negative | Present | → | On | ||
| P-i | Absent | → | On | ||
| Present | → | On | |||
| P | Absent | → | On | ||
| Positive | Present | → | Off | ||
| Presence | P-i | Absent | → | Off | |
| Present | → | On | |||
| P | Absent | → | Off | ||
| Negative | Present | → | On | ||
| P-i | Absent | → | On | ||
| Present | → | Off |
This table describes the rules determining gene expression when a single protein regulates genes. The rules show the regulatory protein complexes (protein and effector-metabolite), when present. We distinguish two types of conformation of regulators notated as P when they bind to their DNA-binding site with no effector, and as P-i regulators when they bind to DNA with the effector present. In the case of an absent protein, we assume that the expression state depends on the promoter strength.
Consistency in Expression Between Simple and Complex Regulons
Regulatory proteins govern the expression of groups of genes beyond their specific simple regulons (Neidhardt and Savageau 1996). As described previously, we identified which regulators are on and off, as well as which ones are active or inactive to exert their function. These can be considered the initial conditions of the network, in the sense that they apply only to simple regulons and depend on the experimental values for a given condition. Because these same regulators also work coordinately regulating the expression of a diversity of complex regulons, we can ask whether all of these interconnected complex regulons are expressed consistently by comparing the derived expression values with those from the experiment. An additional required ingredient to answer this question is a set of rules that define the level of expression of genes subject to the action of multiple regulators. Traditionally, logical rules have been used when modeling multiple interactions (Thomas and D'Ari 1990). We evaluated consistency on the basis of two types of rules, logical rules and rules derived from an extensive literature search of the few well-studied complex regulons in E. coli. Logical rules stipulate that for multiple repression, one repressor is sufficient to repress, and for multiple activators, one activator is also sufficient to activate. As a result of a literature survey summarized in Table 4, we identified a combination of general rules and specific rules for the case of CRP. CRP-dependent promoters in the case of positive systems dominate well-studied complex regulons. For positively regulated complex regulons, we define a separate CRP-dependent rule (Lawley and Pittard 1994; Goosen and van de Putte 1995; Darwin et al. 1998; Jacobson and Fuchs 1998; Ishihama 2000; Pilpel et al. 2001), stipulating that both activators have to be active for genes to be expressed. In the case of CRP independent promoters, we propose that the presence of one of the activators in its active conformation is enough for gene expression. Complex regulons subject to negative regulation or dual systems follow the expected logical rule, stipulating that the presence of an active repressor is enough to cause the genes to be turned off. Finally, for systems regulated by two repressors, we observed that the proteins show independent repression on the promoters, but maximal repression is obtained when both repressors are active (analyzed cases can be seen in Table 4). Thus, within our Boolean model, the presence of an active repressor is enough to turn genes off. We did not find enough experiments performed in complex systems involving three or more regulatory proteins in a single promoter with sufficient detail to dissect the role of each regulator separately. We therefore extended the rules proposed here for regulons with more than two regulators, as explained in the Methods section. It is clear that these rules are in some cases excessive generalizations that could, as more knowledge accumulates in these complex mechanisms, be improved in this process of putting things together. Nothing prevents our methodology from having ultimately precise rules for every complex regulon, as long as they are well defined.
Table 4.
An Example of the TU's Used to Infer the Behavior of Multiple Regulated Systems
| Protein function | Coregulated group | Regulated promoter(s) | Evidence | Reference | Hypothesis |
|---|---|---|---|---|---|
| Activators | CRP-MalT | malk, malE | CRP triggers MalT repositioning to an appropriate activating position | (Richet et al. 1991) | Both activators are required |
| CRP-FNR | ansB | Both proteins make independent contacts with RNApol-activating transcription | (Busby and Ebright 1997) | Both activators are required | |
| CRP-MelR | melAB | MelR and CRP bind cooperatively at the promoter forming a complex that activates transcription in a codependent manner | (Wade et al. 2001) | Both activators are required | |
| IHF-NR(l), IHF-Xy1R, IHF-NarL | sigma54 promoters and narG | The first protein bends DNA in such a way that the second protein can interact directly with RNApol. Both are required for maximal activation. The presence of IHF alone (bending protein) does not activate transcription | (Goosen and van de Putte 1995) | One specific activator is sufficient | |
| FNR-NarL | napF | Both are required for maximal activation. NarL (proximal site) can activate transcription a few folds compared with the control, but FNR is unable to do it. | (Darwin et al. 1998) | One specific activator is sufficient | |
| DnaA-FIS | nrd | Both are required for maximal activation. DnaA causes more activation than FIS. | (Jacobson and Fuchs 1998) | One specific activator is sufficient | |
| Dual | CRP-CytR | deo | CytR functions as CRP corepressor, causing CRP to repressed transcription | (Rasmussen et al. 1996) | One repressor is sufficient |
| CRP-Lacl | lacZYA | When Lacl is bound, CRP is unable to reach its regulatory site | (Lewis et al. 1996; Matthews, 1996) | One repressor is sufficient | |
| CRP-AraC | araBAD | When AraC is bound, CRP is unable to activate transcription. | (Lee et al. 1981, 1992) | One repressor is sufficient | |
| FNR-FIS-HNS | nirB | FNR anaerobic activation is repressed by the presence of FIS and HNS. | (Wu et al. 1998; Browning et al. 2000) | One repressor is sufficient | |
| NarL-NarP, -IHF | nirB | NarL-NarP complex displaces IHF repression, permitting RNApol to initiate transcription. | (Wu et al. 1998; Browning et al. 2000) | The activator displaces the repressor activity | |
| Repressor | CRP-GalR | gal | The GalR repressor could bind alone to its operators' sites, generating repression. When the repressor HU binds, the repression of gal promoter increases. The proteins repress independently, but maximal repression is reached when both are present | (Aki and Adhya 1997) | One repressor is sufficient |
| FNR-ArcA(over two different promoters) | cyoAB, sdh | The proteins show independent repression, but maximal repression is reached when both are present. | (Cotter and Gunsalus 1992) | One specific repressor is sufficient | |
| ArcA-Betl | betT | The proteins repress independently, but maximal repression is reached when both are present | (Lamark et al. 1996) | One specific repressor is sufficient | |
| TyrR-TrpR | aroLM | TyrR represses in absence of TrpR, but TrpR is unable to repress alone | (Lawley and Pittard 1994) | One specific repressor is sufficient |
The application of these rules, together with the defined active or inactive state of regulator proteins enables us to predict the expression of complex regulons. We assessed the consistency of individual strict complex regulons, comparing our predictions with the observed expression state in the experiments. For example, ArcA and FNR are two repressors predicted to be repressing five genes. These five genes appeared off under heat shock, a state that is consistent with both repressors being in an active state. In this case, the experimental and the predicted expression state coincide, so we define the (ArcA, FNR) complex regulon as a case of direct consistency.
All of the combinations of multiple interactions were considered, and different cases and types of rules associated with consistency of complex regulons are listed in the last column of Table 5. We found the highest level of consistency (87%) in heat shock, compared with that of stationary phase, in which we obtained 70% of consistent regulons. These values are similar to our previous estimates, on the basis of a small set of well-studied genes under these different conditions. We obtained a consistency of 76% and 71% for minimal medium and anaerobiosis growth, respectively. The random estimations of consistency on the basis of an average of 1000 simulated values, given the network and the rules (see the Methods section), are 45% for heat shock, 49% for stationary phase, 54% for minimal medium, and 53% in anaerobiosis. The corresponding Z scores of 4.48, 2.49, 2.54, and 2.24 clearly suggest that the observed values are significantly different from random values.
Table 5.
Rules for Multiple Regulated Systems
| Regulator1 | Regulator2 | Conformation of regulator1 | Conformation of regulator2 | Predicted discrete state | Experimental discrete state | Consistency |
|---|---|---|---|---|---|---|
| Any two activators | ||||||
| activator | activator | activating | activating | on | on | direct |
| activator | activator | not activating | not activating | off | on | incongruent |
| activator | activator | not activating | activating | on | on | activator rule |
| activator | activator | not activating | activating | on | off | incongruent |
| activator | activator | not activating | not activating | off | off | direct |
| One activator is CRP | ||||||
| Crp | activator | not activating | activating | off | off | crp rule |
| Crp | activator | not activating | activating | off | on | incongruent |
| Crp | activator | activating | activating | on | on | direct |
| Crp | activator | not activating | not activating | off | on | incongruent |
| Repressors | ||||||
| repressor | repressor | repressing | not repressing | off | on | incongruent |
| repressor | repressor | repressing | repressing | off | off | direct |
| repressor | repressor | not repressing | repressing | off | off | repressor rule |
| Dual groups | ||||||
| activator | repressor | activating | repressing | off | on | incongruent |
| activator | repressor | activating | not repressing | on | on | direct |
| activator | repressor | not activating | not repressing | ??? | on | basal level |
| activator | repressor | activating | repressing | off | off | repressor rule |
| activator | repressor | not activating | repressing | off | off | direct |
Example of multiple interaction rules extracted from literature for positive, negative, and dual regulated. The first two columns describe the regulator function. Columns 3 and 4 represent the state of the regulator. Column 5 describes the predicted state. Column 6 gives the microarray discrete state, and column 7 represents the consistency between columns 5 (predicted discrete state) and 6 (experimental discrete state).
Re-evaluating Consistency: Alternative Model and Error Propagation
The evaluation of consistency involves several assumptions and operational definitions that function as abstractions of our knowledge on gene regulation. Our approach enables us to evaluate how different components of the model, and knowledge involved, affect consistency. It is interesting to compare the results we obtained with a simpler model that does not take into account the different conformations of regulatory proteins. In such a model, if an activator is on, it is activating, and if a repressor is on, it is repressing. Accordingly, an off activator is not activating and an off repressor is not repressing.
As shown in Figure 1, we found that in all conditions tested the difference between the maximum and minimum level of consistency of NC (no conformation) and NCRP (no conformation and no CRP rule) ranges from around 18% to 40%, with heat shock and anaerobiosis being the most affected conditions. This dramatic decrease illustrates how sensitive these numbers are to different definitions of rules.
Figure 1.
Summary of consistency in each condition. The figure describes the consistency between knowledge extracted from particular experiments written in terms of our multiple rules and microarray data. We show four measures, the first one with the protein state depending on its effector prediction; the second, assuming no CRP rule (NCRP), the third, assuming that all regulatory genes on are active (NC) for no conformation, and the last one considering a combination between the second and the third assumption (NC NCRP). In all cases, off regulatory genes were assumed as inactive.
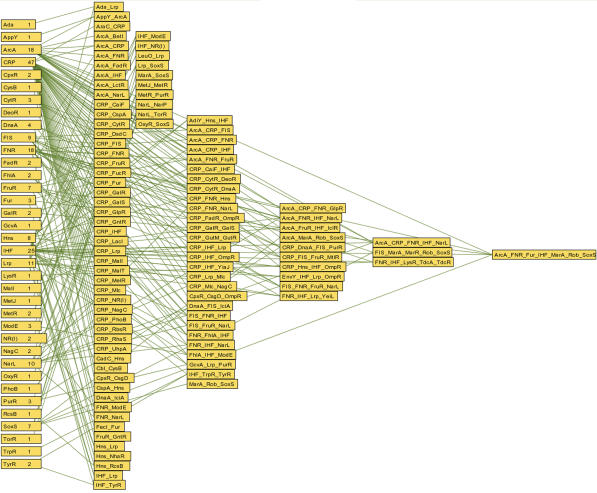
We observe that simple negative autoregulated regulons tend to be less consistent in several conditions than simple negative regulons in which there is no autoregulation. This was observed in the autoregulated regulons governed by PdhR, PurR, and FhlA and OxyR, contrary to simple regulons like ArcA, DnaA, and OmpR. This observation can be rationalized in terms of the oscillations of expression of homeostatic systems (Thomas and D'Ari 1990). Additionally, the identification of low-expressed regulatory proteins, problems in quantification specific to the experiments analyzed, and our partial knowledge of all regulatory interactions affecting the sets of genes analyzed, also affect consistency estimates. On the other hand, the connectivity of regulatory interactions offers a support to restrict the possible outcomes in an important way. CRP participates in 47 complex regulons, FNR in 20, in addition to their simple regulon, whereas AraC participates in only one complex regulon. Figure 2 shows the connectivity across all regulons, simple and complex, with our current knowledge in E. coli. It is this rich structure that was exploited here to evaluate the consistency of expression of global profile experiments in E. coli.
Figure 2.
Connectivity across simple regulons and complex regulons. Each box represents a regulon. Simple regulons are alphabetically ordered at left. Complex regulons are ordered in increasing numbers of regulatory proteins. Connections depart only from simple regulons and arrive at complex regulons that share the same protein. In a few cases, the number of connections of a complex regulon is smaller than the number of its proteins. This is because not every regulator has a simple regulon.
The E. coli network of regulatory interactions follows a power-law distribution (Oosawa and Savageau 2002) similar to what has been observed in other biological networks (Ravasz et al. 2002), that is to say, few nodes are highly connected, whereas many have a low connectivity. One would expect that highly connected regulons, because they are subject to multiple controls, are more sensitive in their expression values than regions with little dependencies/connectivity. On the other hand, local regulators would tend to define distinguishable expression ranges. In this sense, it is interesting to evaluate the impact on overall consistency of a wrongly assigned value of initial conditions to a regulatory protein. Figure 3 shows how a wrong-state assignment of a regulator affects consistency as a function of the connectivity of the regulator. In all of the conditions tested, the most connected protein (CRP) affects consistency less (12% on average) than two regulatory proteins in the middle range of connectivity. FNR connected to 20 complex regulons, and ArcA, with 18 connections, diminish consistency on average 16% and 19%, respectively. The less connected protein FIS, affecting eight complex regulons in our set, decreases consistency only 2%. We suggest that the reason for such a phenomenon is that CRP is a global regulator, with regulatory effects that are redundant compared with those already defined by other regulators. Certainly, global regulators tend to collaborate with other regulators quite extensively (Martínez-Antonio and Collado-Vides 2003).
Figure 3.
Consistency vs. connectivity. The graph shows the way in which an incorrect predicted state on a large connected regulatory protein influences the level of consistency. We show the cases for four proteins in the four conditions tested. Regulators are ordered from the least to the most connected. Fis coregulating with 15 proteins, ArcA with 18, FNR related 20, and CRP coregulating with 47 proteins.
The results presented here suggest that our rule-based approach gives the best levels of congruence, in spite of the noise prevailing in the microarray data and the generalizations made about gene regulation. The results related to each condition are shown in Table 5S in the Supplemental material.
DISCUSSION
The final quantitative result of comparing the consistency between the microarray experiments and the predictions on the basis of the literature is a single number of consistency for each experiment. The range and reproducibility of the high consistency observed shall be strongly dependent on the quality of the experiments. Note that we used a single control as a reference, and all experiments were performed in the same laboratory.
The value of the work presented here is not only the precise degree of consistency, but also an elaborated construction of ideas and knowledge of gene regulation integrated into a rich system whose output is compared with that of the experiment. Furthermore, the virtue of the approach presented here lies in the fact that almost any piece in this construction can be substituted by a different alternative, and can be evaluated by means of the effect on consistency with experimental data. In this sense, this work opens a large window of possibilities for future research. It is also important to emphasize that this setting of ideas and construction is only applicable when the network is known. The large amount of accumulated knowledge on transcription initiation, as well as on operon organization—estimated to represent 25% of the total regulatory network in E. coli—is currently rather unique in this respect.
RegulonDB describes knowledge in a discrete and static way, indicating regulatory interactions and their positive and negative effects. Consistency as described in this work was assigned to a single experimental condition. The expression levels of genes in simple regulons were used to assign the active and inactive state of regulators. We made a comparison, using rules of multiple interactions and gene expression levels within complex regulons. In this way, the comparison of these two levels (state assignment and multiple interaction comparison), determine our final measurements.
Given the rules of multiple interactions, one could think that positive regulators have more redundant interactions than negative regulators. However, we did not observe a difference in consistency when comparing repressors and activators; in all of the conditions analyzed, we found, on average, that 51% of the complex regulons regulated only by activators were consistent, and 49% of those regulated only by repressors were consistent also. One would expect that the total connectivity of nonredundant interactions would exhibit a linear relationship with error propagation. If this were the case, the confidence associated with the experimental determination of the expression of genes could depend on their place within the network.
Estimation of consistency put together three ingredients, the knowledge of the network and precise interactions, the setting of the initial conditions of the state of regulatory proteins as derived from the experiment, and the rules determining the outcome of multiple interactions. As genome projects, modeling of regulatory networks (Covert et al. 2001), and the associated bioinformatic tools progress, such as predictions of regulatory interactions in upstream regions in a large number of bacterial and eukaryotic genomes (Tao et al. 1999; Dombrecht et al. 2002; Halfon et al. 2002; Ravasz et al. 2002), the construction and approach implemented here could, in principle, be applicable and expandable to many other organisms beyond E. coli.
The ability to perform these comparisons opens questions to future research in order to precisely address and improve the adequate level of representation in the modeling of regulatory network interactions, and to integrate our understanding of the regulatory mechanisms as a function of the large set of interconnected regulatory interactions.
METHODS
Growth Conditions
For all experiments, a single colony of E. coli strain MG1655 was inoculated into MOPS minimal medium supplemented with 0.2% glucose (Neidhardt et al. 1974) as initial condition. Three control cultures were grown to mid-logarithmic phase in Erlenmeyer flasks at 37°C with constant aeration. The heat-shock experiment was performed by moving a flask with a mid-logarithmic phase culture from a 37°C water bath to a 50°C bath for 5 min. For the anaerobic growth experiment (or perhaps more correctly called microaerobic growth), the culture was grown in a sealed and evacuated flask to mid-logarithmic phase. For the stationary-phase experiment, a culture was grown until cells reached a stable optical density (600 nm) of 1.5.
Microarray Experiments
Total RNA was prepared from cells using QIAGEN RNeasy columns. Each control RNA sample was labeled with Cy3, and the corresponding experimental RNA sample labeled with Cy5 and cohybridized to a microarray as described previously (Richmond et al. 1999). High-density glass microarrays containing spots corresponding to nearly all E. coli ORFs were prepared as described previously (Richmond et al. 1999; Tao et al. 1999).
Data Treatment
Microarrays were scanned using a Packard Scanarray microarray scanner. The resulting images files were analyzed by determining the average pixel density (intensity) for each spot in the array using Quantarray detection software. A grid of individual ellipses corresponding to each spot was overlaid on the image to identify each spot to be quantified. Spot-specific and local background intensity values were exported to a Microsoft Access database, where background was subtracted from each intensity value. These processed signals between Cy3 and Cy5 channels were normalized, dividing each intensity value by the total intensity of all the spots on the DNA array.
Because the three conditions tested used the same control, we determined the Pearson correlation coefficient of the logarithmic percentage intensities individually for each gene as a measure of reproducibility on these repeated initial conditions. These varied from 0.79 to 0.87. All analyses performed were restricted to the set of genes whose expression values, defined as (Intensity-background)/background were ≥2 in each of the three experimental conditions. A total of 2157 genes satisfied this requirement.
We used the normalized intensities to generate logarithmic expression ratios, as shown in the equation
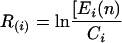 |
in which Ci is the mean normalized expression value of the three repetitions, and Ei(n) is the normalized intensity value for gene i in the stress condition n. We used these values to create a relative expression scale for each gene. We included in our scale the logarithmic expression ratio of the control condition, being zero for all of the genes. Even if we know that this value reports no change in expression, its position in the scale describes the discrete state of the genes in the control condition. The relative expression error RE(i) for each gene i, in each condition n, was computed as
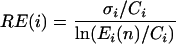 |
in which σ is the standard deviation estimated from the normalized expression values for gene i in the three control repetitions.
Discretization in on and off values was performed for each gene by taking the midpoint of the maximum and minimum of the relative values (in which the control is also included with a value of zero). Only those genes whose values and relative errors did not touch the midline were considered as either on if above, or off if below such midline. Note that in this way, we could discretize values for all four conditions, including the control.
Homogeneity in Strict Regulons
Once each gene had an assigned on or off value for each experiment, we grouped the on and off values in regulons strictly coregulated by the same set of proteins having the same function as an activator or a repressor. We accepted or rejected homogeneous strict regulons using a binomial probability on the basis of the total fraction of genes labeled as off. This measurement assumes independence between the results of each experiment, and independence for each gene within a strict regulon. The probability that k genes show an off state in a strict regulon of N members follows a binomial distribution. Now, we define an interval given by
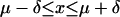 |
in which µ = Np and δ = Np (1 - p) are the expected value and standard deviation, respectively, in the binomial distribution, and p is the global frequency of genes in an off state. For a particular strict regulon, frequencies k/N within this interval were rejected, and those outside of the interval were accepted. Values below µ - δ are considered off, and values above µ + δ are considered on.
Prediction of Conformation and State of Regulators
We used the discrete and homogeneous expression values of strict regulons of a single protein to infer the presence or absence of the allosteric effector of the corresponding regulatory protein; we called this process conformation assignment. In the cases in which a regulator has no simple regulon, but the regulator participates in a complex regulon, their state was deduced after defining that of the other coregulators.
The effector prediction was performed automatically using a program implemented in Prolog, which uses as inputs, (1) the set of on and off values of homogeneous strictly coregulated simple regulons, (2) the known conformations obtained from RegulonDB, and (3) the rules from Table 3 under “Regulator Presence”.
Consistency Evaluation
We determined the consistency for each strict complex regulon in each condition. This is done performing a prediction of its discrete expression state given the conformations and states of the regulatory proteins involved, and the rules for multiple interactions. When this prediction matches the observed homogeneous expression of the strict regulon, it is considered as consistent, otherwise, it is inconsistent. Table 5 gives examples of the combinations for two protein-regulated systems. Taking the rules for two regulators and combining their effects, we defined rules for systems involving more than two regulators. In the case of negative and dual complex regulons, the presence of one active repressor is enough to turn genes off, regardless of the number and state of additional regulators affecting the regulon. For complex regulons coregulated only by activators and excluding CRP, the presence of an active positive regulator is sufficient to turn the genes on. For CRP-dependent strict complex regulons, all of the positive regulators have to be active to turn the regulon on; otherwise the genes are off.
We generated 1000 arrays with on and off entries selected randomly. Each of these randomized arrays of complex regulons were compared with the original arrays of complex regulons in such a way that we were able to generate the distribution of matches for each condition tested. With this information, we calculated the expected value of consistent entries and their standard deviation.
Acknowledgments
R.M.G. had been supported by a Ph.D. fellowship from DGEPUNAM. This work has been supported by grant 0028 from Conacyt-México, and by grant GM62205-02 from NIH. We thank Edgar Díaz-Peredo, Julio Freyre, Heladia Salgado, César Bonavides, Delfino García, and Víctor del Moral for their computer support, and Alejandro Garcíarrubio, Jaques vanHelden, Socorro Gama-Castro, Agustino Matínez-Antonio, and Gabriel Moreno-Hagelsieb and for fruitful discussions during the performance of this work. We acknowledge the useful comments of an anonymous referee.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1387003.
[Supplemental material is available online at www.genome.org. The data set and supplemental material are available at http://www.cifn.unam.mx/Computational_Genomics/Consistency/.]
References
- Aki, T. and Adhya, S. 1997. Repressor induced site-specific binding of HU for transcriptional regulation. EMBO J. 16: 3666-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, M.P., Grundy, W.N., Lin, D., Cristianini, N., Sugnet, C.W., Furey, T.S., Ares Jr., M., and Haussler, D. 2000. Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc. Natl. Acad. Sci. 97: 262-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning, D.F., Cole, J.A., and Busby, S.J. 2000. Suppression of FNR-dependent transcription activation at the Escherichia coli nir promoter by Fis, IHF and H-NS: Modulation of transcription initiation by a complex nucleo-protein assembly. Mol. Microbiol. 37: 1258-1269. [DOI] [PubMed] [Google Scholar]
- Busby, S. and Ebright, R.H. 1997. Transcription activation at class II CAP-dependent promoters. Mol. Microbiol. 23: 853-859. [DOI] [PubMed] [Google Scholar]
- Cotter, P.A. and Gunsalus, R.P. 1992. Contribution of the fnr and arcA gene products in coordinate regulation of cytochrome o and d oxidase (cyoABCDE and cydAB) genes in Escherichia coli. FEMS Microbiol. Lett. 70: 31-36. [DOI] [PubMed] [Google Scholar]
- Covert, M.W., Schilling, C.H., Famili, I., Edwards, J.S., Goryanin, I.I., Selkov, E., and Palsson, B.O. 2001. Metabolic modeling of microbial strains in silico. Trends Biochem. Sci. 26: 179-186. [DOI] [PubMed] [Google Scholar]
- Darwin, A.J., Ziegelhoffer, E.C., Kiley, P.J., and Stewart, V. 1998. Fnr, NarP, and NarL regulation of Escherichia coli K-12 napF (periplasmic nitrate reductase) operon transcription in vitro. J. Bacteriol. 180: 4192-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht, B., Marchal, K., Vanderleyden, J., and Michiels, J. 2002. Prediction and overview of the RpoN-regulon in closely related species of the Rhizobiales. Genome Biol. 3: RESEARCH0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. 95: 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosen, N. and van de Putte, P. 1995. The regulation of transcription initiation by integration host factor. Mol. Microbiol. 16: 1-7. [DOI] [PubMed] [Google Scholar]
- Halfon, M.S., Grad, Y., Church, G.M., and Michelson, A.M. 2002. Computation-based discovery of related transcriptional regulatory modules and motifs using an experimentally validated combinatorial model. Genome Res. 12: 1019-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama, A. 2000. Functional modulation of Escherichia coli RNA polymerase. Annu. Rev. Microbiol. 54: 499-518. [DOI] [PubMed] [Google Scholar]
- Jacobson, B.A. and Fuchs, J.A. 1998. Multiple cis-acting sites positively regulate Escherichia coli nrd expression. Mol. Microbiol. 28: 1315-1322. [DOI] [PubMed] [Google Scholar]
- Kauffman, S. 1974. The large scale structure and dynamics of gene control circuits: An ensemble approach. J. Theor. Biol. 44: 167-190. [DOI] [PubMed] [Google Scholar]
- Lamark, T., Rokenes, T.P., McDougall, J., and Strom, A.R. 1996. The complex bet promoters of Escherichia coli: Regulation by oxygen (ArcA), choline (BetI), and osmotic stress. J. Bacteriol. 178: 1655-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley, B. and Pittard, A.J. 1994. Regulation of aroL expression by TyrR protein and Trp repressor in Escherichia coli K-12. J. Bacteriol. 176: 6921-6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D.H., Huo, L., and Schleif, R. 1992. Repression of the araBAD promoter from araO1. J. Mol. Biol. 224: 335-341. [DOI] [PubMed] [Google Scholar]
- Lee, N.L., Gielow, W.O., and Wallace, R.G. 1981. Mechanism of araC autoregulation and the domains of two overlapping promoters, Pc and PBAD, in the L-arabinose regulatory region of Escherichia coli. Proc. Natl. Acad. Sci. 78: 752-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, M., Chang, G., Horton, N.C., Kercher, M.A., Pace, H.C., Schumacher, M.A., Brennan, R.G., and Lu, P. 1996. Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science 271: 1247-1254. [DOI] [PubMed] [Google Scholar]
- Maas, W.K. and Clark, A.J. 1964. Studies on the mechanism of repression of arginine biosynthesis in E. coli.II. Dominance of repressibility in diploids. J. Mol. Biol. 8: 365-370. [DOI] [PubMed] [Google Scholar]
- Martínez-Antonio, A. and Collado-Vides, J. 2003. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 6: 1-8. [DOI] [PubMed] [Google Scholar]
- Matthews, K.S. 1996. The whole lactose repressor. Science 271: 1245-1246. [DOI] [PubMed] [Google Scholar]
- Neidhardt, F.C., Bloch, P.L., and Smith, D.F. 1974. Culture medium for enterobacteria. J. Bacteriol. 119: 736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt, F.S. and Savageau, M.A. 1996. Regulation beyond the operon. In Escherichia coli and Salmonella: Cellular and molecular biology, 2nd ed. (eds. F. Neidhardt et al.), vol. 2, pp. 1310-1324. ASM Press, Washington, DC. [Google Scholar]
- Oh, M.K. and Liao, J.C. 2000. Gene expression profiling by DNA microarrays and metabolic fluxes in Escherichia coli. Biotechnol. Prog. 16: 278-286. [DOI] [PubMed] [Google Scholar]
- Oosawa, C. and Savageau, M.A. 2002. Effects of alternative connectivity on behavior of randomly constructed Boolean networks. Physica D: Nonlinear Phenomena 170: 143-161. [Google Scholar]
- Palsson, S. 2001. The effects of deleterious mutations in cyclically parthenogenetic organisms. J. Theor. Biol. 208: 201-214. [DOI] [PubMed] [Google Scholar]
- Perez-Rueda, E. and Collado-Vides, J. 2000. The repertoire of DNA-binding transcriptional regulators in Escherichia coli K-12. Nucleic Acids Res. 28: 1838-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilpel, Y., Sudarsanam, P., and Church, G.M. 2001. Identifying regulatory networks by combinatorial analysis of promoter elements. Nat. Genet. 29: 153-159. [DOI] [PubMed] [Google Scholar]
- Rasmussen, P.B., Holst, B., and Valentin-Hansen, P. 1996. Dual-function regulators: The cAMP receptor protein and the CytR regulator can act either to repress or to activate transcription depending on the context. Proc. Natl. Acad. Sci. 93: 10151-10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasz, E., Somera, A.L., Mongru, D.A., Oltvai, Z.N., and Barabasi, A.L. 2002. Hierarchical organization of modularity in metabolic networks. Science 297: 1551-1555. [DOI] [PubMed] [Google Scholar]
- Richet, E., Vidal-Ingigliardi, D., and Raibaud, O. 1991. A new mechanism for coactivation of transcription initiation: Repositioning of an activator triggered by the binding of a second activator. Cell 66: 1185-1195. [DOI] [PubMed] [Google Scholar]
- Richmond, C.S., Glasner, J.D., Mau, R., Jin, H., and Blattner, F.R. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27: 3821-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado, H., Santos-Zavaleta, A., Gama-Castro, S., Millan-Zarate, D., Diaz-Peredo, E., Sanchez-Solano, F., Perez-Rueda, E., Bonavides-Martinez, C., and Collado-Vides, J. 2001. RegulonDB (version 3.2): Transcriptional regulation and operon organization in Escherichia coli K-12. Nucleic Acids Res. 29: 72-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savageau, M.A. 1998. Rules for the evolution of gene circuits. Pac. Symp. Biocomput. 54-65. [PubMed]
- Tao, H., Bausch, C., Richmond, C., Blattner, F.R., and Conway, T. 1999. Functional genomics: Expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 181: 6425-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieffry, D., Huerta, A.M., Perez-Rueda, E., and Collado-Vides, J. 1998. From specific gene regulation to genomic networks: A global analysis of transcriptional regulation in Escherichia coli. Bioessays 20: 433-440. [DOI] [PubMed] [Google Scholar]
- Thomas, R. and D'Ari, R. 1990. Biological Feedback. CRC Press, Boston, MA.
- Wade, J.T., Belyaeva, T.A., Hyde, E.I., and Busby, S.J. 2001. A simple mechanism for co-dependence on two activators at an Escherichia coli promoter. EMBO J. 20: 7160-7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H., Tyson, K.L., Cole, J.A., and Busby, S.J. 1998. Regulation of transcription initiation at the Escherichia coli nir operon promoter: A new mechanism to account for co-dependence on two transcription factors. Mol. Microbiol. 27: 493-505. [DOI] [PubMed] [Google Scholar]