Abstract
The apicomplexan Cryptosporidium parvum is one of the most prevalent protozoan parasites of humans. We report the physical mapping of the genome of the Iowa isolate, sequencing and analysis of chromosome 6, and ∼0.9 Mbp of sequence sampled from the remainder of the genome. To construct a robust physical map, we devised a novel and general strategy, enabling accurate placement of clones regardless of clone artefacts. Analysis reveals a compact genome, unusually rich in membrane proteins. As in Plasmodium falciparum, the mean size of the predicted proteins is larger than that in other sequenced eukaryotes. We find several predicted proteins of interest as potential therapeutic targets, including one exhibiting similarity to the chloroquine resistance protein of Plasmodium. Coding sequence analysis argues against the conventional phylogenetic position of Cryptosporidium and supports an earlier suggestion that this genus arose from an early branching within the Apicomplexa. In agreement with this, we find no significant synteny and surprisingly little protein similarity with Plasmodium. Finally, we find two unusual and abundant repeats throughout the genome. Among sequenced genomes, one motif is abundant only in C. parvum, whereas the other is shared with (but has previously gone unnoticed in) all known genomes of the Coccidia and Haemosporida. These motifs appear to be unique in their structure, distribution and sequences.
Cryptosporidium parvum is an apicomplexan parasite, conventionally placed in the class Coccidia, order Eimeriida, family Cryptosporidiidae. This is in the same order as the parasites Toxoplasma, Sarcocystis, and Eimeria, and in the same phylum (Apicomplexa) as Plasmodium, Babesia, and Theileria (http://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html/). Speciation within the genus is still controversial but ten species, infecting a range of vertebrate hosts, have been named (Chappell and Okhuysen 2002). On the basis of a large number of polymorphic loci, at least eight genotypes of C. parvum have been identified (Sulaiman et al. 2002; Xiao et al. 2002), of which genotypes 1 and 2 are responsible for most human infections (Guyot et al. 2001; McLauchlin et al. 2000). Genotype 1 is confined almost exclusively to humans, whereas genotype 2 infects a range of wild and domesticated mammals in addition to humans. The taxonomic status of the genotypes is unclear, but C. parvum genotype 1 was recently renamed as Cryptosporidium hominis (Morgan-Ryan et al. 2002). The Iowa isolate used in this study was previously identified as genotype 2, based on RFLP analysis (Carraway et al. 1997).
C. parvum is an obligate intracellular parasite, transmitted as highly durable oocysts in feces. Ingested oocysts excyst in the ileum, releasing sporozoites which infect the intestinal epithelium. Subsequent development includes both a cyclic asexual reproduction and the production of gametes giving rise to further oocysts, which are either excreted or reinfect the host. Symptoms are mainly gastrointestinal (diarrhea, abdominal cramps, vomiting), coupled with fever and headache. Human cryptosporidiosis was first reported as recently as 1976 (Meisel et al. 1976; Nime et al. 1976), but is now recognized as highly prevalent (Casemore et al. 1997; Fayer et al. 1997). Although the parasite is normally eradicated from the gut after 1–2 weeks, infection persists in immunosuppressed patients, and can be life-threatening. There is currently no effective therapy. In addition to sporadic cases, massive outbreaks occur periodically, typically through contamination of water supplies or of food (e.g., Glaberman et al. 2002; Howe et al. 2002; Rose et al. 2002).
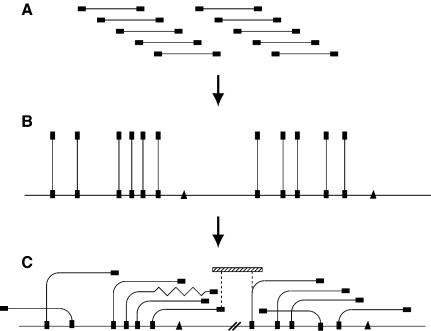
Conventional approaches to genome sequencing—whether based on physical mapping or on shotgun sequencing—are critically dependent upon the fidelity and representation of the clone libraries on which they are based. Dependence on a cloned resource both as a sequencing substrate and for determining the long-range order of the sequence data makes these approaches vulnerable to gaps, cloning artefacts, and large, well conserved repeat elements. We therefore set out to develop an alternative strategy which integrated complementary mapping techniques to overcome these drawbacks.
Electrophoretic analysis suggests that the C. parvum genome contains eight chromosomes of between 0.9 and 1.5 Mbp, giving a total genome size of ∼10.4 Mbp (Blunt et al. 1997; Caccio et al. 1998). We had already made (Piper et al. 1998a) a complete medium-resolution map of the genome of the Moredun isolate by HAPPY mapping (Dear and Cook 1993; Dear 1997). Also, a library of PAC clones made from this isolate (Piper et al. 1998b) showed that the majority of such clones were stable in culture, suggesting that an integrated mapping/sequencing strategy which combined the HAPPY and PAC-based approaches would be feasible. The strategy which we devised for the present study uses HAPPY mapping to define the positions of cloned fragments in the genome, before confirming their overlaps to create a robust, `HAPPily anchored' physical map (Fig. 1). This approach greatly accelerates physical mapping and, by combining clone-independent (HAPPY) with clone-dependent (overlap) data, is robust against artefacts and repeats. Where gaps remain in the physical map, adjacent contigs are oriented by virtue of the HAPPY data, aiding directed gap-closure.
Figure 1.
Construction of HAPPily-anchored physical map. (A) Members of a genomic PAC library are end-sequenced (filled rectangles). (B) One end-sequence of each clone is HAPPY-mapped to establish its position in the genome; additional sequence-tagged sites (triangles) are also mapped. (C) Overlaps between nearby clones are determined, by using PCR to test each clone for its content of nearby mapped markers, creating contigs. Chimeric clones or deletions (zigzag line) become apparent. The orientations of contigs which are separated by uncloned portions (heavy parallel diagonal lines) are known from the HAPPY map, and additional linking clones (hatched rectangle) can be sought by screening the same or other libraries with the markers adjacent to the gap (dashed vertical lines).
Based on our experiences with Cryptosporidium, we feel that this integrated approach overcomes some of the limitations of established strategies, and may be relevant to the sequencing of other genomes.
RESULTS
Construction of a `HAPPily Anchored' Physical Map of the Genome
We first constructed a genomic PAC library of the Iowa isolate of C. parvum (mean insert size ∼35 kbp). The end-sequences of 1172 clones, chosen at random from this library, were determined, and the positions in the genome of one end-sequence from each of 827 clones (as well as a further 269 STS markers, taken from previously published sequences) were determined using HAPPY mapping. This is an in vitro technique which maps sequence-based markers directly on native genomic DNA (see Methods).
To convert the HAPPY map into a robust physical map, each anchored clone was screened by PCR for the presence of the seven markers which, according to the HAPPY map, lay on either side of its mapped end-sequence (Fig.1). This enabled the direction and extent of overlaps between clones to be established, and the local order of the mapped markers to be refined (e.g., where the marker spacing was below the 5–10-kbp resolution limit of the HAPPY map). Construction of the HAPPY map of the ∼10.4 Mbp genome (Fig. 2) and its conversion to a `HAPPily anchored' physical map took a total of about four person-weeks.
Figure 2.
Genome-wide HAPPily anchored physical map. For clarity, markers are shown equally spaced. For each chromosome, HAPPY marker names are given below the map; names beginning `cr' and `CpG' indicate markers taken from Piper et al. (1998a) and from database sequences, respectively. Other names represent PAC end-sequences in condensed form; `q' = SP6 end, `p' = T7 end; for example, 06f03q is the SP6 end of the PAC clone pica_0006_f03. PAC clones are shown above each chromosome; the clone end represented by the mapped marker is indicated by a triangle; a short vertical line at the opposite end of a clone indicates that it ends within the interval shown; lack of this vertical line indicates that the clone may extend further than shown. Additional PAC or BAC clones identified during preliminary gap closure are indicated by dashed lines with the condensed clone name above them (names prefixed with `b' are BAC clones). Physical gaps are indicated by paired diagonal lines; the order of segments between these gaps is determined by the HAPPY data; the orientation of some of the smallest segments is not rigorously determined. Chromosome 6 is depicted in two parts; clone b3b01 is shown on both parts to illustrate the overlap. Solid triangles indicate deletions in clones; the solid black rectangle indicates a region of chromosome 6 which was sequenced by PCR amplification based on the corresponding part of the Moredun isolate genome (see text).
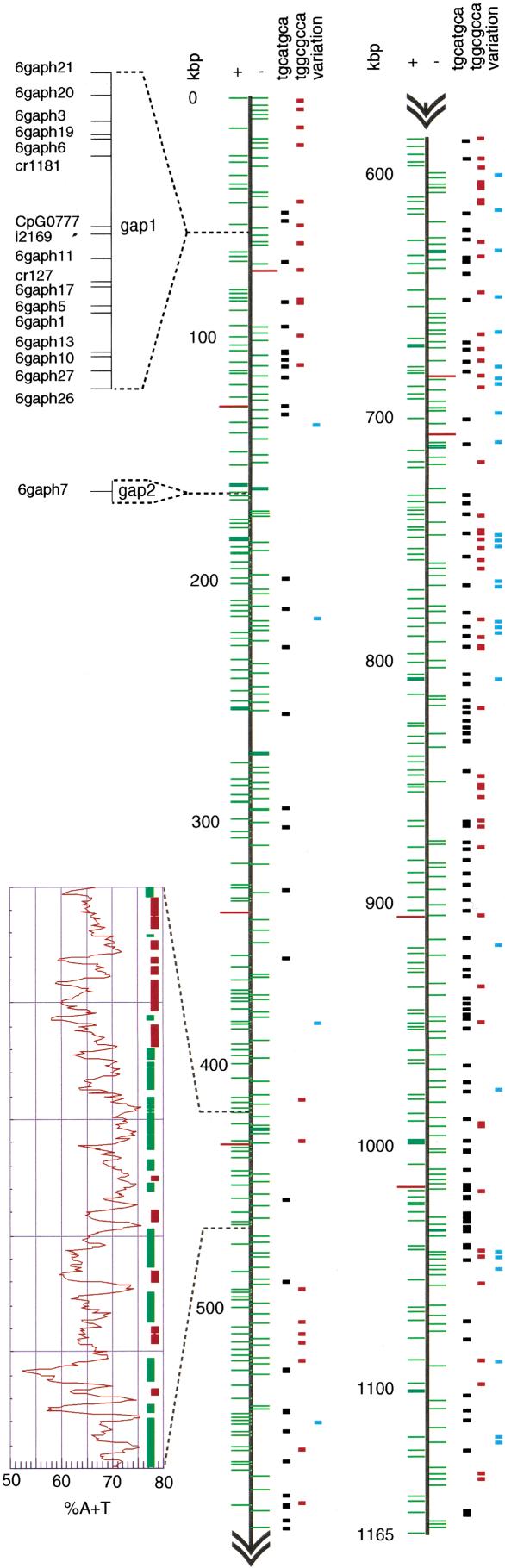
Finally, gaps in the genome-wide physical map were closed by screening the remainder of the PAC library (and, in some cases, a BAC library; see Methods) for the terminal markers of adjacent contigs. Only two regions on chromosome 6 remained intractable (i.e., physical gaps unspanned by any large clones; Fig. 3). Although an ApaI restriction fragment spanning both these gaps and the flanking sequence was isolated, subclones of this fragment showed an extreme bias in their distribution, most originating from the already known parts of the fragment (data not shown). However, the minority of the subclones which did fall within the gaps were HAPPY mapped at high resolution to produce dense local maps of the unsequenced regions (see Methods).
Figure 3.
C. parvum chromosome 6. The chromosome is shown in two halves (heavy vertical lines). Start points of predicted coding sequences on the + and - strands of the chromosome are indicated by short green horizontal lines to the left and right, respectively (red: tRNA genes). Octamer palindrome motifs are indicated by short black (TGCATGCA) and red (TGGCGCCA) bars, respectively, polymorphisms by blue bars. Two gaps in the sequence are indicated, and the STS markers mapped within them are shown in expanded form (upper left). The A+T content (sliding window of 100 bp) of a representative 50-kbp segment of the chromosome is shown in expanded form (graph, lower left), aligned with protein-coding regions on the + (green bars) and - (red bars) strands.
Comparison of this map with the previous, medium-resolution HAPPY map of the Moredun isolate of C. parvum (Piper et al. 1998a) did not reveal any differences in marker order, beyond those expected given the resolution of the earlier map.
Sequencing of Chromosome 6
A tiling path of 54 PAC clones (plus four BAC clones found during gap-closure; see Methods) was selected, and each of these clones was sequenced independently prior to alignment with consensus sequences from the overlapping clones (see Methods). The mean coverage of the tiling path with large insert clones was 1.6-fold. One gap was spanned by a previously sequenced PAC clone from an earlier library of the Moredun isolate of C. parvum (Piper et al. 1998b); overlapping PCR primer pairs were designed at intervals to amplify the corresponding region of the Iowa isolate, and these PCR products were sequenced directly.
Gross Sequence Characteristics
The largely complete sequence of the chromosome comprises 1,162,753 basepairs, and includes two gaps which have been indicated in the EMBL sequence submission (Accession no. BX526834) as blocks of 300 Ns. The total size of these gaps is estimated at 159 kbp, based on restriction fragment (ApaI) sizes; the local high-density HAPPY maps of the gap regions (above) indicate that the second gap is <10 kb in size. One of the telomeres was obtained (below), whereas restriction analysis indicates that approximately 10 kbp of sequence is missing from the other telomere (data not shown). Hence, the total length of the chromosome is predicted to be 1.33 Mbp. This compares well with the previously published estimate of 1.44 Mbp (Blunt et al. 1997) and with our own PFG-based estimate (mean of several determinations) of 1.31 Mbp (data not shown).
The overall G+C content of chromosome 6 is 31.1%, being approximately uniform along the chromosome on a gross scale. Analysis of the 867 kbp of PAC end-sequences scattered throughout the rest of the genome reveals a similar figure (31.9%). Although this is comparable to that of another apicomplexan parasite, Theileria annulata (32.9%), other apicomplexans display a wide range of G+C contents (e.g., Plasmodium falciparum, 18.2% [Gardner et al. 2002]; Eimeria tenella, 53.2% [http://www.sanger.ac.uk/Projects/E_tenella/], and Toxoplasma gondii, 53.6%; [http://www.tigr.org/tdb/e2k1/tga1/]).
Centromere and Telomeres
The presumed centromeres in the most complete apicomplexan genome, that of P. falciparum, are tracts of 2–3 kbp which are extremely A+T-rich (>97% A+T) and contain imperfect tandem repeats (Gardner et al. 2002). No comparable feature is seen on C. parvum chromosome 6 (Fig. 3); it is possible that a centromere lies within one of the gaps in our sequence, though the additional STSs mapped within these gaps are not unusual in base composition.
A total of 17 nonredundant sequences containing more than two tandem copies of the telomere repeat hexamer (CCTAAA) were obtained (Methods). The largest telomere repeat motif contained 51 copies of the hexamer, interspersed with a small number of imperfect or incomplete copies but, because the repeat-primer used to obtain these fragments can prime at any point in a long tandem array of telomere repeats, an upper limit on the number of repeats cannot be set.
Only one sequence, cptel29, was assigned unambiguously to chromosome 6 by digital blotting (Methods), and PCR was used to generate a sequencing template linking this to the main PAC/BAC contig. None of the other putative telomeric sequences was assigned experimentally to chromosome termini, and clearly at least one must be an internal sequence or an artefact.
Simple Tandem Repeats
Chromosome 6 was analyzed (P.H. Dear, unpubl. software) for the presence of tandem repeats of significant lengths (Table 1). Homopolymer tracts of ≥10 bp are abundant but, strikingly, are almost exclusively poly-A or poly-T—only four of the 236 tracts are poly-G or poly-C. Dinucleotide repeats are also abundant (again biased strongly towards A/T sequences), with progressively smaller numbers of repeats as the number of dinucleotide units increases. The units of most of the longest repeats appear themselves to contain imperfect trinucleotide repeats, implying reduplication of an earlier repeat region. A broadly similar picture is seen in the 867 kbp of PAC end-sequences from elsewhere in the genome.
Table 1.
Tandem Repeat Motifs (Including Single-Base Tracts) in C. parvum Chromosome 6 (1.163Mbp of sequence) and in the PAC End-Sequences Lying in the Remainder of the Genome (867kbp of Sequence)
| Repeat unit length | n≥ | Number (Chr6) | Number (PAC ends) |
|---|---|---|---|
| 1 | 10 | 236 | 170 |
| 2 | 7 | 25 | 23 |
| 3 | 5 | 82 | 33 |
| 4 | 4 | 11 | 8 |
| 5 | 3 | 11 | 4 |
| 6 | 3 | 26 | 15 |
| 7 | 3 | 1 | 0 |
| 8 | 3 | 1 | 0 |
| 9 | 3 | 5 | 0 |
| 10 | 3 | 1 | 0 |
| 11-15 | 2 | 26 | 17 |
| 16-20 | 2 | 8 | 3 |
| 21-25 | 2 | 2 | 3 |
| 26-30 | 2 | 3 | 1 |
| ≥31 | 2 | 0 | 1 |
For each repeat unit length, repeats consisting of ≥n repeat units were scored
Hyperabundant Palindromic Octamer Motifs
We find two striking and highly repeated octamer motifs in the genome of C. parvum. The first, TGGCGCCA, is palindromic; the second, TGCATGCA, is doubly palindromic (i.e., a palindromic octamer composed of two repeats of a palindromic tetramer). Both occur at very high frequencies compared to that predicted for a random sequence of bases with the same overall nucleotide composition. TGGCGCCA occurs 71 times on chromosome 6 (as opposed to an expected frequency of around two, an overabundance of 35-fold), whereas TGCATGCA occurs 166 times (expected frequency around nine, overabundance of 18-fold). Both motifs are present almost exclusively in the ∼20% of the chromosome which is noncoding, and are scattered fairly uniformly along the chromosome apart from a region of ∼300 kbp in which TGCATGCA is scarce and TGGCGCCA is completely absent (Fig. 3). The bases flanking the motifs are not strongly conserved, apart from a preference for A or T in the two bases on either side of both types of octamer. Analysis of the 867 kbp of PAC end-sequences from the remainder of the genome reveals a similar overabundance of these two octamer sequences (data not shown).
These octamer motifs may also reflect the phylogeny of Cryptosporidium and the other apicomplexans, and are considered in this context below.
Gene Density, Number, and Organization
In the absence of a large data set of annotated C. parvum genes, a variety of complementary methods was used for gene identification, supplemented by extensive manual analysis (see Methods). All analyses were integrated using the Artemis software package (Rutherford et al. 2000).
A total of 474 protein-coding genes are predicted, giving a mean density of one per 2.46 kbp. Only 122 of the genes (25.7%) have predicted introns—delimited by the usual eukaryotic GT...AG motifs at either end—and these have an average of 2.7 exons per gene. The mean size of exons is 1277 bases, whereas that of introns is 154 bases; 78% of the sequence appears to be coding (including introns). Introns and intergenic regions generally have a higher AT content than protein-coding sequence, though the difference is not clear or consistent enough to serve as a reliable predictor of gene organization (Fig. 3). Six loci contain potentially overlapping pairs of coding sequences. In addition, eight tRNA genes are scattered across the chromosome.
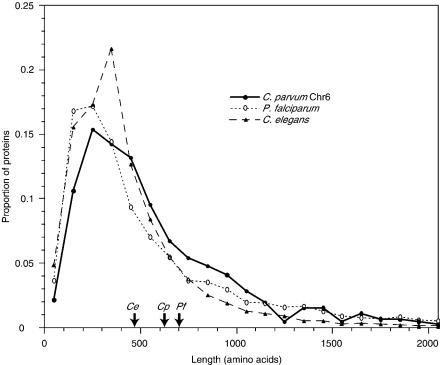
One unusual feature of the predicted genes is that their mean coding length (616 amino acids) is considerably greater than in other eukaryotic genomes (e.g., about 470 amino acids for Saccharomyces cerevisiae or 475 for Caenorhabditis elegans). Comparison reveals that in general, each C. parvum sequence is of similar length to its orthologs in C. elegans and S. cerevisiae, but that there is an apparent underrepresentation of smaller genes and a relative overabundance of proteins longer than 700 amino acids (Fig. 4). These larger proteins are not preferentially those with one or multiple predicted transmembrane helices, nor do they appear to be significantly biased in their amino acid composition or repetitive nature.
Figure 4.
Distribution of protein lengths. Predicted lengths of proteins on chromosome 6 of C. parvum and in the complete genomes of P. falciparum and C. elegans were sorted into size-bins of 100 amino acids, and the proportion of proteins in each bin were plotted for each species. Proteins longer than 2100 amino acids are not shown; the arrows on the x-axis indicate the arithmetic mean length of all proteins in each of the three species.
Comparisons with Previously Sequenced C. parvum Genes
Only 13 of the predicted genes on chromosome 6 have previously been sequenced in any Cryptosporidium species, the majority being from C. parvum. In most (10) cases, we find that our predicted protein sequence matches perfectly either the only published counterpart, or one of several sequences of a protein known to vary among isolates. In the remaining three cases (histone deacetylase 56k.08, the sporozoite cysteine-rich protein 1MB.07, and the ATP-binding cassette 1MB.703), we find only one or two amino acid differences (most of which are conservative) compared to the previously published C. parvum sequences.
Structural and Functional Gene Classification
A range of tools was used to identify functional and structural motifs in the predicted genes (see Methods). The domains in the predicted coding sequences were characterized by matches against the hidden Markov models of the Pfam (Bateman et al. 2002) and SUPERFAMILY (release 1.59; Gough and Chothia 2002) databases. The SUPERFAMILY database contains a library of hidden Markov models for all domains of known three-dimensional structure, as defined in the Structural Classification of Proteins (SCOP 1.59) database (Lo Conte et al. 2000).
A total of 371 Pfam domains of 174 different types were identified in 206 (43%) of the 474 predicted proteins. SUPERFAMILY analysis found a total of 265 SCOP structural domains in 196 (41%) of the predicted proteins. In both the Pfam and SCOP analyses, the majority of domain families are represented by only one or two examples, whereas a small number of domain families are more abundant. The predominant domains include P-loop-containing nucleotide triphosphate hydrolases (41 instances), RNA-binding domains (22), and protein kinase domains (11).
A small number of proteins contain uncommon or unique combinations of domains. 1MB.635, a putative aminoacyl tRNA synthetase, contains a four-domain combination to date seen only in Plasmodium spp. and S. pombe, whereas 1MB.33 contains a combination of two domains seen only in C. parvum.
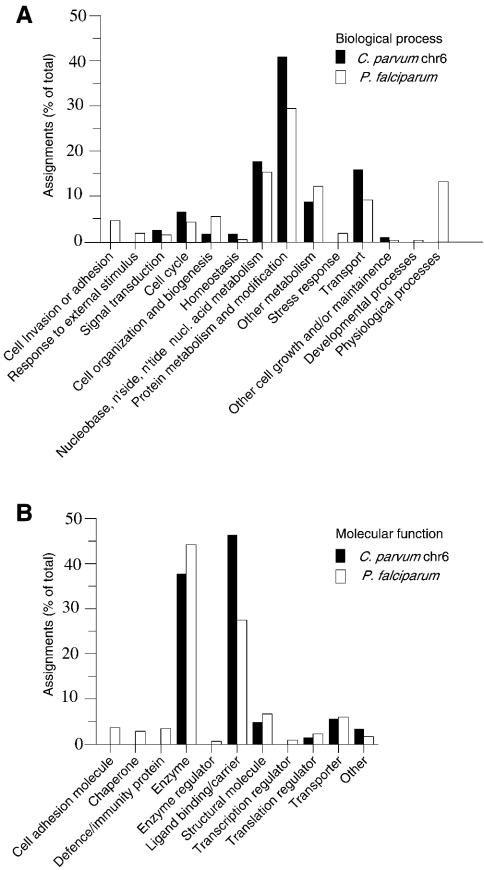
The results of these analyses were integrated using Interpro (Apweiler et al. 2000), and this output was in turn used to assign GO (Gene Ontology) classification terms (Ashburner et al. 2000) to the predicted proteins. The distribution of functions and biological processes is broadly similar to that seen in Plasmodium (Fig. 5).
Figure 5.
Gene Ontology (GO) classifications of proteins. Classification of predicted genes on C. parvum chromosome 6 is compared with that of P. falciparum genes under (A) `Biological process' and (B) `Molecular function' ontologies. Classification of Plasmodium proteins is based on Gardner et al. (2002).
Of the 474 predicted proteins, only 215 have significant similarity to any protein of known or inferred function. Therefore, no function can be inferred for the remaining 259 (55%) of the predicted chromosome 6 proteins.
Genes of Potential Relevance to Infectivity, Diagnosis, or Therapy
A variety of protein types are of interest for the identification and characterization of Cryptosporidium, or as potential therapeutic targets. Such genes include those predicted to encode membrane, extracellular, or cell-surface proteins, transport proteins, and mitochondrial or plastid proteins.
Of the 474 predicted genes, 118 were predicted (see Methods) to have at least one transmembrane domain, with 26 containing more than six such domains. Approximately half (52) of the putative membrane-spanning proteins also have predicted signal peptides. In addition, the 11 genes which carry protein kinase domains may warrant further investigation as potential regulators of signal transduction or cell–cell interactions.
Thirteen of the predicted membrane-spanning proteins have sequence similarities to known transport proteins from other species, including four ABC transporters (genes 1MB.703, 1MB.800, 1MB.816, and 1MB.836). In addition, we find a potential homolog (1MB.736) of the putative chloroquine resistance transporter found in P. falciparum (Q9N623). Chloroquine has hitherto been found to be only moderately effective as an anti-cryptosporidial agent in in vitro tests (Armson et al. 1999), and the vulnerability of C. parvum to this drug may be modulated by this gene, just as the Plasmodium homolog renders chloroquine ineffective against some forms of malaria (Su et al. 1997; Nomura et al. 2001).
SUPERFAMILY analysis reveals 10 candidate extracellular or cell-surface proteins carrying domains typically found in such proteins (Table 2). These proteins could be involved in parasite–host interactions, and hence may be of interest for further experimental characterization.
Table 2.
Predicted Proteins With Cell-Surface Protein-Associated Domains
| Gene | Domains | Transmembrane region(s) | Signal peptide |
|---|---|---|---|
| 1MB.09 | EGF/laminin | Yes | No |
| 1MB.07 | Trombospondin, EGF/laminin | Yes | Yes |
| 1MB.145 | Thrombospondin | No | Yes |
| 1MB.513 | Immunoglobulin, transglycosidase | No | Yes |
| 1MB.748 | Immunoglobulin, kinase | No | No |
| 1MB.312 | FKBP-like | No | Yes |
| 1MB.491, 1MB.495 | Ankyrin repeat | Yes | Yes |
| 1MB.779 | Concanavalin A-like lectins/glucanase | Yes | Yes |
We find several genes encoding oocyst wall proteins and other surface antigens, known to vary in detail both between and within species of Cryptosporidium (e.g., Spano et al. 1997; McLauchlin et al. 2000; Akiyoshi et al. 2002). Predicted protein 1MB.63 encodes the previously known sporozoite antigen glycoprotein GP15 (Q9U521). Gene 1MB.13 encodes a large (3007 amino acids) protein, and is a perfect match for a known partial C. parvum oocyst wall protein sequence (41 kD oocyst wall protein [fragment], Q9U4U4). Gene 1MB.208 (1623 amino acids) differs by two amino acids from a previously identified oocyst wall protein precursor (Q06550). Another of the predicted genes (56k.19) has no matches to known genes, but much of its sequence has significant identity (22%, over 1081 amino acids) to the oocyst wall protein precursor Q06550 and, like the previous two predicted wall proteins, also contains repetitive amino acid tracts. This gene therefore may encode a new oocyst wall protein.
C. parvum had been reported to lack mitochondria (Current 1989; Tetley et al. 1998) though, in contrast, there are reports of their presence in at least some developmental stages (Riordan et al. 1999; Beyer et al. 2000). As in an earlier EST-based gene survey (Strong and Nelson 2000), we find several genes which appear to encode mitochondrial proteins. Predicted proteins 1MB.254 and 1MB.530 show convincing sequence similarities to mitochondrial carrier proteins, but apparently lack a signal for export to mitochondria (http://mips.gsf.de/cgi-bin/proj/medgen/mitofilter; Claros and Vincens 1996). It is possible that the relevant signals in Cryptosporidium are unusual, and hence are not detected by this software.
Also of interest are genes whose proteins may be associated with the apicoplast, a relict plastid characteristic of the Apicomplexa, which has been proposed as a vulnerable target for drugs against parasites of this phylum (Fichera and Roos 1997). The question of whether C. parvum possesses an apicoplast has been debated (e.g., Blunt et al. 1997; Tetley et al. 1998; Zhu et al. 2000). In Plasmodium falciparum, 551 nuclear-encoded proteins (about 10% of the total) were identified, with varying degrees of confidence, as being potentially targeted to the apicoplast (Gardner et al. 2002). Forty-five of the 474 predicted proteins of C. parvum chromosome 6 were identified as potentially apicoplast-targeted, using an algorithm trained on Plasmodium sequences (Zuegge et al. 2001; http://gecco.org.chemie.uni-frankfurt.de/pats/pats-index.ph). However, in the absence of experimental evidence, these predictions must be considered tentative.
Phylogeny and Comparison with Other Genomes
Among the apicomplexans, Eimeria and Toxoplasma both fall within the Coccidia, whereas Plasmodium is within the Haemosporida and Theileria is within the Piroplasmida. The gregarines (including Monocystis and Gregarina spp.) are a fourth major group of apicomplexans, believed to have diverged early from these other groups, and for which little sequence data exist. Cryptosporidium has long been placed in the Coccidia, but comparisons of small-subunit ribosomal RNA (ssrRNA) and β-tubulin gene sequences suggested that it is more closely related to the early-branching gregarines than to the other apicomplexans (Carreno et al. 1999; Leander et al. 2003).
On a gross level, the predicted proteins of C. parvum chromosome 6 reflect a greater similarity to other apicomplexans than to non-apicomplexan species, though not by as great a margin as one might expect. For example, based on a FASTA expectation value of e < 0.01, 237 (50%) of the predicted proteins have matches to those in P. falciparum, whereas 184 (39%) have matches to those of S. cerevisiae; both of these latter genomes are fully sequenced, and all three genomes have comparable numbers of genes. If the criteria for declaring matches are made more stringent, the number of matches between species decreases and the number of matches to P. falciparum becomes more similar to the number of matches to S. cerevisiae.
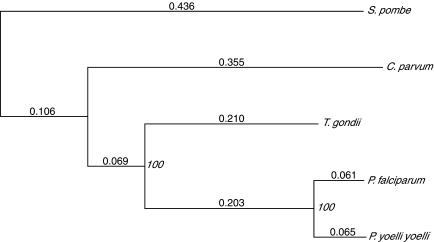
To try to resolve relationships among the apicomplexa, we sought to compare the protein sequences of those genes on C. parvum chromosome 6 with their homologs in T. gondii, P. falciparum, and P. yoelii; Schizosaccharomyces pombe was used as an outgroup. To identify homologs of C. parvum proteins among the other available apicomplexan protein sequences, the 474 predicted chromosome 6 proteins were searched against the Non-Redundant Protein Database (Jenuth 2000) with FASTA (Pearson and Lipman 1988). In addition to β-tubulin (analyzed in earlier phylogenetic studies, Leander et al. 2003), there were four other proteins which had matches to both T. gondii and Plasmodium sequences at expectation values lower than that for the best S. pombe match, and hence were considered likely to be orthologs. Putative orthologs were aligned using CLUSTAL W (Thompson et al. 1994), and the four alignments were concatenated. A phylogenetic tree was calculated from the concatenated alignment using distances based on the PAM matrices (Dayhoff et al. 1978) from 1833 positions in the alignments, and the neighbor-joining and bootstrapping options in PHYLO_WIN (Galtier et al. 1996). As can be seen in Figure 6, Cryptosporidium is an outgroup to both the coccidian Toxoplasma and the haemosporidan Plasmodium. Inclusion of the β-tubulin sequence in the calculation lead to an essentially identical tree, as did calculations based on each of the five proteins individually (data not shown). In all cases, the bootstrap support (a measure of robustness of the tree) was 100%, providing strong additional evidence for the deep-branching position of Cryptosporidium within the Apicomplexa. Analysis of the same sequences using more sophisticated algorithms (maximum likelihood analysis with heterogeneity corrections, and probabilistic neighbor-joining algorithms; Schmidt et al. 2002; see also Teichmann and Mitchison 1999) yielded an identical pattern of relationships between the species, again with strong statistical support (data not shown).
Figure 6.
Protein sequence-based phylogenetic tree. The tree was calculated from the aligned and concatenated sequences of four proteins from the five species indicated. Branch lengths are distances calculated using PAM matrices; bootstrap values are indicated at nodes. The proteins used and (in brackets) the gene name on C. parvum chromosome 6 and the GenBank identifiers for their sequences from P. falciparum, P. yoelii yoelii, S. pombe, and T. gondii, respectively, are as follows: protein disulphide isomerase (56K11, 23612738, 23481103, 19113783, 14494995); glyceraldehyde-3-phosphate dehydrogenase (1MB519, 23509820, 23491258, 19112028, 13377044); heat shock protein 60 (1MB751, 23507957, 23479768, 19113806, 5052052); and protein phosphatase 2b (1MB598, 23612977, 23489838, 19112970, 22535354).
The two octamer DNA motifs noted above as being highly abundant in the C. parvum genome may also shed some light on the phylogenetic relationships of the apicomplexans. As far as we are aware, such motifs have not hitherto been noticed in any of the previously sequenced apicomplexan genomes. Our analysis finds that the first such motif, TGCATGCA, is common not only in C. parvum but also in the coccidians E. tenella and T. gondii (being about 18-fold overrepresented in the first two genomes, and about 11-fold in the third), somewhat less common in the haemosporidian P. falciparum (about sevenfold overrepresented), and rare in the piroplasmid T. annulata. The second motif, TGGCGCCA, is abundant only in C. parvum, and occurs no more than expected by chance in the other genomes. There were insufficient sequence data to analyze representatives of the other apicomplexan group, the Gregarina.
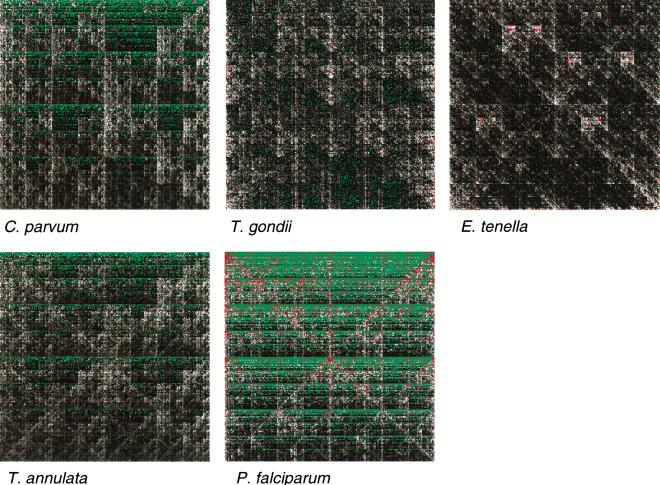
To compare the overall character of these genomes, we made chaos plots (Jeffrey 1990), which allow the frequency of every possible octamer in a sequence to be visualized. Because the genomes have very different G+C contents, we used a modified program (P.H. Dear, unpubl.) which plots the frequency of each octamer relative to the frequency expected based only on the G+C content of the sequence. As can be seen in Figure 7, the two coccidians Toxoplasma and Eimeria have broadly similar characters (once normalized for base composition), whereas chromosome 6 of C. parvum is significantly different from these and more similar to that of the piroplasmid Theileria; the haemosporidian Plasmodium is significantly different again. A similar picture is seen when considering only the coding or noncoding regions of the genomes, or when examining the 867kbp of nonchromosome 6 sequence from C. parvum (data not shown). Numerical comparison of the chaos plots (the root-mean-square difference in relative frequencies of each octamer between two genomes to be compared) supports the visual interpretation of the plots (data not shown).
Figure 7.
Normalized chaos plots for apicomplexan genomes. Each pixel represents the frequency of a given octamer sequence in the genome, relative to the frequency expected in a randomly ordered sequence with the same base composition as the genome in question (log scale; green <10-6; grayscale black through white =10-6 through 5; red >5). In each plot, the octamers [G]8, [C]8, [A]8, and [T]8 are represented at the top left, top right, bottom left, and bottom right corners, respectively.
Finally, we looked for syntenic relationships between C. parvum and P. falciparum by comparing the positions, in the two genomes, of the 237 C. parvum chromosome 6 genes for which homologs with FASTA expectation values of <0.01 could be identified in P. falciparum. No substantial region of the C. parvum chromosome appears to have homologs preferentially from any one region of the P. falciparum genome. There were four instances of microsynteny (which we define as two genes separated by four or fewer intervening genes in one genome, with their orthologs also separated by four or fewer genes in the other), but this is not appreciably more than would be expected by chance alone, if the gene order were randomly scrambled between the two genomes.
SNPs and Other Polymorphisms Within the Iowa Isolate
Because it is not possible to initiate infection from a single sporozoite of C. parvum, isolates of this species cannot be guaranteed to be clonal. We therefore looked for possible polymorphisms within the Iowa isolate by comparing the overlapping regions of independently sequenced PACs and BACs; such overlaps cover approximately 57% of the chromosome 6 sequence. We found 39 clear differences between overlapping clones; in all such cases, the consensus sequence for each of the overlapping large-insert clones was unambiguous, thereby excluding the possibility of sequencing errors. Such inter-clone differences could in principle arise as artefacts within the clones, and indeed one (a 54-bp insertion/deletion) was confirmed by PCR as being such an artefact. The remaining 38 differences were single-base substitutions or small insertions/deletions (often in repeat tracts). Cloning artefacts should occur more or less independently of the coding structure of the cloned fragments, but we find that most (25/38) of them are confined to the small part of the genome (20%) which is noncoding. The few found in predicted coding regions exhibit a bias (relative to random mutation) in favor of silent substitutions or insertions/deletions which do not cause frame-shifts (Table 3). Only one apparent polymorphism disrupts a predicted gene, causing a frame-shift in the first of three exons of 1MB.764; this may be a true instance of a mutation arising within the PAC clones, or might reflect a mis-prediction in the start-point of the gene (such that the polymorphism, in a homopolymer tract, is not actually within the gene). With this possible exception, we believe that most of the differences represent true polymorphisms.
Table 3.
DNA Sequence Polymorphisms Found in Regions of Overlap Between PAC Clones of the Iowa Isolate of C. parvum
| Location | Type | Number |
|---|---|---|
| Noncoding | Insertion/deletion | 1 |
| Single-base substitution | 8 | |
| Simple-sequence repeat length variation | 16 | |
| Total noncoding | 25 | |
| Coding | Silent base substitution | 6 |
| Nonsilent base substitution | 5 | |
| In-frame insertion/deletion | 1 | |
| Out-of-frame insertion/deletion | 1 | |
| Total coding | 13 | |
| Total | 38 |
The polymorphisms we observe are distributed nonrandomly along the chromosome (Fig. 3), most falling in the region between 500 and 800 kb (Fig. 3). This may reflect a true bias in the distribution of polymorphisms along the chromosome or may be because, in this ∼300-kb region, more of the overlaps between the clones are fortuitously represented by PACs from two different genotypes within the Iowa isolate.
DISCUSSION
Mapping and Sequencing Strategy
Our strategy was intended to overcome the limitation inherent in both shotgun sequencing and conventional physical mapping: the dependence on the integrity and representation of clone libraries for both local sequence data and long-range contiguity. The initial HAPPY mapping enabled the extremely rapid production of an anchored physical map which was robust against clone artefacts, and allowed directed gap closure to be initiated at an early stage. Of our two remaining gaps, one is quite large, but our attempts to create small-insert subclones of the restriction fragment spanning these gaps suggest that a shotgun sequencing strategy would still have left this region fragmented and unordered. Overall, the strategy was successful; we would expect it to offer an advantage in sequencing the many genomes in which extreme biases in base composition or other features make the production of well ordered sequence by conventional means technically challenging.
Our experience in mapping other genomes (e.g., Dear et al. 1998; Konfortov et al. 2000) suggests that this approach is completely scaleable. The number and sizes of clones to be mapped, and the resolution with which their end-sequences are HAPPY-mapped in the first instance, can all be varied over a wide range.
Sequence Analysis
Given the absence of a comprehensive set of gene models for C. parvum, it is reasonable to ask how effective and accurate our gene predictions are. The high density of predicted genes means that there is little room on the chromosome for short `hidden' genes. The number of genes predicted on the chromosome 6 sequence (∼10% of the genome) implies a total gene number of around 4500–5000, comparable to that of P. falciparum (∼5300; Gardner et al. 2002). This again suggests that large numbers of genes have not been missed by the predictions. Given the absence of biological data, unambiguous assignment of the starting methionine of each gene cannot be made; in some cases, therefore, the true gene lengths or exon numbers may differ from those predicted. One must also ask whether chromosome 6 is representative of the remainder of the C. parvum genome. None of our analyses (including comparison with ∼867 kbp of clone-end sequences from the remainder of the genome) reveals any obvious differences in character between chromosome 6 and the remainder, nor are gross variations seen between the different chromosomes in the genomes of Plasmodium spp. or of most other fully sequenced genomes. We therefore think it likely that chromosome 6 of C. parvum is fairly typical of the genome.
With a mean density of one predicted gene per 2.46 kb, chromosome 6 of C. parvum is particularly gene-dense. In comparison, the P. falciparum genome (approximately twice the size of that of C. parvum) contains one predicted gene per 4.3 kb (Gardner et al. 2002) and, among the fully sequenced eukaryotes, only S. cerevisiae and Encephalitozoon cuniculi have higher densities of predicted genes (one per 2 kb and one per 1.5 kb, respectively; Goffeau et al. 1996; Katinka et al. 2001).
Analysis of the predicted genes on chromosome 6 reveals three unusual features, which collectively suggest that the gene set of C. parvum is different in character both from that of P. falciparum and from those of most other eukaryotes. First, the average size of the predicted proteins (616 amino acids, about 30% longer than yeasts or C. elegans) is surprisingly large (Fig. 4). P. falciparum also has an apparent bias toward larger proteins (mean size 700 amino acids), though at least some of this bias is due to the presence of extensive nonglobular domains (simple sequence repeats) in otherwise conventional P. falciparum proteins; this does not appear to be the case in C. parvum. The larger C. parvum proteins taken as a whole do not appear to have any distinguishing feature other than their size (e.g., they are not preferentially membrane proteins; nor are they more likely to have homologs in the Plasmodium genome). The significance of this size bias, therefore, remains unclear. Second, a large number (118; 25%) of the predicted proteins contain predicted transmembrane domains, and 26 (5%) contain more than six such domains, typical of channels or receptors. The corresponding proportions for P. falciparum are 18% and only 0.8%, respectively. Third, a significantly lower proportion (43%) of the predicted proteins have recognizable structural motifs (Pfam domains) than is the case for most genomes (mean 55%, standard deviation 6% for all fully sequenced genomes). A similar situation is seen in P. falciparum, where only 37% of the predicted proteins have matches to known Pfam domains.
The two abundant octamer palindromes (TGCATGCA and TGGCGCCA) are unlike other repeat classes in their structure and distribution. They differ in character both from simple tandem repeats and from more complex (e.g., transposon-related) elements, each of which arises and persists in genomes through partly understood mechanisms regardless of selective advantage. The TGCATGCA motif is common in the binding sites for homeotic proteins in some Drosophila enhancers (Manak et al. 1995), but it is not clear whether there is any functional equivalence to the C. parvum octamer. Neither octamer has a consistent positional relationship with coding sequences or other obvious sequence features. The distribution and palindromic nature of these motifs suggest a possible role in chromatin packaging or dynamics and, given their prevalence in C. parvum, E. tenella, and T. gondii (all of whose life cycles involve a stable dormant phase), we speculate that they may be involved in stabilizing the DNA during prolonged dormancy.
The phylogeny of the apicomplexans remains controversial. Our analysis of protein sequences strongly supports earlier suggestions (Carreno et al. 1999; Leander et al. 2003) based on the comparison of β-tubulin and rRNA sequences, that Cryptosporidium diverged early during the evolution of the apicomplexans, before the divergence of Plasmodium (a haemosporidian) from Toxoplasma (a coccidian). There were insufficient sequences from other apicomplexan groups to provide new data on the relative ages of other divergences (e.g., between the gregarines and other apicomplexans). Though a comparison based on a larger range of sequences would be desirable, the fact that each of our protein alignments alone led to the same relationship as the concatenated alignment of all four proteins (and that the same relationship was inferred in previous studies based on β-tubulin sequences; Carreno et al. 1999) argues that the proteins chosen are reasonably representative of the remainder. The lack of synteny between C. parvum and P. falciparum also argues for a distant relationship between these two species, though this cannot be placed in context until extensive genome sequences become available for other apicomplexans. A further hint at the remoteness of these two species comes from the relatively low degree of similarity between their predicted proteomes. The distribution of the TGCATGCA motif, conversely, suggests that Cryptosporidium is more closely related to the Eimeriida (coccidians) than to the Haemosporida or Piroplasmida, although lack of data on other apicomplexans (particularly the gregarines) again obscures the complete picture. On a more optimistic note, it is clear that protein sequences are sufficiently conserved between the apicomplexans to retain a strong phylogenetic signal, and hence the phylogeny of this group is likely to be resolved once more sequences from other species become available for comparison.
The profiles of normalized octamer frequencies (which are more similar to those of Theileria than to the other apicomplexans considered) appear to reflect concerted changes in genome architecture which consistently accompany changes in G+C content, rather than phylogenetic relationships. This raises the more general question of why even closely related genomes can differ markedly in base composition, and how the overall character of the genome—the normalized frequencies of dimers and longer sequences—varies in concert.
METHODS
Genomic DNA and Large-Insert Cloning
Purified oocysts of C. parvum (Iowa) were obtained from the Sterling Parasitology Laboratory. Genomic DNA was prepared in agarose strings (2×108 oocysts/mL) as described (Piper et al. 1998a). Libraries of genomic DNA, size-selected after partial digestion with Sau3A or EcoRI, were made in the vectors pCYPAC2 (Ioannou and de Jong 1996) or pBACe3.6 (Frengen et al.1999), respectively, in ElectroMAX DH10B Competent Cells (Life Technologies), using essentially standard techniques (Piper et al. 1998b).
Mapping
HAPPY mapping was performed essentially as described (Piper et al. 1998a). Briefly, 10 μL of agarose-embedded DNA was melted in 1×PCR bufferII (Applied Biosystems) at 65°C for 8 min, the DNA sheared by gentle inversion, diluted 2300-fold in water, and 2-μL aliquots were dispensed into a 96-well thermocycler plate (88 samples plus 8 × 2 μL water as negative controls). Samples were preamplified by primer-extension pre-amplification (PEP) as described (Zhang et al. 1992; Piper et al. 1998a) in a total volume of 5 μL. Each PEP product was diluted to 150 μL with water, and 5-μL subfractions were dispensed into 30 replica mapping plates, overlaid with 30 μL of mineral oil, and stored at -80°C until needed. PCR primers (forward, internal, and reverse) were designed for one end-sequence of each PAC clone, plus additional sequences taken from Piper et al. (1998a) or from database sequences, essentially as described (Piper et al. 1998a; Konfortov et al. 2000). For mapping, the forward and reverse primers for between 96 and 200 markers were used in a Phase1 PCR reaction with one of the replica mapping plates (total volume 10 μL per well, comprising 5 μL of diluted PEP product, PCR Gold Buffer [Applied Biosystems], 2mM MgCl2, 0.2 μM each primer, 2U Taq Gold [Applied Biosystems], 200 μM each dNTP; 93°C × 9 min, then 25 cycles of 94°C × 20 sec, 55°C × 30 sec, 72°C × 1 min). Phase 1 products were then diluted to 600 or 1200 μL, and 5 μL of each product was used in a Phase 2 heminested PCR reaction for each marker in turn (10 μL total volume containing PCR Gold Buffer [Applied Biosystems], 1.5 mM MgCl2, 1 μM each of forward and reverse primer for the marker in question, 0.25 U Taq Gold [Applied Biosystems], 200 μM each dNTP; 93°C × 9 min, then 33 cycles of 94°C × 20 sec, 52°C × 30 sec, 72°C × 1 min). PCR products were supplemented with 8 μL of loading dyes (4× SyBr Green1, 15% w/v Ficoll 400, 0.1 mg/mL bromophenol blue), resolved by brief electrophoresis, and imaged under UV illumination. Analysis was as described (Dear 1997), with markers being sorted into linkage groups at an LOD threshold of 6. Linkage groups were assigned to chromosomes based on their content of previously mapped markers (Piper et al. 1998a); assignment of some groups was verified by digital blotting (below).
For conversion of the HAPPY map into a physical map, both PAC clones and primers were rearrayed robotically according to their positional order defined by the HAPPY map. Each PAC clone was screened using a standard single-phase PCR for the presence of the seven markers which lay to either side of it according to the HAPPY map. The marker content of each clone was then used to deduce the clone overlaps (P.H. Dear, unpubl.).
Gap Closure
Gaps in the physical map were closed in the first instance by screening the PAC and BAC libraries by PCR for further clones, using the HAPPY markers on either side of the gap. In some cases, the end-sequences of clones extending into the larger gaps were used to design new markers with which to rescreen the libraries for clones, closing the gaps by a `walking' approach. Both of the gaps which remained uncloseable by this approach on chromosome 6 were found to lie in a 350-kbp ApaI restriction fragment; this fragment was purified using standard procedures, digested with Sau3A, and subfragments were cloned into pBluescriptIISK+ and M13mp19 and sequenced using standard procedures. Sequences not matching the sequence of the known part of the ApaI fragment were confirmed as originating from the fragment by digital blotting (below), and were used to design further HAPPY markers which were mapped as described above.
PAC and BAC Sequencing
Large-insert clone DNA (∼20 μg) was isolated using the double acetate method (http://www.genome.ou.edu/BAC_isoln_200ml_culture.html), then sheared, concentrated, and desalted using standard protocols. DNA was then end-repaired (30 min, 15°C, 100 μL reaction: 20 μg sheared DNA, 15 U T4 DNA polymerase, 10 U Klenow DNA polymerase [both MBI Fermentas, Vilnius, Lithuania], 500 μM each dNTP, 10 μL Yellow Tango Buffer [MBI Fermentas]), desalted, and tailed with an extra A residue (30 min, 50°C, 100 μL reaction: 20 μg sheared DNA, 50 μM each dCTP, dGTP, dTTP, 2mM dATP, 20 U Taq polymerase [MBI Fermentas], 10 μL Yellow Tango buffer). A-tailed DNA was then size-fractionated by electrophoresis, and the 1.0–1.5-kbp fraction was isolated and purified using essentially standard methods before cloning into pGEM-T (Promega). Plasmid clones were sequenced from both ends with standard primers using the Big Dye terminator chemistry on ABI 3700 capillary sequencers (Applied Biosystems), to give a mean coverage of approximately eightfold for each PAC or BAC. The Gap4 program (Bonfield et al. 1995) was used first to assemble complete BAC and PAC clone sequences, and then to assemble contigs of these.
Telomere Cloning
Terminal EcoRI and BamHI restriction fragments carrying telomere sequences from the whole genome were obtained using a combination of linker-ligation and telomere-repeat-specific PCR, followed by cloning in pPCR-Script (Stratagene), and identification of telomere-containing clones by probing with a telomere-repeat-specific oligonucleotide ([AAACCT]5); positive clones were sequenced by standard methods. For each putative telomere, PCR primers were designed against the unique (nontelomere-repeat) part of the sequence and used to assign the sequence to one of the five chromosomal bands resolvable by Pulsed Field Gel Electrophoresis (PFGE), by digital blotting (below).
Digital Blotting
Digital blotting was devised as an alternative to Southern blotting for assigning sequences to discrete DNA bands resolved by PFGE. Briefly, chromosomes or restriction fragments are resolved by PFGE in low-melting-point agarose; each band is excised and melted in an equal volume of water (typically 10 ml for bands containing 100–500 ng of DNA), and 12 serial threefold dilutions are made for each. Five microliters of each dilution of each band are then screened using standard PCR conditions for the STS to be assigned. Typically, all bands give a positive result at their highest concentration (because PFGE does not perfectly resolve the DNA, and hence each band is contaminated with material from each of the others) but, over successive threefold dilutions, the amount of product decreases; the band in which the STS lies continues to give a strong product for two or three more dilutions than the other bands.
Sequence Analysis
All analyses were integrated in the Artemis software package (Rutherford et al. 2000).
Several complementary methods of gene prediction were used, including BLASTX analysis against the SWALL (Swiss-Prot + TrEMBL) protein database, mapping of the 567 EST sequences to the genome sequence, and application of the GeneID prediction software, using a training set for Dictyostelium discoideum (which has a similar G+C genome content) and a minimum gene length of 120 coding bases. Other gene prediction tools (e.g., HMMGENE and Genefinder) were also tested with the same training set, but were found to be `correct' for a lower proportion of predicted genes.
Fasta analyses of the predicted genes were performed; first-pass assignment of a function to the predicted CDS was primarily based on the extent and degree of Fasta similarity (>40% identity and opt score >100 OR >20% percent identity and opt score >200); those exhibiting weaker similarities were initially classified as hypothetical predicted proteins. Gene models and putative functions were refined manually for each CDS, based on a close inspection of the similarity data and/or domain information (e.g., Pfam).
Predicted transmembrane domains and signal peptides were identified using TMHMM (Sonnhammer et al. 1998) and SignalP (Nielsen et al. 1997), respectively.
Acknowledgments
We thank J. Pachebat and J. McLauchlin for helpful comments in the preparation of this manuscript, G. Nyakatura for advice on sequencing, and J. Gough for providing SUPERFAMILY assignments to our proteins.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1555203.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to EMBL. The sequence of Chromosome 6 appears under accession number BX526834; the end-sequences of the PAC clones appear under accession numbers AJ561222–AJ563278.]
Article published online before print in July 2003.
References
- Akiyoshi, D.E., Feng, X., Buckholt, M.A., Widmer, G., and Tzipori, S. 2002. Genetic analysis of a Cryptosporidium parvum human genotype 1 isolate passaged through different host species. Infect. Immun. 70: 5670-5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler, R., Attwood, T.K., Bairoch, A., Bateman, A., Birney, E., Biswas, M., Bucher, P., Cerutti, L., Corpet, F., Croning, M.D., et al. 2000. InterPro—An integrated documentation resource for protein families, domains and functional sites. Bioinformatics 16: 1145-1150. [DOI] [PubMed] [Google Scholar]
- Armson, A., Meloni, B.P., Reynoldson, J.A., and Thompson, R.C.A. 1999. Assessment of drugs against Cryptosporidium parvum using a simple in vitro screening method. FEMS Microbiol. Lett. 178: 227-233. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., Ball, C.A., Blake, J.A., Botstein, D., Butler, H., Cherry, J.M., Davis, A.P., Dolinski, K., Dwight, S.S., Eppig, J.T., et al. 2000. Gene ontology: Tool for the unification of biology. Nat. Genet. 25: 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S.R., Griffiths-Jones, S., Howe, K.L., Marshall, M., and Sonnhammer, E.L. 2002. The Pfam protein families database. Nucleic Acids Res. 30: 276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer, T.V., Svezhova, N.V., Sidorenko, N.V., and Khokhlov, S.E. 2000. Cryptosporidium parvum (Coccidia, apicomplexa): Some new ultrastructural observations on its endogenous development. Eur. J. Protistol. 36: 151-159. [Google Scholar]
- Blunt, D.S., Khramstov, N.V., Upston, S.J., and Montelone, B.A. 1997. Molecular karyotype analysis of Cryptosporidium parvum: Evidence for eight chromosomes and a low-molecular-size molecule. Clin. Diagn. Lab. Immunol. 4: 11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfield, J.K., Smith, K.F., and Staden, R. 1995. A new DNA sequence assembly program. Nucleic Acids Res. 23: 4992-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccio, S., Camilli, R., La Rosa, G., and Pozio, E. 1998. Establishing the Cryptosporidium parvum karyotype by NotI and SfiI restriction analysis and Southern hybridizations. Gene 219: 73-79. [DOI] [PubMed] [Google Scholar]
- Carraway, M., Tzipori, S., and Widmer, G. 1997. A new restriction fragment length polymorphism from Cryptosporidium parvum identifies genetically heterogeneous parasite populations and genotypic changes following transmission from bovine to human hosts. Infect. Immun. 65: 3958-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreno, R.A., Martin, D.S., and Barta, J.R. 1999. Cryptosporidium is more closely related to the gregarines than to coccidia as shown by phylogenetic analysis of apicomplexan parasites inferred using small-subunit ribosomal RNA gene sequences. Parasitol. Res. 85: 899-904. [DOI] [PubMed] [Google Scholar]
- Casemore, D., Wright, S., and Coop, R. 1997. Cryptosporidiosis —Human and animal epidemiology. In Cryptosporidium and cryptosporidiosis (ed. R. Fayer), pp. 65-92. CRC Press, Boca Raton, Florida.
- Chappell, C.L. and Okhuysen, P.C. 2002. Cryptosporidiosis. Curr. Opin. Infect. Dis. 15: 523-527. [DOI] [PubMed] [Google Scholar]
- Claros, M.G. and Vincens, P. 1996. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241: 779-786. [DOI] [PubMed] [Google Scholar]
- Current, W.L. 1989. Cryptosporidium spp. In Parasitic infections of the immunocompromised host (eds. P.W. Walzer and R.M. Genta), pp. 251-341. Marcel Dekker, NY.
- Dayhoff, M.O., Schwartz, R.M., and Orcutt, B.C. 1978. A model of evolutionary change in proteins. In Atlas of protein sequence and structure 5, Suppl. 3. pp. 345-352. National Biomedical Research Foundation, Washington, DC. [Google Scholar]
- Dear, P.H. 1997. HAPPY mapping. In Genome mapping: A practical approach (ed. P.H. Dear). pp. 95-123. IRL Press, Oxford, UK.
- Dear, P.H. and Cook, P.R. 1993. HAPPY mapping—Linkage mapping using a physical analog of meiosis. Nucleic Acids Res. 21: 13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dear, P.H., Bankier, A.T., and Piper, M.B. 1998. A high-resolution metric HAPPY map of human chromosome 14 Genomics 48: 232-241. [DOI] [PubMed] [Google Scholar]
- Fayer, R., Speer, C.A., and Dubey, J.P. 1997. The general biology of Cryptosporidium. In Cryptosporidium and cryptosporidiosis (ed. R. Fayer), pp. 1-42. CRC Press, Boca Raton, FL.
- Fichera, M.E. and Roos, D.S. 1997. A plastid organelle as a drug target in apicomplexan parasites. Nature 390: 407-409. [DOI] [PubMed] [Google Scholar]
- Frengen E., Weichenhan D., Zhao B., Osoegawa K., van Geel M., and de Jong P.J. 1999. A modular, positive selection bacterial artificial chromosome vector with multiple cloning sites. Genomics 58: 250-253. [DOI] [PubMed] [Google Scholar]
- Galtier, N., Gouy, M., and Gautier, C. 1996. SEAVIEW and PHYLO_WIN: Two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12: 543-548. [DOI] [PubMed] [Google Scholar]
- Gardner, M.J., Hall, N., Fung, E., White, O., Berriman, M., Hyman, R.W., Carlton, J.M., Pain, A., Nelson, K.E., Bowman, S., et al. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419: 498-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaberman, S., Moore, J.E., Lowery, C.J., Chalmers, R.M., Sulaiman, I., Elwin, K., Rooney, P.J., Millar, B.C., Dooley, J.S., Lal, A.A., et al. 2002. Three drinking-water-associated cryptosporidiosis outbreaks, Northern Ireland. Emerg. Infect. Dis. 8: 631-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau, A., Barrell, B.G., Bussey, H., Davis, R.W., Dujon, B., Feldmann H., Galibert, F., Hoheisel, J.D., Jacq, C., Johnston, M., et al. 1996. Life with 6000 genes. Science 274: 546-567. [DOI] [PubMed] [Google Scholar]
- Gough, J. and Chothia, C. 2002. SUPERFAMILY: HMMs representing all proteins of known structure. SCOP sequence searches, alignments and genome assignments. Nucleic Acids Res. 30: 268-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyot, K., Follet-Dumoulin, A., Lelievre, E., Sarfati, C., Rabodonirina, M., Nevez, G., Cailliez, J.C., Camus, D., and Dei-Cas, E. 2001. Molecular characterization of Cryptosporidium isolates obtained from humans in France. J. Clin. Microbiol. 39: 3472-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, A.D., Forster, S., Morton, S., Marshall, R., Osborn, K.S., Wright, P., and Hunter, P.R. 2002. Cryptosporidium oocysts in a water supply associated with a cryptosporidiosis outbreak. Emerg. Infect. Dis. 8: 619-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannou, P.A. and de Jong, P.J. 1996. Construction of bacterial artificial chromosome libraries using the modified P1 (PAC) System. Curr. Protocols Hum. Genet. 5: 1-24. [DOI] [PubMed] [Google Scholar]
- Jeffrey, H.J. 1990. Chaos game representation of gene structure. Nucleic Acids. Res. 18: 2163-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuth, J.P. 2000. The NCBI. Publicly available tools and resources on the Web. Meth. Mol. Biol. 132: 301-312. [DOI] [PubMed] [Google Scholar]
- Katinka, M.D., Duprat, S., Cornillot, E., Metenier, G., Thomarat, F., Prensier, G., Barbe, V., Peyretaillade, E., Brottier, P., Wincker, P., et al. 2001. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414: 450-453. [DOI] [PubMed] [Google Scholar]
- Konfortov, B.A., Cohen, H.M., Bankier, A.T., and Dear, P.H. 2000. A high-resolution HAPPY map of Dictyostelium discoideum chromosome 6. Genome Res. 10: 1737-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leander, B.S., Clopton, R.E., and Keeling, P.J. 2003. Phylogeny of gregarines (Apicomplexa) as inferred from SSU rDNA and β-tubulin. Int. J. Syst. Evol. Microbiol. 53: 345-354. [DOI] [PubMed] [Google Scholar]
- Lo Conte, L., Ailey, B., Hubbard, T.J.P., Brenner, S.E., Murzin, A.G., and Chothia, C. 2000. SCOP: A Structural classification of proteins database. Nucleic Acids Res. 28: 257-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manak, J.R., Mathies, L.D., and Scott, M.P. 1995. Regulation of a decapentaplegic midgut enhancer by homeotic proteins. Development 120: 3605-3619. [DOI] [PubMed] [Google Scholar]
- McLauchlin, J., Amar, C., Pedraza-Diaz, S., and Nichols, G.L. 2000. Molecular epidemiological analysis of Cryptosporidium spp. in the United Kingdom: Results of genotyping Cryptosporidium spp. in 1705 fecal samples from humans and 105 fecal samples from livestock animals. J. Clin. Microbiol. 38: 3984-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel, J.L., Perera, D.R., Meligro, C., and Rubin, C.E. 1976. Overwhelming watery diarrhea associated with a Cryptosporidium in an immunosuppressed patient. Gastroenterol. 70: 1156-1160. [PubMed] [Google Scholar]
- Morgan-Ryan, U.M., Fall, A., Ward, L.A., Hijjawi, N., Sulaiman, I., Fayer, R., Thompson, R.C., Olson, M., Lal, A., and Xiao, L. 2002. Cryptosporidium hominis n. sp. (Apicomplexa: Cryptosporidiidae) from Homo sapiens. J. Eukaryot. Microbiol. 49: 433-440. [DOI] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10: 1-6. [DOI] [PubMed] [Google Scholar]
- Nime, F.A., Burek, J.D., Page, L.D., Holscher, M.A., and Yardley, J.H. 1976. Acute enterocolitis in a human being infected with the protozoan Cryptosporidium. Gastroenterol. 70: 592-598. [PubMed] [Google Scholar]
- Nomura, T., Carlton, J.M., Baird, J.K., del Portillo, H.A., Fryauff, D.J., Rathore, D., Fidock, D.A., Su, X., Collins, W.E., McCutchan, T.F. et al. 2001. Evidence for different mechanisms of chloroquine resistance in 2 Plasmodium species that cause human malaria. J. Infect. Dis. 183: 1653-1661. [DOI] [PubMed] [Google Scholar]
- Pearson, W.R. and Lipman, D.J. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. 85: 2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper, M.B., Bankier, A.T., and Dear, P.H. 1998a. A HAPPY map of Cryptosporidium parvum. Genome Res. 8: 1299-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper, M.B., Bankier, A.T., and Dear, P.H. 1998b. Construction and characterization of a genomic PAC library of the intestinal parasite Cryptosporidium parvum. Mol. Biochem. Parasitol. 95: 147-151. [DOI] [PubMed] [Google Scholar]
- Riordan, C.E., Langreth, S.G., Sanchez, L.B., Kayser O., and Keithly, J.S. 1999. Preliminary evidence for a mitochondrion in Cryptosporidium parvum: Phylogenetic and therapeutic implications. J. Eukaryotic Microbiol. 46 52-55. [PubMed] [Google Scholar]
- Rose, J.B., Huffman, D.E., and Gennaccaro, A. 2002. Risk and control of waterborne cryptosporidiosis. FEMS Microbiol. Rev. 26: 113-123. [DOI] [PubMed] [Google Scholar]
- Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M.A., and Barrell, B. 2000. Artemis: Sequence visualization and annotation. Bioinformatics 16: 944-945. [DOI] [PubMed] [Google Scholar]
- Schmidt, H.A., Strimmer, K., Vingron, M., and von Haeseler, A. 2002. TREE-PUZZLE: Maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18: 502-504. [DOI] [PubMed] [Google Scholar]
- Sonnhammer, E.L., von Heijne, G., and Krogh, A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6: 175-182. [PubMed] [Google Scholar]
- Spano, F., Putignani, L., McLauchlin, J., Casemore, D.P., and Crisanti, A. 1997. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol. Lett. 150: 209-217. [DOI] [PubMed] [Google Scholar]
- Strong, W.B. and Nelson, R.G. 2000. Preliminary profile of the Cryptosporidium parvum genome: An expressed sequence tag and genome survey sequence analysis. Mol. Biochem. Parasitol. 107: 1-32. [DOI] [PubMed] [Google Scholar]
- Su, X., Kirkman, L.A., Fujioka, H., and Wellems, T.E. 1997. Complex polymorphisms in an approximately 330kDa protein are linked to chloroquine-resistant P. falciparum in Southeast Asia and Africa. Cell 91: 593-603. [DOI] [PubMed] [Google Scholar]
- Sulaiman, I.M., Lal, A.A., and Xiao, L. 2002. Molecular phylogeny and evolutionary relationships of Cryptosporidium parasites at the actin locus. J. Parasitol. 88: 388-394. [DOI] [PubMed] [Google Scholar]
- Teichmann, S.A. and Mitchison, G. 1999. Is there a phylogenetic signal in prokaryote proteins? J. Mol. Evol. 49: 98-107. [DOI] [PubMed] [Google Scholar]
- Tetley, L., Brown, S.M.A., McDonald, V., and Coombs, G.H. 1998. Ultrastructural analysis of the sporozoite of Cryptosporidium parvum. Microbiol. 144: 3249-3255. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, L., Sulaiman, I.M., Ryan, U.M., Zhou, L., Atwill, E.R., Tischler, M.L., Zhang, X., Fayer, R., and Lal, A.A. 2002. Host adaptation and host-parasite coevolution in Cryptosporidium: Implications for taxonomy and public health. Int. J. Parasitol. 32: 1773-1785. [DOI] [PubMed] [Google Scholar]
- Zhang, L., Cui, X., Schmitt, K., Hubert, W., Navidi, W., and Arnheim, N. 1992. Whole genome amplification from a single cell: Implications for genetic analysis. Proc. Natl. Acad. Sci. 87: 5487-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, G., Marchewka, M.J., and Keithly, J.S. 2000. Cryptosporidium parvum appears to lack a plastid genome. Microbiol. 146: 315-321. [DOI] [PubMed] [Google Scholar]
- Zuegge, J., Ralph, S., Schmuker, M., McFadden, G.I., and Schneider, G. 2001. Deciphering apicoplast targeting signals—Feature extraction from nuclear-encoded precursors of Plasmodium falciparum apicoplast proteins. Gene 280: 19-26. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html/; NCBI Taxonomy Homepage.
- http://www.sanger.ac.uk/Projects/E_tenella/; The Sanger Institute Eimeria tenella Genome Project.
- http://www.tigr.org/tdb/e2k1/tga1/; The TIGR Toxoplasma gondii Genome Project.
- http://mips.gsf.de/cgi-bin/proj/medgen/mitofilter; MITOP—Description of MITOP.
- http://gecco.org.chemie.uni-frankfurt.de/pats/pats-index.ph; Modlab—The Molecular Design Laboratory.
- http://www.genome.ou.edu/BAC_isoln_200ml_culture.html; Cleared Lysate Method.