Abstract
Many protein features useful for prediction of protein function can be predicted from sequence, including posttranslational modifications, subcellular localization, and physical/chemical properties. We show here that such protein features are more conserved among orthologs than paralogs, indicating they are crucial for protein function and thus subject to selective pressure. This means that a function prediction method based on sequence-derived features may be able to discriminate between proteins with different function even when they have highly similar structure. Also, such a method is likely to perform well on organisms other than the one on which it was trained. We evaluate the performance of such a method, ProtFun, which relies on protein features as its sole input, and show that the method gives similar performance for most eukaryotes and performs much better than anticipated on archaea and bacteria. From this analysis, we conclude that for the posttranslational modifications studied, both the cellular use and the sequence motifs are conserved within Eukarya.
Biological systems modeling at the molecular level normally requires knowledge about the functionality of the interacting components. The determination of protein function is an essential requirement for many types of systems biology. It is a fundamental axiom that the structure of a protein determines its function. However, whether this is true or not depends very strongly on the level at which one defines “function.” A close relationship between structure and function is observed if the detailed biochemical function is studied, such as which reaction is catalyzed by an enzyme. This type of functionality is often termed the “molecular function,” and it is highly conserved within superfamilies, members of which, according to the SCOP definitions, are required to be related in sequence, structure, and function (Todd et al. 2001).
When studying the much broader “cellular role” categories, the relationship between structure and function becomes much less clear. For example, predicted protein secondary structure is much more useful for predicting enzyme class membership than cellular roles (Jensen et al. 2002). Several examples exist in which proteins have different cellular roles although they belong to the same superfamily. The reverse is also true: Proteins from many different superfamilies are involved in each of the particular cellular role categories.
Even for the chemically related EC classification, the relationship to structure is unclear. For example, the α/β-hydrolase superfamily—contrary to what the name indicates—contains not only hydrolases but also oxidoreductases, transferases, and lyases (Todd et al. 2001). Another example is the zinc peptidase superfamily, which includes a nonenzymatic receptor. Still, conservation of the enzyme class is seen for the majority of the enzyme superfamilies.
Typically, in any genome the function of only half the proteins can be assigned by sequence similarity search methods, whereas the rest remain unassigned. Some of these sequences of unknown function do not resemble any other known protein sequence; others have homologs, but the function of these is also unknown. In any case, it is very difficult to suggest a function for these proteins.
For a long time, the paradigm behind solving this daunting task has been based on protein structure determination and prediction. The rationale has been that the structure of a protein is what determines its function, for which reason the function could be predicted via the structure, for example, by homology building.
To be able to do this, several structural genomics initiatives have been started. These initiatives will be very useful for gaining new insight into the detailed chemical function of proteins that are today poorly understood. But given the relatively weak correlation between protein structure and cellular role combined with the vast number of unrelated proteins of unknown function, we believe that a different approach to predicting the cellular role of these proteins should be taken.
Instead, we have attempted to predict protein function based on predicted properties of proteins, such as physico-chemical properties, predicted posttranslational modifications, and subcellular localization signals (Gupta et al. 2002; Jensen et al. 2002). Although predicted from sequence, they are more conserved among orthologs than paralogs, given the same degree of sequence conservation. This is in contrast to three-dimensional structure, which is conserved for paralogs as well as orthologs.
We furthermore demonstrate that the sequence-derived protein properties characterize proteins of different cellular roles in ways that are conserved not only within Eukarya, but in several cases within all three domains of life: Eukarya, Archaea, and Bacteria. These discoveries have been made through a cross-species analysis of the performance of the ProtFun prediction method (Jensen et al. 2002) for a wide variety of organisms covering mammals, invertebrates, plants, and fungi as well as Crenarchaeota, Euryarchaeota, and Eubacteria.
RESULTS AND DISCUSSION
Features Are More Conserved Among Orthologs Than Paralogs
It is well known and often used in function assignment that orthologs more often have identical function than paralogs (Jensen 2001). If the sequence-derived protein features we use are, indeed, indicative of protein function, they should then be expected to be more conserved within pairs of orthologous proteins than within pairs of paralogous proteins. However, as most of the predicted features used as input by ProtFun have not been experimentally verified, such comparison must be based on the predicted features. The ProtFun method is used as a similarity measure for this comparison, as similarity in feature space will lead to similar function predictions. Orthologous proteins should more often be predicted to have the same function than paralogous proteins.
We have verified this on a data set consisting of all orthologs and paralogs between the complete genomes of Homo sapiens and Drosophila melanogaster. Because orthologs typically are more similar than paralogs at the sequence level, we have examined the feature similarities as a function of the sequence identity (see Fig. 1). It is clear that the functional similarity as predicted from sequence-derived features is most conserved for orthologs over the entire range of sequence similarity studied.
Figure 1.
Estimated probability for same cellular role as function of similarity for orthologs and paralogs. These probabilities were estimated as the overlap integral of the ProtFun predictions for H. sapiens and D. melanogaster proteins involved in each pair. The probabilities could not be reliably estimated outside the range 30%-80% identity as orthology versus paralogy cannot be reliably predicted for distant homologs and because very closely related paralogs are likely predicted to be orthologs.
The ProtFun predictions rely exclusively on sequence-derived features as input. The ability of the method to discern between orthologs and paralogs, therefore, has the implication that the protein features are selectively conserved for orthologs. This is consistent with the observation that O-glycosylation sites are often not conserved between orthologs on a site-by-site basis but rather as a bulk property (K. Julenius, R. Gupta, K. Rapacki, L.J. Jensen, and S. Brunak, unpubl.).
Cross-Species Comparison
The ProtFun function prediction method has been trained on human protein sequences of known function only (Jensen et al. 2002), but given that it relies on protein features that occur in all eukaryotes, it should be expected to be able to generalize to other organisms as well. Considering the results above, it would appear that the method is able to generalize at least to metazoans.
To investigate this further, we evaluated the performance of ProtFun on the complete genomes of 48 organisms. When doing this kind of comparative analysis of genomes/proteomes from many different species, there are several potential sources of artifacts. Because the genes have been annotated using different gene-finding methods or different similarity cutoffs, there can be vast differences in the quality of the annotations (Skovgaard et al. 2001). Also, because genomes have been annotated by different groups, inconsistencies in the functional annotations are likely to occur. To compare protein function across multiple genomes, one has to make sure that the annotation is consistent.
We address these problems by reannotating the function of all proteins based on sequence similarity using the EUCLID method (Tamames et al. 1998; Andrade et al. 1999), restricting ourselves to use proteins for which a function could be assigned reliably. Because questionable ORFs that might have been annotated as genes are very unlikely to display significant sequence similarity to proteins in SWISS-PROT, these will automatically be rejected. The fully automated assignments into functional classes ensure comparability across organisms, but are likely to be less accurate than the original annotations. The fact that not only our own predictions will contain errors, but also the labeling to which we compare, means that we will obtain a conservative estimate of the ProtFun performance.
Good Performance on All Eukaryotes
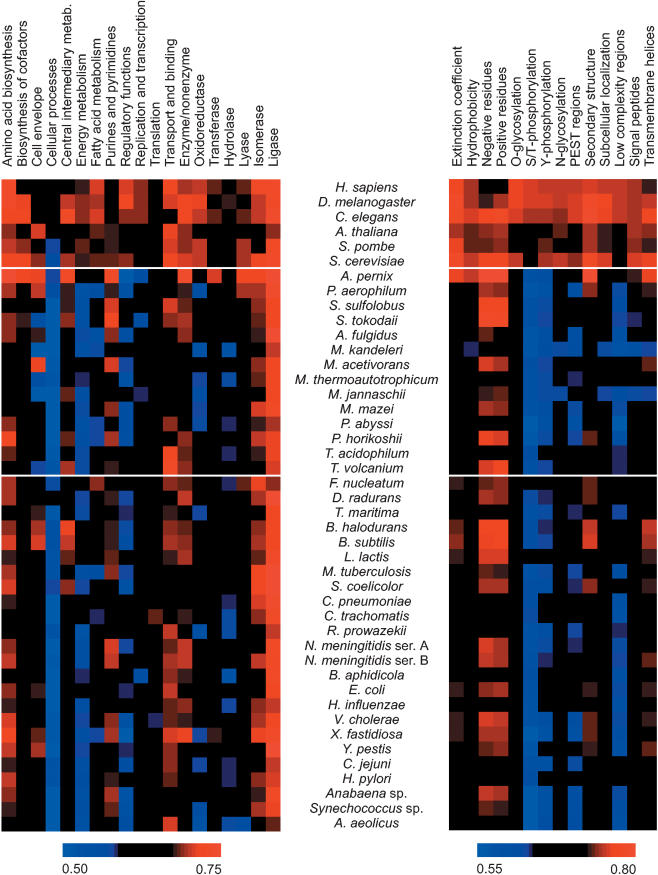
To our surprise, the ProtFun method performs almost equally well on all other eukaryotes tested including yeasts (see Fig. 2). This ability to generalize across very different phyla shows that the trends found by the artificial neural networks not only hold for human proteins but have, in fact, been conserved throughout the eukaryotic domain of life.
Figure 2.
ProtFun performance for functional classes and performance contributions from input features. For 44 organisms the area under the receiver output characteristic (ROC) curve has been plotted for all cellular role categories and enzyme classes (left panel). These performances were mapped into input features based on the feature usage matrix (see Fig. 1 in Jensen et al. 2002).
Sequence-Derived Input Features
Our approach to function prediction relies on sequence-derived input features. These represent physical/chemical and functional biological properties of the protein that can be either calculated or predicted from the amino acid sequence alone. These features include predicted protein secondary structure, transmembrane helices, subcellular localization, and posttranslational modifications.
Although all of these features make biological sense for eukaryotes in general, many of the feature predictors had been trained on mammalian or vertebrate data sets. Their performance on other eukaryotes was therefore unknown.
In the case of prokaryotes, some of the features make no sense at all. For instance, the lack of compartmentation in prokaryotes means that prediction of most subcellular localization makes little sense. How well other features like posttranslational modifications (PTMs) will work for prokaryotes is even less clear: The functional role of a modification may be different from that in eukaryotes, the motif may be different, or the modification may not take place at all.
We have analyzed this in a systematic and quantitative fashion. The performances obtained for cellular roles were mapped according to feature importance. The resulting values represent the performance contributed by each sequence-derived feature. Figure 2 shows these values visualized in the same way as the functional category performances. The general trends are as follows: Features representing structural properties like predicted secondary structure and membrane-spanning helices as well as more general physico-chemical properties of the proteins generalize well to prokaryotes. On the other hand, most of the features representing predicted PTMs and protein sorting signals are of limited value in archaeal and bacterial genomes.
There are certain organisms that deviate from the patterns described above. One example is Buchnera aphidicola, which belongs to the γ subdivision of proteobacteria. In contrast to most other organisms, even the correlations between simple physical/chemical properties (extinction coefficient, hydrophobicity, and number of negative/positive residues) appear to break down (see Fig. 2). All of these features reflect different aspects of the amino acid composition. The lack of correlation is thus likely to result from the unusual amino acid composition of B. aphidicola proteins, which is reflected in the predicted isoelectric points of B. aphidicola proteins (Shigenobu et al. 2000, 2001).
Many Features Fail on Prokaryotes
It was anticipated, because of the very different organization of the eukaryotic and prokaryotic cells, that the predicted protein subcellular localization (according to PSORT) would be of little use in prokaryotes. Still, one could have expected N-terminal signal peptide prediction to work, as the signal peptides not only exist in prokaryotes but can be accurately predicted by the SignalP method that we use in ProtFun (Nielsen et al. 1997).
The problem is that signal peptides do not play the exact same role in eukaryotes and in prokaryotes. Also, eukaryotes have several types of similar N-terminal targeting sequences, which can all be detected from the SignalP scores. For example, eukaryotic proteins targeted for the mitochondria will have mitochondrial targeting peptides, whereas their prokaryotic counterparts would be expected to be cytoplasmic and thus not have signal peptides. This difference in the meaning of similar biological motifs in prokaryotes and eukaryotes explains the very poor performance of the energy metabolism predictor for prokaryotes.
Two types of predicted glycosylation sites, both targeting secreted and membrane-associated proteins, are being used by ProtFun. One is N-linked β-GlcNAc glycosylation of asparagines, which takes place in the endoplasmic reticulum. The other is O-linked α-GalNAc glycosylation of serines and threonines, which takes place in the Golgi. Possibly because glycosylation has not been studied nearly as much in prokaryotes as in eukaryotes, only one of the two types (N-linked β-GlcNAc glycosylation) has been observed in prokaryotes (Spiro 2002). As with signal peptides, the consensus sequence for this modification appears to be the same in prokaryotes and eukaryotes. It is thus reasonable to expect the NetNGlyc predictor to work on prokaryotes even though it was trained on eukaryotic sequences.
Glycosylation also seems to play much the same role in prokaryotes and eukaryotes, but much fewer proteins appear to be glycosylated in prokaryotes (Spiro 2002). The small number of glycoproteins may explain why glycosylation predictions appear to be of limited value for predicting functional classes in prokaryotes, despite both the consensus sequence and function being conserved.
Similar to glycosylation, phosphorylation is known to play an important role in prokaryotes, where it is involved in regulation as in eukaryotes. This makes phosphorylation sites a biologically relevant feature, which could be used for function prediction in prokaryotes. However, it was questionable if predicted phosphorylation sites could be used because the NetPhos predictor was trained solely on eukaryotic data. This depends entirely on whether the specificities of some of the prokaryotic kinases are sufficiently close to those of eukaryotic kinases. In our cross-species analysis, we find that predicted phosphorylation sites contribute little to the performance on prokaryotic proteins, which indicates that the specificities of prokaryotic kinases are quite different from those of eukaryotic kinases.
Considering that so many of the input features used by ProtFun make little or no sense for prokaryotic organisms (the predictors are most often based on mammalian data), it is somewhat surprising that the method works at all for them. Figure 2 shows that the features mainly responsible for this are the physical/chemical properties, in particular the size and charge of the protein represented by the number of negative/positive residues. The only other features that contribute significantly are those related to structure, that is, secondary structure and transmembrane helix prediction, which also are somewhat species unspecific. In this sense, our work reconfirms very early work by de Lisi and coworkers (Klein et al. 1984).
Universal Feature Usage and Consensus in Eukarya
An interesting implication of the ability to generalize across species is that the different posttranslational modifications apparently serve the same purposes for most if not all eukaryotes. Not only do eukaryotes have the gene repertoire for making the same modifications, they also use them in a consistent manner.
The fact that all feature-function correlations hold within Eukarya has one further implication. It indirectly indicates that most (if not all) of the predictors that are used by ProtFun can be expected to work with reasonable accuracy for all eukaryotes. As mentioned above, this could not be taken for granted as some of them have been trained on data sets consisting exclusively of human proteins.
Same Structure—Different Function
In the Introduction, several examples were presented of SCOP superfamilies containing enzymes from entirely different enzyme classes and superfamilies containing both enzymes and nonenzymes. These cases show that conservation of structure at the superfamily level is not sufficient to guarantee that function is also conserved.
With respect to enzyme classification, the Cupredoxin superfamily is one of the most diverse, containing an almost equal proportion of enzymes and nonenzymes. Table 1 shows the enzyme probabilities predicted by ProtFun along with the experimental assignment (Todd et al. 2001). Although all the proteins have the same conserved three-dimensional structure, our approach is able to correctly discriminate between the enzymatic and nonenzymatic members of the Cupredoxin superfamily. It should be pointed out, however, that all the enzymatic members of the superfamily belong to the same protein family, even though some of them are <30% identical at the amino acid level.
Table 1.
Predictions for Members of the Cupredoxin Superfamily
| PDB identifier | Chain | Enzyme prob. | Experimental assignment |
|---|---|---|---|
| 1 NWP | A | 0.257 | Nonenzyme |
| 1 NWP | B | 0.257 | Nonenzyme |
| 2 CBP | 0.289 | Nonenzyme | |
| 1 AAC | 0.301 | Nonenzyme | |
| 1 PLC | 0.310 | Nonenzyme | |
| 1 RCY | 0.325 | Nonenzyme | |
| 2 CUA | B | 0.354 | Nonenzyme* |
| 2 CUA | A | 0.368 | Nonenzyme* |
| 1 JER | 0.404 | Nonenzyme | |
| 1 PAZ | 0.416 | Nonenzyme | |
| 1 CYW | 0.483 | Nonenzyme* | |
| 1 A65 | A | 0.652 | Enzyme |
| 1 NIF | 0.688 | Enzyme | |
| 1 AOZ | A | 0.773 | Enzyme |
| 1 AOZ | B | 0.773 | Enzyme |
| 1 KCW | 0.792 | Enzyme |
For each member of the superfamily, the enzyme probability score from ProtFun is listed along with the experimental enzyme/nonenzyme assignment (Todd et al. 2001). The nonenzymes marked with an asterisk are part of enzymatic complexes, but do not contain active sites.
Evolution of members within the same superfamily of proteins both with and without enzymatic activity is likely to have happened through gene duplication events and subsequent adaptation of one of the copies for a new function. An enzymatic and a nonenzymatic member of the same superfamily are thus likely to be paralogs. It is therefore plausible that the stronger conservation of protein features observed for orthologs compared with paralogs is related to the ability to discriminate between structurally similar but functionally dissimilar proteins.
Conclusions
For a long time, there has been a very strong focus on the importance of protein structure for understanding protein function. However, based on our analysis we conjecture that many other protein properties, for example, posttranslational modifications, may in fact be more, or at least, equally important for determining and maintaining the function of a protein. These properties appear to be conserved among proteins of similar function, both in cases in which the evolutionary relationship can be detected by sequence similarity and in more distantly related proteins of similar structure.
METHODS
Generation of the Data Set
A set of 23,740 protein sequences corresponding to predicted human genes was downloaded from the Ensembl database (Hubbard et al. 2002). Similarly, a set of 14,334 D. melanogaster protein sequences was obtained from FlyBase (Rechsteiner and Rogers 1996), 20,263 Caenorhabditis elegans sequences from the protein database WormBase, and 25,617 Arabidopsis thaliana protein sequences from The Arabidopsis Information Resource (TAIR; Huala et al. 2001).
In addition to these eukaryotic data sets, the complete genome sequences of the two yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe were downloaded from GenBank, and all translations of annotated protein coding regions were extracted (Benson et al. 2002). Protein data sets for 14 archaea and 28 bacteria were extracted in the same manner from the complete genome sequences (see Table 2).
Table 2.
Data Sets Used for Cross-Species Evaluation
| Organism | Protein sequences | Assigned by EUCLID |
|---|---|---|
| Homo sapiens | 23,740 | 13,419 |
| Drosophila melanogaster | 14,334 | 7235 |
| Caenorhabditis elegans | 20,263 | 7840 |
| Arabidopsis thaliana | 25,617 | 11,771 |
| Schizosaccharomyces pombe | 4952 | 2786 |
| Saccharomyces cerevisiae | 6329 | 3302 |
| Aeropyrum pernix | 2694 | 684 |
| Pyrobaculum aerophilum | 2605 | 867 |
| Sulfolobus solfataricus | 2977 | 1186 |
| Sulfolobus tokodaii | 2826 | 1045 |
| Archaeoglobus fulgidus | 2407 | 1074 |
| Methanobacterium thermoautotrophicum | 1869 | 867 |
| Methanococcus jannaschii | 1715 | 781 |
| Methanosarcina mazei | 3371 | 1420 |
| Methanopyrus kandleri | 1691 | 653 |
| Methanosarcina acetivorans | 4540 | 1850 |
| Pyrococcus abyssi | 1765 | 855 |
| Pyrococcus horikoshii | 2064 | 786 |
| Thermoplasma acidophilum | 1031 | 783 |
| Thermoplasma volcanium | 1499 | 792 |
| Anabaena sp. | 5366 | 2444 |
| Aquifex aeolicus | 1522 | 926 |
| Borrelia burgdorferi | 850 | 461 |
| Bacillus halodurans | 4066 | 2223 |
| Bacillus subtilis | 4100 | 2240 |
| Buchnera sp. | 564 | 469 |
| Campylobacter jejuni | 1654 | 975 |
| Chlamydia pneumoniae | 1052 | 530 |
| Chlamydia trachomatis | 894 | 498 |
| Deinoccocus radiodurans | 2937 | 1332 |
| Escherichia coli | 4289 | 2883 |
| Fusobacterium nucleatum | 2068 | 1083 |
| Haemophilus influenzae | 1709 | 1183 |
| Helicobacter pylori | 1566 | 815 |
| Lactococcus lactis | 2266 | 1229 |
| Mycoplasma genitalium | 480 | 332 |
| Mycoplasma pneumoniae | 677 | 441 |
| Mycobacterium tuberculosis | 3918 | 1973 |
| Neisseria meningitides ser. A | 2121 | 1132 |
| Neisseria meningitides ser. B | 2025 | 1088 |
| Rickettsia prowazekii | 834 | 548 |
| Streptomyces coelicolor | 7848 | 3625 |
| Synechocystis sp. | 3169 | 1598 |
| Thermotoga maritima | 1846 | 1064 |
| Treponema pallidum | 1031 | 507 |
| Vibrio cholerae | 3828 | 2054 |
| Xylella fastidiosa | 2766 | 1184 |
| Yersinia pestis | 4008 | 2566 |
The column “protein sequences” lists the number of protein-coding regions annotated in the genomes, with the exception of the organisms H. sapiens, D. melanogaster, C. elegans, and A. thaliana (see text for details on these data sets). The protein sequences that could be assigned to a cellular role by the EUCLID method (last column) show the amount of data available for validation of the ProtFun method for each organism.
To ensure comparable function annotation among these many genomes, all existing (if any) information on protein function was discarded and the proteins were automatically reassigned to cellular role categories by using the EUCLID method (Tamames et al. 1998; Andrade et al. 1999) and to enzyme categories based on the following criteria.
Based on BLAST matches to known proteins in SWISS-PROT, EUCLID collected keywords that are used in an additive scoring system to calculate a Z-score for each cellular role category. We annotated each category separately based on the EUCLID Z-scores with the same rules used to label the training examples used for development of the ProtFun method (Jensen et al. 2002). Sequences were labeled as “positive examples” if their Z-scores were above 3 whereas sequences with a Z-score <0 were labeled as “negative” examples. Any sequences having a Z-score from 0 to 3 were left out of the analysis for the category in question.
Sequences were assigned to enzyme classes based on the same BLAST matches used for selecting keywords above. As SWISS-PROT provides enzyme class information for most enzymes in the description field, this information was extracted for all BLAST matches identified by running EUCLID. Labeling of enzyme versus nonenzyme as well as the major enzyme class was decided by voting among the matches. At least two-thirds majority for either “yes” or “no” was required for a sequence to be labeled.
Performance Evaluation
One of the best commonly used performance evaluation criteria is the correlation coefficient, which we have used as the main criterion during development of the ProtFun method. However, the correlation coefficient cannot be used for the problem at hand because of its dependence on the relative frequency of positive and negative examples in the data set (Baldi et al. 2000). Correlation coefficients can thus not be used for comparing the performance of our prediction method across genomes with different breakdown on functional categories.
Instead, we opt for using the area under the receiver output characteristic (ROC) curve, a plot of true negative rate versus true positive rate. The area under this curve will be 1 for a perfect predictor and 0.5 for a predictor performing no better than random. Like the correlation coefficient, this performance measure is balanced, taking into account the tradeoff between high sensitivity and low rate of false positives. In addition to this, it is also independent of the data set composition in terms of positive and negative examples.
Feature Mapping
The ROC area performances functional classes were mapped onto the sequence-derived features. These sequence-derived features were the prediction methods NetNGlyc (data not shown), NetOGlyc (Hansen et al. 1998), NetPhos (Blom et al. 1999), PEST regions (Rechsteiner and Rogers 1996), PSIPRED (Jones 1999), PSORT (Nakai and Horton 1999), SEG filter (Wootton 1994), SignalP (Nielsen et al. 1999), and TMHMM (Krogh et al. 2001), as well as the number of calculated features: extinction coefficient, grand average hydrophobicity, and the numbers of positively and negatively charged residues.
For each organism, the performance of each of these 14 input features was calculated as a weighted average of the ROC areas of the 12 cellular role categories. Each cellular role category entered with a weight corresponding to the number of neural networks in its ensemble of predictors that make use of the feature in question (see Fig. 1 in Jensen et al. 2002). We decided not to include the enzyme classifiers in this mapping procedure because all of the neural network ensembles make use of a large number of sequence-derived features. This makes it very difficult to correctly attribute the predictive performance to the right features for these classifiers.
Obtaining Sets of Orthologs and Paralogs
Assignment of orthologs versus paralogs is far from being a trivial problem. To obtain a large data set of orthologs/in-paralogs and out-paralogs, we have made use of the INPARANOID tool to classify the pairs of homologous proteins between the H. sapiens and D. melanogaster data sets described above (Remm et al. 2001). Paralogs were assigned based on BLAST matches covering at least 50% of the sequence length, which were not listed as orthologs by INPARANOID. By this approach, we predicted 13,562 pairs of orthologous proteins and 151,923 pairs of paralogous proteins. To ensure comparability of the two data sets, only pairs of paralogs consisting of one H. sapiens and one D. melanogaster protein were included.
Acknowledgments
The authors thank Ulrik de Lichtenberg and Thomas Skøt Jensen for valuable discussions and ideas. We also thank Marie Skovgaard for comments on the manuscript. This work was supported by the Danish National Research Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1190803. Article published online before print in October 2003.
References
- Andrade, M.A., Brown, N.P., Leroy, C., Hoersch, S., de Daruvar, A., Reich, C., Franchini, A., Tamames, J., Valencia, A., Ouzounis, C., et al. 1999. Automated genome sequence analysis and annotation. Bioinformatics 15: 391-412. [DOI] [PubMed] [Google Scholar]
- Baldi, P., Brunak, S., Chauvin, Y., Andersen, C.A.F., and Nielsen, H. 2000. Assessing the accuracy of prediction algorithms for classification: An overview. Bioinformatics 16: 412-424. [DOI] [PubMed] [Google Scholar]
- Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J., Rapp, B.A., and Wheeler, D.L. 2002. GenBank. Nucleic Acids Res. 30: 17-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom, N., Gammeltoft, S., and Brunak, S. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294: 1351-1362. [DOI] [PubMed] [Google Scholar]
- Gupta, R., Jensen, L.J., and Brunak, S. 2002. Orphan protein function and its relation to glycosylation. In Ernst Schering Research Foundation Proceedings (eds. H.-M. Mewes et al.), Vol. 38, pp. 275-294. Springer-Verlag, Berlin, Germany. [PubMed] [Google Scholar]
- Hansen, J.E., Lund, O., Tolstrup, N., Gooley, A.A., Williams, K.L., and Brunak, S. 1998. NetOglyc: Prediction of mucin type O-glycosylation sites based on sequence context and surface accessibility. Glycoconj. J. 15: 115-130. [DOI] [PubMed] [Google Scholar]
- Huala, E., Dickerman, A., Garcia-Hernandez, M., Weems, D., Reiser, L., LaFond, F., Hanley, D., Kiphart, D., Zhuang, J., Huang, W., et al. 2001. The Arabidopsis Information Resource (TAIR): A comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 29: 102-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard, T., Barker, D., Birney, E., Cameron, G., Chen, Y., Clark, L., Cox, T., Cuff, J., Curwen, V., Down, T., et al. 2002. The Ensembl genome database. Nucleic Acids Res. 30: 38-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, L.J., Gupta, R., Blom, N., Devos, D., Tamames, J., Kesmir, C., Nielsen, H., Stærfeldt, H.H., Rapacki, K., Workman, C., et al. 2002. Prediction of human protein function from post-translational modifications and localization features. J. Mol. Biol. 319: 1257-1265. [DOI] [PubMed] [Google Scholar]
- Jensen, R.A. 2001. Orthologs and paralogs—We need to get it right. Genome Biol. 2: interactions1002.1-1002.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D.T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292: 195-202. [DOI] [PubMed] [Google Scholar]
- Klein, P., Kanehisa, M., and DeLisi, C. 1984. Prediction of protein function from sequence properties. Discriminant analysis of a data base. Biochim. Biophys. Acta 787: 221-226. [DOI] [PubMed] [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E.L. 2001. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305: 567-580. [DOI] [PubMed] [Google Scholar]
- Nakai, K. and Horton, P. 1999. PSORT: A program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24: 34-36. [DOI] [PubMed] [Google Scholar]
- Nielsen, H., Brunak, S., Engelbrecht, J., and von Heijne, G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10: 1-6. [DOI] [PubMed] [Google Scholar]
- Nielsen, H., Brunak, S., and von Heijne, G. 1999. Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein Eng. 12: 3-9. [DOI] [PubMed] [Google Scholar]
- Rechsteiner, M. and Rogers, S.W. 1996. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21: 267-271. [PubMed] [Google Scholar]
- Remm, M., Storm, C.E.V., and Sonnhammer, E.L.L. 2001. Automatic clustering of orthologs and in-paralogs from pairwise species comparison. J. Mol. Biol. 314: 1041-1052. [DOI] [PubMed] [Google Scholar]
- Shigenobu, S., Watanabe, H., Hattori, M., Sakaki, Y., and Ishikawa, H. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407: 81-86. [DOI] [PubMed] [Google Scholar]
- Shigenobu, S., Watanabe, H., Sakaki, Y., and Ishikawa, H. 2001. Accumulation of species-specific amino acid replacements that cause loss of particular protein functions in Buchnera, an endocellular bacterial symbiont. J. Mol. Evol. 53: 377-386. [DOI] [PubMed] [Google Scholar]
- Skovgaard, M., Jensen, L.J., Brunak, S., Ussery, D., and Krogh, A. 2001. On the total number of genes and their length distribution in complete microbial genomes. Trends Genet. 17: 425-428. [DOI] [PubMed] [Google Scholar]
- Spiro, R.G. 2002. Protein glycosylation: Nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12: 43R-56R. [DOI] [PubMed] [Google Scholar]
- Tamames, J., Ouzounis, C., Casari, G., Sander, C., and Valencia, A. 1998. EUCLID: Automatic classification of proteins in functional classes by their database annotations. Bioinformatics 14: 542-543. [DOI] [PubMed] [Google Scholar]
- Todd, A.E., Orengo, C.A., and Thornton, J.M. 2001. Evolution of function in protein superfamilies, from a structural perspective. J. Mol. Biol. 307: 1113-1143. [DOI] [PubMed] [Google Scholar]
- Wootton, J.C. 1994. Non-globular domains in protein sequences: Automated segmentation using complexity measures. Comput. Chem. 18: 269-285. [DOI] [PubMed] [Google Scholar]