Abstract
Objective
To identify Candida species in asymptomatic subjects in Bobo-Dioulasso (Burkina Faso) by the matrix-assisted laser desorption ionization-time of flight mass spectrometry.
Methods
A cross-sectional study was conducted from January to February 2013 in Bobo-Dioulasso to collect fecal and urine specimens from voluntary donors. Fungal strains were isolated on Sabouraud dextrose agar and analyzed using matrix-assisted laser desorption ionisation-time-of-flight mass spectrometry.
Results
A total of 135 samples including stools (78.5%, 106/135) and urine (21.5%, 29/135) were analyzed. The results revealed that fecal specimens contained mainly Candida krusei (C. krusei) (42.5%) followed by Candida albicans (29.3%), Candida glabrata (18.0%) and Candida tropicalis (C. tropicalis) (4.7%). C. krusei (34.6%) was also found to be the most frequently identified in urine samples followed by Candida albicans (27.0%), C. tropicalis (15.4%) and Candida parapsilosis. However, uncommon species such as Candida nivariensis, Candida kefyr, Candida norvegensis, Candida parapsilosis, Candida lusitaniae and Candida robusta were also identified from fecal and urines samples.
Conclusions
This study noted the emergence of species such as C. krusei, Candida glabrata, Candida parapsiolosis, C. tropicalis, Candida nivariensis, Candida norvegensis, and others. It is an imperative to take into account the existence of these species in the therapeutic management of patients in Bobo-Dioulasso.
Keywords: Candida, Mass spectrometry, Matrix-assisted laser desorption ionization-time of flight, Bobo-Dioulasso
1. Introduction
Invasive fungal infections caused by Candida spp. remain a major cause of morbidity and mortality in the immunocompromised individuals, and more than 150 species of yeast have now been associated with human pathologies[1]–[3]. Although Candida albicans (C. albicans) remained the predominant agent of nosocomial infections, an increasing number of infections are being attributed to non-albicans species, such as Candida glabrata (C. glabrata), Candida parapsilosis (C. parapsilosis), Candida tropicalis (C. tropicalis), Candida lusitaniae, and Candida krusei (C. krusei) that emerged over recent years as significant opportunistic pathogens[4],[5].
Recent studies concerning the epidemiology of invasive mycoses showed a significant change in the profile of Candida species involved in human pathology[2]. Many strains of C. albicans are still susceptible to antifungals; however species such as C. krusei, C. glabrata, Candida bracarensis, Candida nivariensis (C. nivariensis), C. parapsilosis and Candida guilliermondii have developed intrinsic resistance to triazoles, amphotericin B or to echinocandins[6],[7]. It is therefore essential to consider these changes in fungal ecology upon the therapeutic management of patients, particularly for empirical prescription.
Given the inherently variable antifungal susceptibility profiles of Candida spp., the correct identification of the species is often critical for efficient therapeutic decisions. The identification of the species therefore remains a crucial element in prediction of the response to antifungal treatment[8]. The main Candida spp. associated with human disease are readily identified by conventional mycological methods, which rely upon a combination of morphological features coupled with the abilities of the organisms to metabolize selected sugars or assimilate a variety of carbon and nitrogen sources. This requires two to five days or more in the case of uncommon species[9]. These phenotypic methods can sometimes lead to misidentification or imprecise identification, especially for species phylogenetically proximate, such as C. albicans/Candida dubliniensis, Candida inconspicua/Candida norvegensis (C. norvegensis) or C. glabrata/C. nivariensis/Candida bracarensis[9],[10]. Thus, the adequate identification of these species often requires the combination of several phenotypic and molecular approaches to increase the discrimination level. The molecular approaches are based on the analysis of the genes encoding the ribosomal RNA. More recently, molecular methods, including restriction fragment length polymorphism, sequencing of internal transcribed spacer regions, multilocus sequence typing, and barcoding have been used to identify clinical and nonclinical isolates. All these methods are relatively expensive and time-consuming[11],[12]. Recent studies report the use of mass spectroscopy for the identification of yeasts as a serious alternative for conventional identification methods[13],[14].
Burkina Faso is a sub Saharan country, with endemic HIV infection and fungal infections. Despite this, there is no coordination on the question of HIV and opportunistic mycoses. In the country, even though the data exist on the identification and management of therapeutics bacteria, there is limited data on fungal infections. The present study was conducted in Bobo-Dioulasso, a town located in the south-western of Burkina Faso. The main objective was to identify Candida species by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS) in asymptomatic individuals.
2. Materials and methods
2.1. Yeast collection strains
A cross-sectional study was conducted to collect fecal and urine samples from voluntary donors for a total period of two months in the laboratory of Parasitology-Mycology in the university hospital of Bobo-Dioulasso. The collected samples included urines and stool. Fungal strains were isolated from these samples by cultivation on Sabouraud dextrose agar and frozen in sterile water at -20 °C until used[9].
2.2. Sample preparation
Pure fungal strains from frozen stocks were obtained by 24 to 48 h incubation on BBL-CHROME agar™ or CandiSelect4™ or on Sabouraud medium supplemented with antibiotics[15],[16]. For the MALDI-TOF assay, the on-target extraction method was used. In brief, microorganisms from a colony were applied directly to a disposable target slide using 1 µL loop[15],[17]. They were lysed with 1.2 µL of 70% formic acid and dried at room temperature. Subsequently, each sample was overlaid with 1.2 µL of matrix solution, a saturated solution of α-cyano-4-hydroxycinnamic acid (CHCA), and air dried at room temperature.
The CHCA matrix co crystallized with the sample and dried on a metal plate (96-AnchorChip™ spot or spots Polished Steel™ 384, Bruker Daltonics) on the site of a spot. The reading of the plate was made in 24 h by MALDI-TOF system (Bruker Daltonics GmbH, Bremen, Germany).
3. Results
A total of 135 fecal samples (78.5%, 106/135) and urine (21.5%, 29/135) from voluntary donors were analyzed by the MALDI-TOF assay. Candida was the only genus of yeast identified by the MALDI-TOF. A total of 132 samples (97.8%, 132/135) were successfully identified through direct deposit on the target. Three urine samples (2.2%, 3/135) were neither identified after direct deposit nor after complete extraction of proteins. Thus success rate of species identification in MALDI-TOF was 97.8% (132/135).
From 106 species identified from fecal samples, C. krusei (42.5%) was the most frequent followed by C. albicans (29.3%), C. glabrata (18.0%) and C. tropicalis (4.7%) (Figure 1). Uncommon species such as C. nivariensis, Candida kefyr, C. norvegensis, C. parapsilosis, and Candida robusta were also identified.
Figure 1. Candida species identified in stools by MALDI-TOF assay.
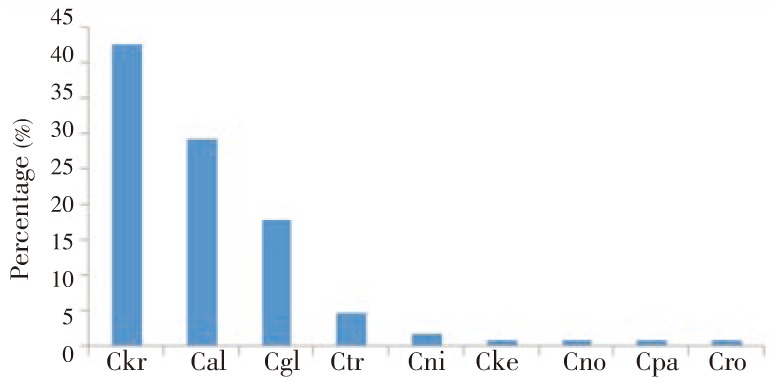
Ckr: C. krusei, Cal: C. albicans, Cgl: C. glabrata, Ctr: C. tropicalis, Cni: C. nivariensis, Cke: Candida kefyr, Cno: C. norvegensis, Cpa: C. parapsilosis, Cro: Candida robusta.
For urine, a total of 26 samples were successfully analyzed by MALDI-TOF system. C. krusei (34.6%) was the most frequent followed by C. albicans (27%), C. tropicalis (15.4%), and C. parapsilosis (Figure 2). C. glabrata, Candida lusitaniae and Candida robusta were the uncommon species identified from urines.
Figure 2. Candida species identified in urine by MALDI-TOF assay.
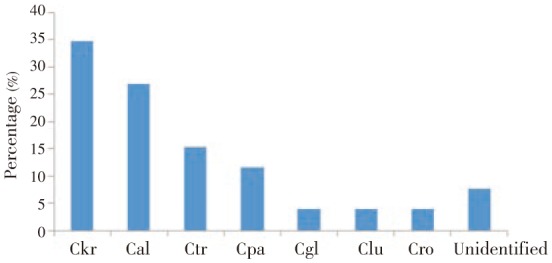
Ckr: C. krusei, Cal: C. albicans, Ctr: C. tropicalis, Cpa: C. parapsilosis, Cgl: C. glabrata, Clu: Candida lusitaniae, Cro: Candida robusta.
4. Discussion
The recent evolution of epidemiology of invasive fungal infections is marked by a spectacular increase in non-albicans species. Diversity of non-albicans species prompted biologists to develop new tools for molecular and immunological characterization of some unusual species whose identification by phenotype methods has proven difficult. More recently, MALDI-TOF MS has been proposed as alternative conventional methods[18]. The present study was designed as identifying some Candida species isolated from stool and urine samples in Bobo-Dioulasso by using MALDI-TOF system. According to our results, C. krusei prevailed in fecal samples and urine samples. Usually, this species is non-pathogenic in the absence of subjacent immunosuppression; this justifies this high prevalence in asymptomatic voluntary donors. In contrast, previous studies showed the predominance of C. albicans in 50% of pathological samples beside the other species emerging such as C. krusei, C. glabrata, C. parapsiolosis, C. tropicalis, C. nivariensis, C. norvegensis, and others[2],[4].
Candida albicans and most of non-albicans yeasts identified in this study such as C. krusei, C. glabrata, C. parapsiolosis, C. tropicalis, C. nivariensis, C. norvegensis are the saprophytes of the digestive tract of humans and many animals. These yeasts are opportunistic which become pathogenic under the influence of general risk factors notably, imbalance of intestinal flora after taking antibiotics or local risk factors including diabetes and deficit of immunity. Identification of these yeasts in this study corroborates their emerging nature in human fungal infections. However, the emergence requires a correct phenotypic and molecular identification for adequate therapeutic management of patients. In our context, further large-scale study in all health regions of Burkina Faso taking into account pathological samples would allow better understanding of their epidemiology.
Furthermore, the identification of some species such as C. nivariensis, C. glabrata, C. parapsilosis which showed intrinsic resistance to usual antifungal should be investigated for antifungal susceptibility[19]. In fact, these strains are from Bobo-Dioulasso, where the prescription of antifungals is almost inexistent compared to antibiotics which are widely prescribed against any infectious. This low usage of antifungals could give another behavior of isolates from Bobo-Dioulasso for the use of antifungal agents. The investigation for antifungal susceptibility would permit a better understanding of the susceptibility of these isolates which showed intrinsic resistance to conventional antifungal such as triazole and amphotericin[19].
Moreover, three strains (2.2%) from this study could not be identified by MALDI-TOF system. In fact, uncommon yeast species are emerging as human pathogens and their identification may pose a challenge when the type strains have not yet been included into the diagnostic databases of identification system. This can lead to misidentification of species. In addition, it is essential that databases designers provide regular updates. This update is major advantage for identification by MALDI-TOF. In fact, two common causes of identification failures may be related to the content of the database[20].
Mass spectrometry MALDI-TOF allows rapid identification of microorganisms isolated in a routine laboratory. The present study is the first conducted in Bobo-Dioulasso. Some emerging species of Candida such as C. krusei, C. glabrata, C. parapsiolosis, C. tropicalis, C. nivariensis, C. norvegensis, have been recorded. These species being isolated from asymptomatic subjects could become pathogenic at any time; it is imperative to take into account the existence of these species in the therapeutic management of patients in Bobo-Dioulasso. However, further large-scale study in all health regions of Burkina Faso taking into account pathological samples would allow better understanding of the epidemiology of Candida yeasts and establish the cartography of these species.
Acknowledgments
Authors thank all the patients who participated in the study and all staff of Laboratory of Parasitology-Mycology, CHU Saint Antoine (Paris).
Comments
Background
Invasive infections by Candida spp. are one of the major threads particularly for immuno-compromised patients. In candidiasis, Candida albicans has long been a major pathogen, but clinical reports of infections of other Candida species are rapidly increasing in late years. Because different Candida species show different antifungal drug susceptibility, accurate and succinct methods for species identification are required.
Research frontiers
The authors determined Candida species cultured from fecal and urine specimens corrected from asymptomatic subjects in a town of Burkina Faso, using MALDI-TOF method.
Related reports
Recent studies report the use of mass spectroscopy for the identification of yeasts as a serious alternative for conventional identification methods.
Innovations and breakthroughs
Candida krusei and some other Candida species which are not usual main human pathogens are found in fecal and urine samples from asymptomatic voluntary donors. The authors are warning and insisting that establishment of proper and simple method for identifying Candida species are required, because susceptibility of antibiotics is different among Candida species.
Applications
Diagnosis, epidemiology, and preventive measures are applied against candidiasis. The identification of some species such as C. nivariensis, C. glabrata, C. parapsilosis which showed intrinsic resistance to usual antifungal should be investigated for antifungal susceptibility.
Peer review
The paper describes several Candida species and their percentages found in fecal and urine samples of asymptomatic voluntary donors in a town of Burkina Faso. The authors used MALDI-TOF MS method which is supposed to be one of the most accurate and succinct methods for identification of fungi and other microorganisms. Although the present work is performed on asymptomatic donors, the work would help in future diagnosis of patients with candidiasis.
Footnotes
Conflict of interest statement: We declare that we have no conflict of interest.
References
- 1.Awosika S, Olajubu F, Amusa N. Microbiological assessment of indoor air of a teaching hospital in Nigeria. Asian Pac J Trop Biomed. 2012;2:465–468. doi: 10.1016/S2221-1691(12)60077-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karou DS, Djigma F, Sagna T, Nadembega C, Zeba M, Kabre A, et al. Antimicrobial resistance of abnormal vaginal discharges microorganisms in Ouagadougou, Burkina Faso. Asian Pac J Trop Biomed. 2012;2:294–297. doi: 10.1016/S2221-1691(12)60025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marr KA. Invasive Candida infections: the changing epidemiology. Oncology. 2004;14:9–14. [PubMed] [Google Scholar]
- 5.Nucci M, Marr KA. Emerging fungal diseases. Clin Infect Dis. 2005;41:521–526. doi: 10.1086/432060. [DOI] [PubMed] [Google Scholar]
- 6.Espinel-Ingroff A. In vitro antifungal activities of anidulafungin and micafungin, licensed agents and the investigational triazole posaconazole as determined by NCCLS methods for 12,052 fungal isolates: review of the literature. Rev Iberoam Micol. 2003;20:121–136. [PubMed] [Google Scholar]
- 7.Miceli MH, Díaz JA, Lee SA. Emerging opportunistic yeast infections. Lancet Infect Dis. 2011;11:142–151. doi: 10.1016/S1473-3099(10)70218-8. [DOI] [PubMed] [Google Scholar]
- 8.Pappas PG, Rex JH, Sobel JD, Filler SG, Dismukes WE, Walsh TJ, et al. Guidelines for treatment of candidiasis. Clin Infect Dis. 2004;38:161–189. doi: 10.1086/380796. [DOI] [PubMed] [Google Scholar]
- 9.Pincus DH, Orenga S, Chatellier S. Yeast identification-past, present, and future methods. Med Mycol. 2007;45:97–121. doi: 10.1080/13693780601059936. [DOI] [PubMed] [Google Scholar]
- 10.Borman AM, Petch R, Linton CJ, Palmer MD, Bridge PD, Johnson EM. Candida nivariensis, an emerging pathogenic fungus with multidrug resistance to antifungal agents. J Clin Microbiol. 2008;46:933–938. doi: 10.1128/JCM.02116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montero CI, Shea YR, Jones PA, Harrington SM, Tooke NE, Witebsky FG, et al. Evaluation of pyrosequencing (R) technology for the identification of clinically relevant non-dematiaceous yeasts and related species. Eur J Clin Microbiol Infect Dis. 2008;27:821–830. doi: 10.1007/s10096-008-0510-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seyfarth F, Ziemer M, Sayer HG, Burmester A, Erhard M, Welker M, et al. The use of ITS DNA sequence analysis and MALDI-TOF mass spectrometry in diagnosing an infection with Fusarium proliferatum. Exp Dermatol. 2008;17:965–971. doi: 10.1111/j.1600-0625.2008.00726.x. [DOI] [PubMed] [Google Scholar]
- 13.Alanio A, Beretti JL, Dauphin B, Mellado E, Quesne G, Lacroix C, et al. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for fast and accurate identification of clinically relevant Aspergillus species. Clin Microbiol Infect. 2011;17:750–755. doi: 10.1111/j.1469-0691.2010.03323.x. [DOI] [PubMed] [Google Scholar]
- 14.Stevenson LG, Drake SK, Shea YR, Zelazny AM, Murray PR. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of clinically important yeast species. J Clin Microbiol. 2010;48:3482–3486. doi: 10.1128/JCM.00687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bader O, Weig M, Taverne-Ghadwal L, Lugert R, Gross U, Kuhns M. Improved clinical laboratory identification of human pathogenic yeasts by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Microbiol Infect. 2011;17:1359–1365. doi: 10.1111/j.1469-0691.2010.03398.x. [DOI] [PubMed] [Google Scholar]
- 16.Sendid B, Ducoroy P, Francois N, Lucchi G, Spinali S, Vagner O, et al. Evaluation of MALDI-TOF mass spectrometry for the identification of medically-important yeasts in the clinical laboratories of Dijon and Lille hospitals. Med Mycol. 2013;51:25–32. doi: 10.3109/13693786.2012.693631. [DOI] [PubMed] [Google Scholar]
- 17.Bille E, Dauphin B, Leto J, Bougnoux ME, Beretti JL, Lotz A, et al. MALDI-TOF MS Andromas strategy for the routine identification of bacteria, mycobacteria, yeasts, Aspergillus spp. and positive blood cultures. Clin Microbiol Infect. 2012;18:1117–1125. doi: 10.1111/j.1469-0691.2011.03688.x. [DOI] [PubMed] [Google Scholar]
- 18.Claydon MA, Davey SN, Edwards-Jones V, Gordon DB. The rapid identification of intact microorganisms using mass spectrometry. Nat Biotechnol. 1996;14:1584–1586. doi: 10.1038/nbt1196-1584. [DOI] [PubMed] [Google Scholar]
- 19.Borman AM, Petch R, Linton CJ, Palmer MD, Bridge PD, Johnson EM. Candida nivariensis, an emerging pathogenic fungus with multidrug resistance to antifungal agents. J Clin Microbiol. 2008;46:933–938. doi: 10.1128/JCM.02116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veloo AC, Welling GW, Degener JE. The identification of anaerobic bacteria using MALDI-TOF MS. Anaerobe. 2011;17:211–212. doi: 10.1016/j.anaerobe.2011.03.026. [DOI] [PubMed] [Google Scholar]


