Abstract
Context:
Congenital adrenal hyperplasia (CAH) is an autosomal recessive condition that arises from mutations in CYP21A2 gene, which encodes for the steroidogenic enzyme 21-hydroxylase. To prevent genital ambiguity in affected female fetuses, prenatal treatment with dexamethasone must begin on or before gestational week 9. Currently used chorionic villus sampling and amniocentesis provide genetic results at approximately 14 weeks of gestation at the earliest. This means that mothers who want to undergo prenatal dexamethasone treatment will be unnecessarily treating seven of eight fetuses (males and three of four unaffected females), emphasizing the desirability of earlier genetic diagnosis in utero.
Objective:
The objective of the study was to develop a noninvasive method for early prenatal diagnosis of fetuses at risk for CAH.
Patients:
Fourteen families, each with a proband affected by phenotypically classical CAH, were recruited.
Design:
Cell-free fetal DNA was obtained from 3.6 mL of maternal plasma. Using hybridization probes designed to capture a 6-Mb region flanking CYP21A2, targeted massively parallel sequencing (MPS) was performed to analyze genomic DNA samples from parents and proband to determine parental haplotypes. Plasma DNA from pregnant mothers also underwent targeted MPS to deduce fetal inheritance of parental haplotypes.
Results:
In all 14 families, the fetal CAH status was correctly deduced by targeted MPS of DNA in maternal plasma, as early as 5 weeks 6 days of gestation.
Conclusions:
MPS on 3.6 mL plasma from pregnant mothers could potentially provide the diagnosis of CAH, noninvasively, before the ninth week of gestation. Only affected female fetuses will thus be treated. Our strategy represents a generic approach for noninvasive prenatal testing for an array of autosomal recessive disorders.
Congenital adrenal hyperplasia (CAH) is an autosomal recessive disorder arising from mutations of the CYP21A2 gene that cause a deficiency of 21-hydroxylase. Affecting 1 of 15 000 live births worldwide (1, 2), the disease is characterized by varying levels of impairments in mineralocorticoid and glucocorticoid synthesis, overstimulation of the androgen pathway, and virilization of female fetuses (2, 3). Three clinical phenotypes, namely salt wasting, simple virilizing, and nonclassical CAH, result from differing extents of 21-hydroxylase impairment established through in silico computational modeling (4).
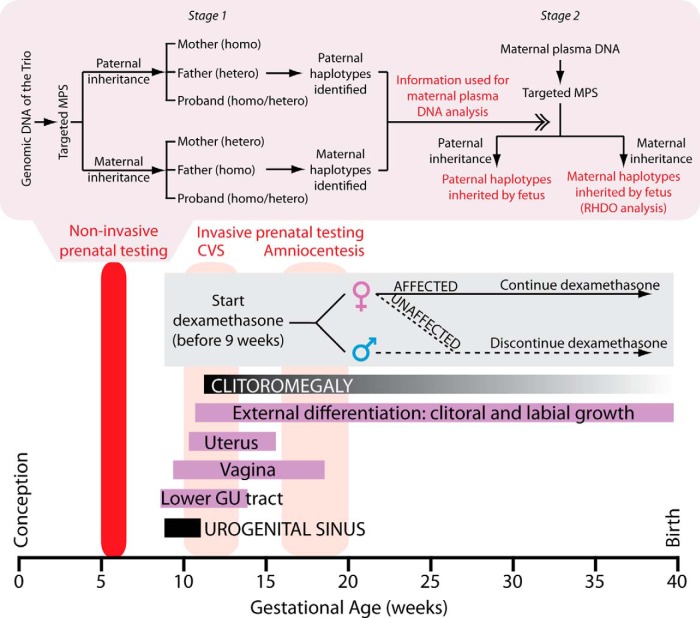
CAH is diagnosed prenatally by chorionic villus sampling (CVS) at approximately 14 weeks of gestation, or later, at approximately 20 weeks, by amniocentesis (Figure 1). However, genital organogenesis begins at approximately 9 weeks of gestation, and excess fetal androgen production causes genital virilization in female fetuses (Figure 1). To prevent genital ambiguity in female fetuses affected with classical CAH, dexamethasone is administered to the mother starting before 9 weeks of gestation (5). Current invasive prenatal diagnosis does not yield genetic results until later (Figure 1). This means that mothers bearing male and unaffected female fetuses will also receive dexamethasone. It should be noted that although CAH is one of the few genetic disorders that can be treated prenatally for phenotypic abnormalities, ie, genital ambiguity in the affected female fetus, there is controversy about prenatal treatment with dexamethasone. The Endocrine Society issued guidelines in 2010 stating that prenatal treatment is not considered the standard of care and should be carried out only as an experimental research procedure under institutional review board approval (6). Furthermore, both amniocentesis and CVS pose a risk to both mother and fetus. There is thus a need for diagnosing CAH before genital organogenesis begins at approximately 9 weeks so that therapy will be given only to mothers with an affected female fetus and not males and unaffected female fetuses.
Figure 1.
Conventional prenatal management and targeted MPS for noninvasive detection of CAH. Temporal relationship between normal genital organ development, detection of CAH mutations by CVS or amniocentesis, and initiation of therapy with dexamethasone are shown. Targeted MPS should provide an early diagnosis before 9 weeks of gestation. In stage 1, genomic DNA samples of the mother, father, and proband (trio) were subjected to targeted MPS of the CYP21A2 region. For paternal inheritance, SNPs that were homozygous (homo) in the mother and heterozygous (hetero) in the father were used. Paternal haplotypes inherited by the proband and absent in the proband were thus identified. For maternal inheritance, SNPs that were heterozygous in the mother and homozygous in the father were used. Maternal haplotypes (linked or not linked with the proband's mutation) were determined. Stage 2 targeted MPS of the CYP21A2 region on plasma cell-free DNA of the pregnant mother. Detection of the paternal-specific alleles in maternal plasma revealed the inheritance of either the paternal haplotype linked or not linked with the proband's mutation from the father. Maternal inheritance was determined by the RHDO analysis (see Supplemental Methods and Reference 7 for details).
Massively parallel sequencing (MPS) of cell-free fetal DNA in maternal plasma has opened new possibilities for the diagnosis of monogenic disorders in utero. Because fetal DNA exists in maternal plasma amid a massive excess of maternal DNA, simple PCR-based analyses cannot be applied. Furthermore, in the case of CAH, because cell-free fetal DNA is highly fragmented, long-range PCR cannot be performed to differentiate between mutations in CYP21A2 and the homologous pseudogene CYP21A1P.
Here we have made the diagnosis of CAH in utero by using 3.6 mL maternal plasma obtained as early as 6 gestational weeks for targeted MPS of the genomic region flanking and including the CYP21A2 gene. We first mapped single-nucleotide polymorphisms (SNPs) linked to the CYP21A2 gene in the parents and proband and then looked for representation of the respective haplotype maps in the plasma of pregnant mothers. This allowed us to elucidate paternal and maternal inheritance of the fetus at the CYP21A2 locus. We noted complete concordance of the CAH diagnosis between invasive diagnosis and noninvasive MPS in all 14 cases. Our approach should permit the diagnosis of CAH before genital development begins, thus restricting dexamethasone therapy to mothers bearing affected females only.
Materials and Methods
CAH pedigrees
Families affected by CAH due to CYP21A2 gene mutations were recruited at Mount Sinai School of Medicine with informed consent, with ethics approval from both Mount Sinai School of Medicine and The Chinese University of Hong Kong Institutional Review Boards. Genetic counseling was provided to the families, and clinical samples (blood, and/or samples from amniocentesis or CVS) were collected. Mothers had to agree to DNA analysis on amniocentesis or chorionic villus samples or on blood from newborns to validate the noninvasive protocol. Plasma and DNA were transferred to The Chinese University of Hong Kong for targeted MPS. Fourteen families were chosen based on the following: 1) having a child affected with classical CAH (proband), 2) both parents carrying at least one mutant CYP21A2 gene, and 3) availability of peripheral blood from parents and proband (trio) and at least 3.6 mL plasma from pregnant mothers.
Targeted massively parallel sequencing and haplotype analysis
Details of the method, as used previously (7, 8), are provided in Supplemental Methods. Briefly, cell-free DNA was isolated from maternal plasma and fetal gender was determined by PCR using a SRY probe (9). Genomic DNA from the trio and plasma from pregnant mothers then underwent massively parallel sequencing. Sequence analysis of the trio established linkage of SNPs in the CYP21A2 locus to the proband's mutational status. It also allowed the deduction of paternal and maternal haplotypes. Notably, to determine inheritance, we carried out relative haplotype dosage analysis (RHDO) (7, 8) to establish overrepresentation of the maternal and paternal haplotypes in the pregnant mother's plasma with reference to the respective noninherited haplotypes. For paternal inheritance determination, we used SNP alleles (comprising a haplotype block) that were homozygous in the mother but heterozygous in the father. To deduce maternal inheritance, we used RHDO to measure the relative concentrations of the maternal haplotypes in the pregnant mother's plasma. For this, SNP alleles in which the mother was heterozygous and the father was homozygous were informative.
Results
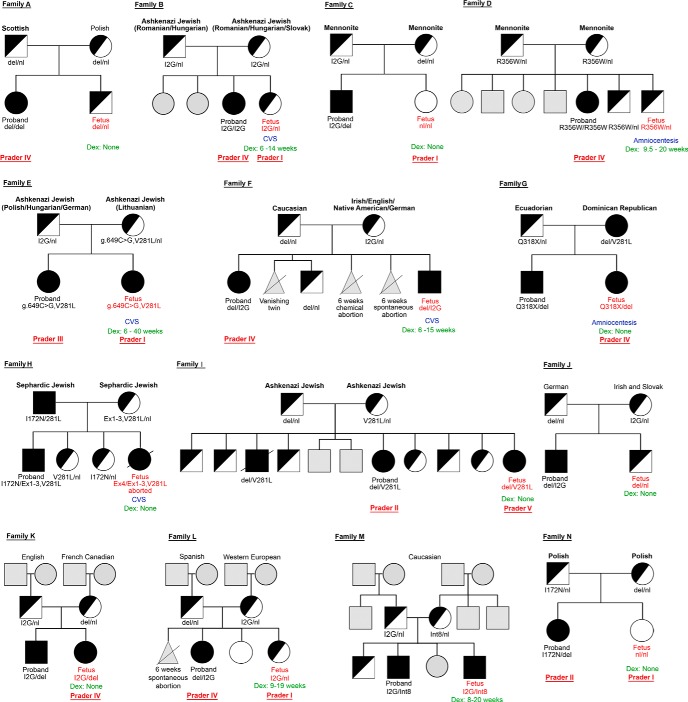
Supplemental Table 1 shows key summary statistics of the sequencing data. Of the 14 fetuses studied, the pedigrees and clinical details of which are shown in Figure 2 and Table 1, respectively, seven had CAH, five were carriers, and two were normal. CVS and amniocentesis were used to establish the diagnosis in five and two cases, respectively, whereas the remaining seven cases were diagnosed in the newborns. Five of the seven mothers, whose fetuses were diagnosed prenatally, received dexamethasone. Of these, dexamethasone was continued to term in just one, whereas in four mothers, treatment was discontinued once the diagnosis was established (two males and two heterozygous females). The one affected female (family E), whose mother continued to receive dexamethasone, was born with normal genitals, whereas the affected females (families G, I, and K), whose mothers did not receive dexamethasone, displayed genital ambiguity at birth. One mother received dexamethasone without invasive diagnosis; treatment was stopped upon male sex determination by ultrasound (family M).
Figure 2.
Pedigree for 14 families with CAH that underwent massively parallel sequencing and haplotype analysis. The parents and proband constituted the trio. Genotype, ethinicity, Prader score (for females), and whether the mother received prenatal dexamethasone are shown. Please read in conjunction with Table 1.
Table 1.
Clinical Information of Studied Families
| Family | Fetal Sex | Mutation Typing |
Invasive Prenatal Diagnosis | Prader Scorea | Dex, wk | Plasma Sample Collection |
CYP21A2 Targeted Sequencing |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Father | Mother | Fetus/Newborn | Fetal, % | Pat Hap | Mat Hap | Fetal Status | ||||||
| A | M | del/nl | del/nl | del/nl | None | None | 8 wk 5 d | 4.8 | Mpa | Nma | C | |
| B | F | I2G/nl | I2G/nl | I2G/nl | CVS | I | 6–14 | 21 wk 3 d | 15.6 | Mpa | Nma | C |
| C | F | I2G/nl | del/nl | nl/nl | None | I | None | 10 wk 2 d | 13.5 | Npa | Nma | N |
| D | M | R356W/nl | R356W/nl | R356W/nl | Amnio | 9.5–20 | 10 wk 4 d | 7.8 | Npa | Mma | C | |
| E | F | I2G/nl | g.649 C>G, V281L/nl | I2G/g.649 C>G, V281L | CVS | I | 6–40 | 21 wk 3 d | 22.3 | Mpa | Mma | A |
| F | M | del/nl | I2G/nl | del/I2G | CVS | 6–10 | 10 wk 3 d | 7.7 | Mpa | Mma | A | |
| G | F | Q318X/nl | del/V281L | Q318X/del | Amnio | IV | None | 24 wk 3 d | 17.6 | Mpa | Mma | A |
| H | F | I172N/V281L | Ex1–3, V281L/nl | I172N/Ex1–3, V281L | CVS | Aborted | None | 6 wk 5 d | 3.8 | Mpa | Mma | A |
| I | F | del/nl | V281L/nl | del/V281L | None | V | None | 9 wk 2 d | 11.5 | Mpa | Mma | A |
| J | M | del/nl | I2G/nl | del/nl | None | None | 7 wk 3 d | 3.4 | Mpa | Nma | C | |
| K | F | I2G/nl | del/nl | I2G/del | None | IV | None | 9 wk 1 d | 3.7 | Mpa | Mma | A |
| L | F | del/nl | I2G/nl | I2G/nl | CVS | I | 9–19 | 8 wk 6 d | 9.0 | Npa | Mma | C |
| M | M | I2G/nl | Int8/nl | I2G/Int8 | None | 8–20 | 12 wk 6 d | 8.6 | Mpa | Mma | A | |
| N | F | I172N/nl | del/nl | nl/nl | None | I | None | 5 wk 6 d | 1.4 | Npa | Nma | N |
Abbreviations: Amnio, amniocentesis; C, carrier; A, affected; del, 30-kb large gene deletion; Dex, dexamethasone treatment; Ex1–3, exons 1–3 cluster conversion; I2G, intron 2 (g.655A/C>G); Int8, IVS8 + 1G>A; Mma, maternal haplotype linked with the proband's mutation; Mpa, paternal haplotype linked with the proband's mutation; Mat Hap, maternal haplotype; N, normal; nl, normal; Nma, maternal haplotype not linked with the proband's mutation; Npa, paternal haplotype not linked with the proband's mutation; Pat Hap, paternal haplotype.
The Prader Scale is a scoring system for grading the degrees (I-V) of female genital masculinization, with I being the least masculinized and V being the most masculinized.
Family A
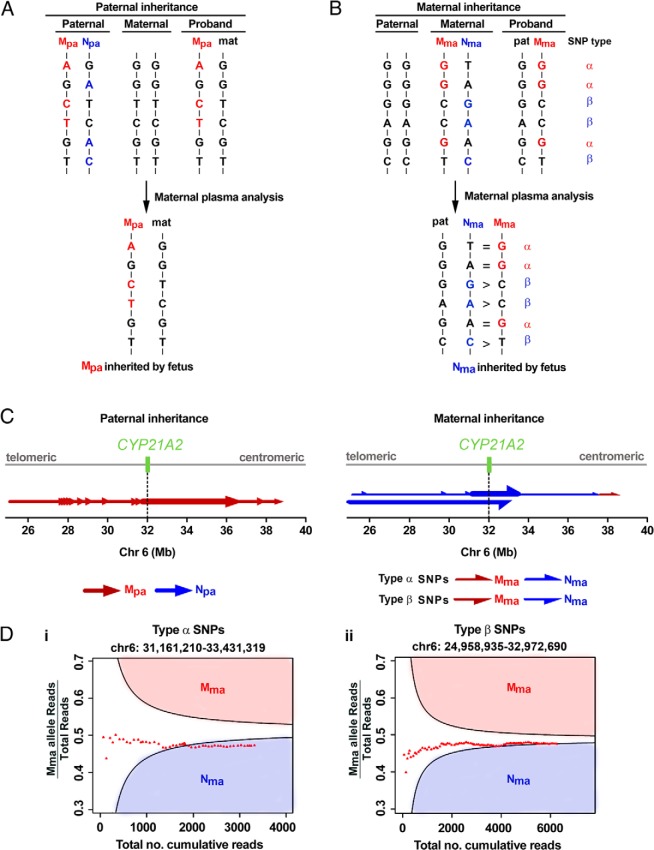
In family A, both parents were carriers of a 30-kb deletion at the CYP21A2 locus (Figure 2 and Table 1). The proband was homozygous and born with ambiguous genitalia. The father's SNP alleles inherited by the proband formed the paternal haplotype linked with the proband's mutation (Mpa). Those alleles absent in the proband formed the paternal haplotype not linked with the proband's mutation (Npa) (Figure 3). We then detected the paternal-specific SNP alleles in the maternal plasma DNA obtained at 8 weeks 5 days, identified to which paternal haplotype they belonged, counted the frequencies of the SNP alleles, and thus determined which paternal haplotype was transmitted to the fetus (Figure 3). We applied the Kolmogorov-Smirnov test to determine whether the difference of allelic counts between the two paternal haplotypes was statistically significant along the targeted region.
Figure 3.
Fetal inheritance of parental mutations in family A. A, For paternal inheritance, SNPs that were homozygous in the mother and heterozygous in the father were selected. The paternal-specific SNP alleles transmitted to the proband belonged to the haplotype linked with the proband's mutation (Mpa, red). The paternal-specific SNP alleles not present in the proband belonged to the haplotype not linked with the proband's mutation (Npa, blue). B, For maternal inheritance, SNPs that were heterozygous in the mother and homozygous in the father were classified into type α- or type β-SNPs. For type α-SNPs, the paternal alleles were the same as those on Mma (red). The equal representation of Mma and Nma alleles in the maternal plasma indicated that the fetus had inherited Nma from the mother. For type β-SNPs, the paternal alleles were the same as those of Nma (blue). The overrepresentation of Nma alleles in the maternal plasma also indicated that the fetus had inherited Nma from the mother. C, Parental haplotype blocks are denoted by arrows along the entire targeted region. The tail and tip of an arrow denote the start and end, respectively, of a RHDO block, established analytically from the telomeric to the centromeric end. Left, Paternal inheritance analysis resulted in 14 paternal haplotype blocks, represented by red arrows, all indicating the inheritance of Mpa. Prenatal determination of the fetal inheritance status was made according to the haplotype block spanning the CYP21A2 gene, indicated with a thick red arrow. Right, Maternal inheritance analysis resulted in six and one SPRT classifications using type α- and type β-SNPs, respectively. The haplotypes established by the type α- and β-SNPs are represented by upper and lower half-arrows, respectively. The fetus had inherited Nma from the mother by both type α- and β-SNPs. The SPRT classification near the centromeric end indicated that a recombination event had occurred. Grey line, targeted region spanning chromosome 6: 24,954,958–38,933,706. Dotted line and green box, CYP21A2 gene on chromosome 6: 32,006,093–32,009,447. D, RHDO analysis showing SPRT classification process using type α-SNPs on chromosome 6: 31,161,210–33,431,319 (i) or type β-SNPs on chromosome 6: 24,958,935–32,972,690 (ii). For details, please refer to Supplemental Methods.
Family A had 1044 informative SNPs for paternal inheritance within the captured CYP21A2 region (Supplemental Table 2). Fourteen paternal haplotype blocks were identified (Figure 3). The haplotype block (chromosome 6:31,713,454–36,394,087) spanning the CYP21A2 gene (chromosome 6: 32,006,093–32,009,447) indicated that the fetus had inherited Mpa, ie, the haplotype linked with the proband's mutation from the father (Figure 3).
For determining maternal inheritance, we assigned informative SNPs as type α or β (7, 8). For type α-SNPs, the paternal alleles were the same as the maternal alleles on the maternal haplotype linked with the proband's mutation (Mma). For type β-SNPs, the paternal alleles were the same as the maternal alleles on the maternal haplotype not linked with the proband's mutation (Nma) (Figure 3). To determine the relative representations of each of the two maternal haplotypes in maternal plasma, we counted the number of plasma DNA molecules carrying SNP alleles corresponding to Mma and Nma, and performed RHDO analyses for type α- and β-SNPs (7, 8), (see Supplemental Methods and Supplemental Table 2). A sequential probability ratio test (SPRT) (10, 11) was used to estimate the balance or imbalance of the dosage of these respective haplotypes (7, 8).
A total of 398 type α-SNPs and 176 type β-SNPs were identified, resulting in six and one SPRT classifications, respectively (Supplemental Table 2). For type α-SNPs, alleles on Mma and Nma were equally represented in five SPRT classifications, indicating that the fetus had inherited Nma. However, an overrepresentation of Mma alleles was observed in the last SPRT classification, indicating that the fetus had inherited Mma at the centromeric end (chromosome 6:37,562,702–38,561,222) (Figure 3). These results could most likely be explained by a recombination event in a major histocompatibility complex class II recombination hot spot (12, 13), which we confirmed with targeted MPS on the newborn's DNA. For type β-SNPs, an overrepresentation of Nma alleles was observed in the SPRT classification, thus indicating that the fetus has inherited Nma, ie, the haplotype not linked with the proband's mutation, from the mother. Thus, the fetus was only a carrier (unaffected).
Families B to M
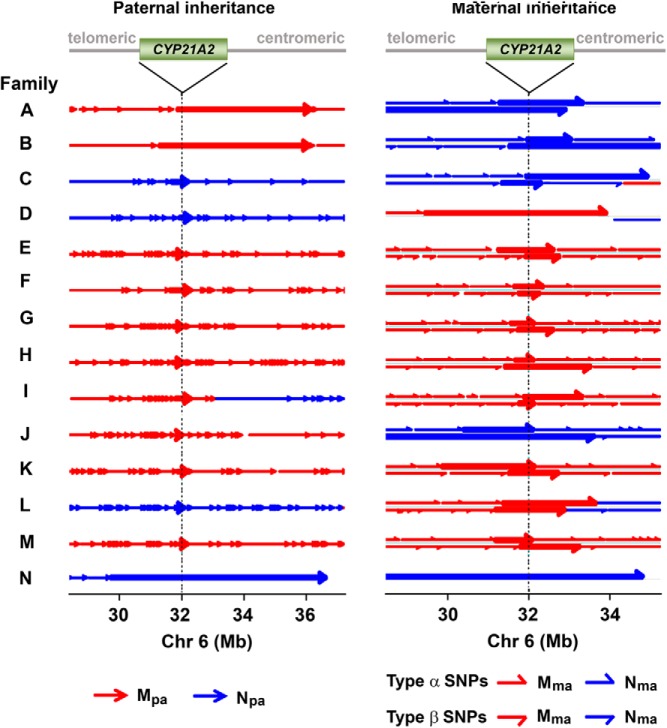
The haplotype blocks of paternal and maternal inheritance were plotted along the targeted region for each case (Figure 4). Overall, we identified seven affected fetuses, five carriers, and two unaffected fetuses in concordance with the clinical data and PCR assays by invasive prenatal diagnosis or newborn blood draw (Table 1). For example, in family G, the father was a carrier of an exon 8 (Q318X) mutation and the mother was heterozygous for an exon 7 (V281L) mutation and a 30-kb deletion. For maternal inheritance analysis by RHDO, we defined the haplotype linked with the proband's 30-kb deletion as Mma, and the haplotype linked with the V281L mutation as Nma (Figure 4). We found that the fetus had inherited the paternal mutation, and the same maternal deletion mutation as the proband, and was hence affected with CAH (Table 1). Likewise, in family H, the father was compound heterozygous for the I172N and V281L mutations, whereas the mother was a carrier of a 30-kb large gene deletion. We defined paternal haplotype linked with the proband's I172N mutation as Mpa (Figure 4) and found that the fetus had inherited the paternal I172N mutation and the maternal deletion and was therefore affected with CAH. Finally, and interestingly, in addition to family A, maternal recombination events were found in families C, D and H, and a paternal recombination was identified in family I (Figure 4). Recombinations in both paternal and maternal alleles were found in family L. The fetal or newborn's DNA analyses confirmed the recombination found in maternal plasma DNA analysis.
Figure 4.
Fetal haplotype analysis in families A to N. A haplotype block is indicated by an arrow. The tail and tip of an arrow denotes the start and end, respectively, of a haplotype block, established analytically from the telomeric to the centromeric end. Left, Fetal inheritance of the paternal haplotype was determined by the presence of paternal-specific SNP alleles in maternal plasma DNA samples. The haplotype block spanning the CYP21A2 gene is denoted by a thick arrow in each family. Right, Fetal inheritance of the maternal haplotype was determined by RHDO analysis. Haplotype blocks within chromosome 6, 28–35 Mb, are shown. Upper and lower half-arrows denote the SPRT classification made by type α- or type β-SNPs, respectively. The SPRT classification spanning the CYP21A2 gene region, denoted by a thick half-arrow, indicates the maternal haplotype inherited by the fetus. Notably, the mother and father of families G and H, respectively, were heterozygous for two CYP21A2 gene mutations themselves. Targeted MPS analysis indicated that the fetuses in both families had inherited the haplotypes linked to the proband's mutations from the affected parents. The dotted lines indicate the location of the CYP21A2 gene (green box). The haplotype blocks of several samples are partially shown, as indicated by a line without an arrow tip toward the centromeric end. In family H, a recombination event in the maternal allele, which occurs outside of the chromosomal region displayed above, is not shown.
Early maternal plasma sampling in family N
DNA analysis of maternal plasma obtained at 5 weeks 6 days (family N) indicated that the fetal DNA concentration was extremely low (1.4%, c.f. 4.8% in family A) (Table 1). Analysis of paternal inheritance showed that the fetus had inherited Npa, ie, the paternal haplotype not linked with the proband mutation. Thus, diagnosis of CAH could be excluded because the fetus had inherited at least one unaffected allele. For maternal inheritance, a total of 506 type α-SNPs and 159 type β-SNPs were identified (Supplemental Table 2). One SPRT classification was made using the first 277 type α-SNPs, whereas the rest of the SNPs did not contribute to any additional classification. The result showed an equal representation of Mma and Nma alleles. Thus, the fetus had inherited Nma, the haplotype not linked with the proband's mutation, from the mother. Notably, the maternal plasma sample of family N made an important point: that the fractional fetal DNA concentration was critical for making an SPRT classification for RHDO analysis for maternal inheritance. We thus proceeded to investigate in silico the effects of varying fractional fetal DNA concentrations on the SPRT analysis (see Supplemental Methods and Supplemental Table 3). The simulation results provide confidence toward making RHDO classifications for cases with extremely low fractional fetal DNA concentrations if the number of SNPs reached 1000 and the sequencing depth was 200-fold.
Discussion
We report the development and validation of targeted MPS for the noninvasive prenatal diagnosis of CAH using maternal plasma. This is the first demonstration of noninvasive prenatal diagnosis in a large number of pregnancies with a monogenic disorder transmitted as a recessive trait. Due to the existence of a highly homologous CYP21A1P pseudogene, and the fact that most CAH mutations are generated by gene conversions that transfer mutations from the pseudogene, it is not possible to directly detect CAH mutations by currently employed methodologies. Instead, our approach is based on the detection of SNPs linked to the active CYP21A2 gene. Here we provide the correct fetal genotype of every case in 14 families. We can draw blood as early as 6 weeks of gestation for the in utero genetic testing of CAH and provide the diagnosis in time to treat the affected female before 9 weeks. Our strategy is also especially applicable to developing countries that lack sterile facilities for CVS and amniocentesis, procedures that can, even in experienced hands, result in miscarriages and hemangiomas (14–17). In principle, because drawing blood does not require sterile facilities, our noninvasive MPS-based protocol will be the first time a prenatal diagnosis can be safely made in developing countries.
The earliest gestational age at which maternal plasma was obtained in our cohort was 5 weeks 6 days (family N). Considering the inherent experimental and analytical time, prenatal diagnosis for CAH families could be available before the ninth week of gestation, at which time fetal genital organogenesis begins (18). As an example, the mother of family B was started on dexamethasone at week 6 (Table 1). However, dexamethasone was discontinued at the 14th week when CVS identified the fetus was heterozygous. Thus, dexamethasone treatment could have been avoided if the genotype was known earlier.
The mother of family I, in contrast, chose not to undergo invasive prenatal diagnosis or be treated with dexamethasone because the female proband, although affected with salt-wasting CAH, had minimally virilized genitalia. However, the pregnancy resulted in a girl with salt-wasting CAH and severe genital ambiguity (Prader V). Remarkably, both children had the same mutation, del/V281L, but had variable phenotypes (2, 4). It is possible that recombination events occurring near the CYP21A2 gene in the paternal allele affected transcriptional regulation (Figure 4), yielding a newborn that was phenotypically distinct from the proband. This genotype-phenotype discordance highlights the importance of caution in predicting clinical phenotypes, even when the mutation is known.
Fifty percent of affected fetuses in autosomal recessive disorders are males. Our protocol uses a PCR assay for gender determination using a SRY probe. A recent publication by Tardy-Guidollet et al (19) demonstrates their success in using the SRY probe for noninvasive prenatal sex determination. Results of the PCR, available within hours after fetal DNA isolation, can be used to eliminate unnecessary prenatal treatment in males. For the male fetuses in families F and M (Figure 2), complete genetic analysis was performed because the parents wished to know whether their child had CAH. Furthermore, genotyping results of male fetuses with severe mutations will prompt physicians to initiate immediate postnatal treatment with glucocorticoids, mineralocorticoids, and dietary salt to prevent adrenal crises and mortality of male newborns with salt-wasting CAH. The early prenatal diagnosis allows ample time for counseling to the parents regarding the postnatal care of a male with a potentially life-threatening type of CAH.
This noninvasive method is applicable not only to CAH but also for other monogenic disorders, such as β-thalassemia (8), 11β-hydroxylase deficiency (20), hemophilia (21), cystic fibrosis (22), and Gaucher disease (23). Unlike the other inherited diseases, our technology is particularly relevant to CAH because the disorder can be effectively treated in utero by administering dexamethasone to the mother to prevent genital ambiguity in affected female fetuses. Furthermore, albeit speculative, it may be possible to design a capture probe set that covers SNPs that flank genes involved in inherited diseases highly prevalent, in particular geographic or ethnic groups, such as CAH and Gaucher disease in Ashkenazi Jews (2, 3, 23).
In conclusion, we report a strategy for the noninvasive prenatal testing for CAH using targeted MPS on fetal cell-free DNA in maternal plasma. Although our data are promising, the number of studied cases, the largest among other studies of monogenic disorders to date, is still relatively small. In three families, cell-free fetal DNA was obtained early enough in gestation to permit starting treatment on only those who would benefit. That these three and the seven between approximately 9 and 10 weeks were also accurately diagnosed would indicate that sufficient fetal DNA is present in maternal plasma by 6 weeks. Sensitivity and specificity of the method will be determined in future large-scale prospective studies. In the long term, however, one would anticipate that, if a safe, noninvasive option for accurate in utero diagnosis is available, families may be more willing to undergo noninvasive prenatal diagnosis and prenatal treatment for affected female fetuses with appropriate genetic counseling.
Acknowledgments
This work was supported by the University Grants Committee of the Government of the Hong Kong Special Administration Region (China), under the Areas of Excellence Scheme (Grant AoE/M-04/06) (to Y.M.L.); a sponsored research agreement with Sequenom (to Y.M.L.); funding from Genesis Foundation and the Maria I. New Children's Hormone Foundation (to M.I.N.); and recruitment from CARES, Inc, a patient advocacy foundation for families affected by CAH. We also gratefully acknowledge the National Institutes of Health for salary and consumable support (Grants DK80459, AG23176, AG40132, and AR65932) (to M.Z. and L.S.).
Disclosure Summary: Y.M.L. and R.W.K.C. are consultants to and hold equities in Sequenom. K.C.A.C. holds equities in Sequenom. Y.M.L., R.W.K.C., Y.K.T., P.J., and K.C.A.C. hold patents or patent applications in noninvasive prenatal testing. The other authors have nothing to disclose.
Footnotes
- CAH
- congenital adrenal hyperplasia
- CVS
- chorionic villus sampling
- MPS
- massively parallel sequencing
- RHDO
- relative haplotype dosage analysis
- SNP
- single-nucleotide polymorphism
- SPRT
- sequential probability ratio test.
References
- 1. Pang SY, Wallace MA, Hofman L, et al. Worldwide experience in newborn screening for classical congenital adrenal hyperplasia due to 21-hydroxylase deficiency. Pediatrics. 1988;81:866–874 [PubMed] [Google Scholar]
- 2. New MI, Abraham M, Gonzalez B, et al. Genotype-phenotype correlation in 1,507 families with congenital adrenal hyperplasia owing to 21-hydroxylase deficiency. Proc Natl Acad Sci USA. 2013;110:2611–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. New MI, Lekarev O, Mancenido D, Parsa A, Yuen T. Congenital adrenal hyperplasia owing to 21-hydroxylase deficiency. In: New MI, Lekarev O, Parsa A, Yuen T, O'Malley BW, Hammer GD, eds. Genetic Steroid Disorders. London: Elsevier; 2014:29–51 [Google Scholar]
- 4. Haider S, Islam B, D'Atri V, et al. Structure-phenotype correlations of human CYP21A2 mutations in congenital adrenal hyperplasia. Proc Natl Acad Sci USA. 2013;110:2605–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. New MI, Abraham M, Yuen T, Lekarev O. An update on prenatal diagnosis and treatment of congenital adrenal hyperplasia. Semin Reprod Med. 2012;30:396–399 [DOI] [PubMed] [Google Scholar]
- 6. Speiser PW, Azziz R, Baskin LS, et al. Congenital adrenal hyperplasia due to steroid 21-hydroxylase deficiency: an Endocrine Society clinical practice guideline. J Clin Endocrinol Metab. 2010;95:4133–4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lo YM, Chan KC, Sun H, et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med. 2010;2:61ra91. [DOI] [PubMed] [Google Scholar]
- 8. Lam KW, Jiang P, Liao GJ, et al. Noninvasive prenatal diagnosis of monogenic diseases by targeted massively parallel sequencing of maternal plasma: application to β-thalassemia. Clin Chem. 2012;58:1467–1475 [DOI] [PubMed] [Google Scholar]
- 9. Costa JM, Benachi A, Gautier E, Jouannic JM, Ernault P, Dumez Y. First-trimester fetal sex determination in maternal serum using real-time PCR. Prenat Diagn. 2001;21:1070–1074 [DOI] [PubMed] [Google Scholar]
- 10. Lo YM, Lun FM, Chan KC, et al. Digital PCR for the molecular detection of fetal chromosomal aneuploidy. Proc Natl Acad Sci USA. 2007;104:13116–13121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou W, Galizia G, Lieto E, et al. Counting alleles reveals a connection between chromosome 18q loss and vascular invasion. Nat Biotechnol. 2001;19:78–81 [DOI] [PubMed] [Google Scholar]
- 12. Kauppi L, Jeffreys AJ, Keeney S. Where the crossovers are: recombination distributions in mammals. Nat Rev Genet. 2004;5:413–424 [DOI] [PubMed] [Google Scholar]
- 13. Taylan F, Altiok E. Meiotic recombinations within major histocompatibility complex of human embryos. Immunogenetics. 2012;64:839–844 [DOI] [PubMed] [Google Scholar]
- 14. Sundberg K, Bang J, Smidt-Jensen S, et al. Randomised study of risk of fetal loss related to early amniocentesis versus chorionic villus sampling. Lancet. 1997;350:697–703 [DOI] [PubMed] [Google Scholar]
- 15. Mujezinovic F, Alfirevic Z. Procedure-related complications of amniocentesis and chorionic villous sampling: a systematic review. Obstet Gynecol. 2007;110:687–694 [DOI] [PubMed] [Google Scholar]
- 16. Kollmann M, Haeusler M, Haas J, Csapo B, Lang U, Klaritsch P. Procedure-related complications after genetic amniocentesis and chorionic villus sampling. Ultraschall Med. 2013;34:345–348 [DOI] [PubMed] [Google Scholar]
- 17. Bauland CG, Smit JM, Bartelink LR, Zondervan HA, Spauwen PH. Hemangioma in the newborn: increased incidence after chorionic villus sampling. Prenat Diagn. 2010;30:913–917 [DOI] [PubMed] [Google Scholar]
- 18. Nimkarn S, New MI. Congenital adrenal hyperplasia due to 21-hydroxylase deficiency: a paradigm for prenatal diagnosis and treatment. Ann NY Acad Sci. 2010;1192:5–11 [DOI] [PubMed] [Google Scholar]
- 19. Tardy-Guidollet V, Menassa R, Costa JM, et al. New management strategy of pregnancies at risk of congenital adrenal hyperplasia using fetal sex determination in maternal serum: French cohort of 258 cases (2002–2011). J Clin Endocrinol Metab. 2014;jc20132895. [DOI] [PubMed] [Google Scholar]
- 20. White PC. Steroid 11β-hydroxylase deficiency and related disorders. In: New MI, Lekarev O, Parsa A, Yuen T, O'Malley BW, Hammer GD, eds. Genetic Steroid Disorders. London: Elsevier; 2014:71–85 [Google Scholar]
- 21. Tsui NB, Kadir RA, Chan KC, et al. Noninvasive prenatal diagnosis of hemophilia by microfluidics digital PCR analysis of maternal plasma DNA. Blood. 2011;117:3684–3691 [DOI] [PubMed] [Google Scholar]
- 22. Nasis O, Thompson S, Hong T, et al. Improvement in sensitivity of allele-specific PCR facilitates reliable noninvasive prenatal detection of cystic fibrosis. Clin Chem. 2004;50:694–701 [DOI] [PubMed] [Google Scholar]
- 23. Mistry PK, Weinthal JA, Weinreb NJ. Disease state awareness in Gaucher disease: a Q&A expert roundtable discussion. Clin Adv Hematol Oncol. 2012;10:1–16 [PubMed] [Google Scholar]