Abstract
The use of DNA sequence-based comparative genomics for evolutionary studies and for transferring information from model species to crop species has revolutionized molecular genetics and crop improvement strategies. This study compared 4485 expressed sequence tags (ESTs) that were physically mapped in wheat chromosome bins, to the public rice genome sequence data from 2251 ordered BAC/PAC clones using BLAST. A rice genome view of homologous wheat genome locations based on comparative sequence analysis revealed numerous chromosomal rearrangements that will significantly complicate the use of rice as a model for cross-species transfer of information in nonconserved regions.
Comparative genomics encompasses cross-genome comparisons of structure and function to estimate similarity of biological organization. Organismal evolution often provides threads of continuity that allow comparative biological analyses to link genes, proteins, genomes, and traits across species and genera. These relational patterns can lead to new knowledge, hypotheses, and predictions about related species. Research ranging from the whole organism to the DNA level has contributed much to our knowledge of genome structure and function due to the complementation of research among scientists from different disciplines studying different species. Comparative genomics research has several goals: (1) to compare the organization of related genomes and infer the basic processes of genome evolution, (2) to transfer information from model species to related organisms, and (3) to integrate information on gene location and expression across species. Crop improvement programs can use comparative genetics to transfer information about genes from model species to their species of interest, to help identify the genes controlling traits of interest, and to assess within-species allelic diversity so that the best alleles can be identified and assembled in superior varieties.
Comparative Mapping of Poaceae
Comparative genomics in the grass family (Poaceae) is of particular importance. The family comprises a number of economically important plants, such as rice (Oryza sativa L.), maize (Zea mays L.), wheat (Triticum aestivum L.), sorghum (Sorghum vulgare L.), barley (Hordeum vulgare L.), rye (Secale cereale L.), and others. Even though Poaceae species diverged over 65 million years ago, comparative mapping studies have indicated that there is a high level of gene order conservation at the macro level (e.g., Hulbert et al. 1990; Ahn et al. 1993; Kurata et al. 1994; Van Deynze et al. 1995a,b,c; Moore et al. 1997; Gale and Devos 1998). For the domesticated grasses, the conserved linkage blocks and their relationships with rice linkage groups have led to hypotheses about the basic organization of the ancestral grass genome (Moore et al. 1995; Gale and Devos 1998; Wilson et al. 1999) and have provided impetus for examining genome conservation in more detail. Conservation of gene content and order at the megabase level is critical for efficient utilization of model species for positional gene cloning (Tanksley et al. 1995), development of molecular markers, and for identifying the region in the model species that might contain candidate genes responsible for a trait of interest. Rice (2n = 24), having a small genome and great economic significance, was the first grass species selected for genome sequencing (Dickson and Cyranoski 2001; Goff et al. 2002; Yu et al. 2002). In contrast, wheat, a polyploid (2n = 6x, AA, BB, DD genomes), with a genome size 40 times larger than that of rice (Argumuganathan and Earle 1991), 25%–30% gene duplication (Anderson et al. 1992; Dubcovsky et al. 1996; Akhunov et al. 2003), and over 80% repeated DNA can clearly benefit from comparative genomics. Hexaploid wheat has a haploid chromosome complement composed of three related genomes, (A, B, and D), each containing seven chromosomes. Chromosomes 4, 5, and 7 are involved in a complex interchange (Naranjo et al. 1987), whereas the rest of the chromosomes in the A, B, and D genomes are largely colinear (Gale and Devos 1998).
Micro-Colinearity
Micro-colinearity has been shown to be conserved in some regions between barley (Dunford et al. 1995) or wheat (Yan et al. 2003) and rice. Investigations of the Sh2/A1 orthologous region in rice, sorghum, and maize (Bennetzen and Ramakrishna 2002), and species in the Triticeae (Li and Gill 2002) showed that the region was largely colinear, but some anomalies were observed: A tandem duplication of an a1 homolog in sorghum could not be detected by linkage mapping; there was a high degree of divergence for intergenic sequences, and intergenic distances were more than sevenfold greater in maize (Bennetzen and Ramakrishna 2002) and 4- to 195-fold greater in the Triticeae (Li and Gill 2002). Furthermore, the colinearity of these loci in wheat and barley was interrupted by intergenic breakages and segmental translocation to nonhomologous chromosomes (Li and Gill 2002). Gene composition and order were conserved in the adh1 region of maize and sorghum, but not in rice (Tikhonov et al. 1999; Tarchini et al. 2000). Duplications of loci separated by large genetic distances in different regions of the same chromosome can complicate comparative mapping, especially when polymorphism levels limit the number of fragments mapped in a given population (Chen et al. 1997). Gene duplication followed by sequence divergence and small translocations of single genes (Tarchini et al. 2000), multigene families (Dubcovsky and Dvorak 1995), the rapidly evolving nature of certain genes, such as disease resistance genes (Leister et al. 1998; Keller and Feuillet 2000), and ectopic recombination to inter- and intrachromosomal sites can all lead to rapid rearrangement of resistance-like genes and nonsyntenic distribution in cereal genomes (Leister et al. 1998).
Clearly, macro-colinearity does not always predict microcolinearity, thus complicating the use of model species for molecular breeding and genetics. Assessment of micro-colinearity requires extensive investment in phenotyping and large population mapping for fine-scale analysis. Accurate characterization of the colinearity of the rice and wheat genomes would considerably improve predictability and efficiency of information transfer.
Whole Genome Comparative Mapping by Sequence Matching
Southern hybridization using anchor probes (Van Deynze et al. 1998) has been the method of choice for evaluating relationships among species and genera and can detect genome fragments estimated to be at least 80% similar. Other methods such as PCR-based fragment amplification may be an all or none reaction (dominant), may amplify nonorthologous loci, or because of primer specificity, inadequately sample sequence variation. The utility of high-density comparative maps is readily apparent in any attempt to identify candidate genes and for marker-assisted selection (MAS). The density of comparative maps using DNA sequence matching is limited by the number of mapped ESTs and/or genomic sequences available for each of the species of interest. By manipulating sequence matching parameters, false hits and paralogs can be identified and analyzed. For those genes that have diverged to the point where it is difficult to identify orthologs using DNA sequence, predicted amino acid sequences and more sophisticated pattern matching methods can be used to search for similarities. Comparative mapping by comparative sequence analysis can be validated by using previously mapped and sequenced genes to estimate predictability in both animals (Band et al. 2000; Rebeiz and Lewin 2000) and plants (Sorrells 2000).
A U.S. National Science Foundation-funded wheat expressed sequence tag (EST) project has been studying the structure and function of the expressed portion of the wheat genome by mapping wheat unigenes to individual chromosome regions. Representative ESTs, each belonging to one of the unigenes (http://wheat.pw.usda.gov/NSF/progress_mapping.html) were used for mapping in the wheat genome utilizing 101 wheat deletion stocks, each of which contain a deletion of a defined part of a chromosome (Endo and Gill 1996), referred to as deletion mapping. As of November 2002, over 100,000 ESTs from various tissues of wheat at different stages of development have been sequenced, and 4485 wheat unigenes have been deletion mapped by this project.
The availability of rice genome DNA sequence data from multiple sources (Dickson and Cyranoski 2001; Goff et al. 2002; Yu et al. 2002) has allowed for an in-depth comparison of genes in the Poaceae and beyond. The portion of the rice genome sequence that could be accurately ordered was used to directly assess the colinearity of genes with those that have been bin-mapped in wheat. This report provides an overview of the structural relationships between the rice and wheat genomes given the present state of knowledge and available data. Data, figures, and supporting analyses for this research can be obtained from GrainGenes (http://wheat.pw.usda.gov/pubs/2003/Sorrells). The purpose of this study was to construct sequence-based comparative maps between rice and wheat using mapped wheat ESTs and rice genome sequence data. High-resolution, sequence-based maps can be used to transfer information from model species to related organisms, integrate information on gene location and expression across species, compare genome structure, and infer evolutionary processes.
METHODS
Source of Sequences
Genetic map and cDNA information was obtained from GrainGenes (http://wheat.pw.usda.gov/), RiceGenes (now Gramene; http://www.gramene.org/), MaizeDB (http://www.agron.missouri.edu/), and the Japan rice genome project (RGP; http://rgp.dna.affrc.go.jp) databases. BAC/PAC sequences available in May 2002 were downloaded from NCBI Entrez (http://www.ncbi.nlm.nih.gov). The sequence and related information of 155,726 wheat ESTs, along with 638 wheat mRNA sequences and 497 sequenced and mapped cDNA clones were downloaded from dbEST/Entrez or from the plant division of GenBank. Two local databases were designed to hold all the wheat EST and rice genomic sequence information, sequences of genetically mapped markers, and all the analysis results. A local mirror of the wEST database (http://wheat.pw.usda.gov/wEST/) contained all the wheat EST deletion mapping results. Only ESTs with known physical locations in wheat were included (Figs. 1, 2). The ESTs were selected from unigene contigs that were based on a Phrap assembly of 7929 contigs using penalty -5, minmatch 50, and minscore 100 as parameters. Because the majority of the ESTs used for generating the unigene set was from 5′ sequencing, clones from putative unigene contigs were 3′ sequenced and submitted to the Cross-Match program for identification of duplicate contigs more than 90% similar over 100 bases or more (http://wheat.pw.usda.gov/NSF/curator/assembly.html). At the time of download, the percentage of each of the rice chromosomes sequenced ranged from 15% to 123% (Table 1). The percent completion included overlapping BAC sequences, thus resulting in numbers exceeding 100%.
Figure 1.
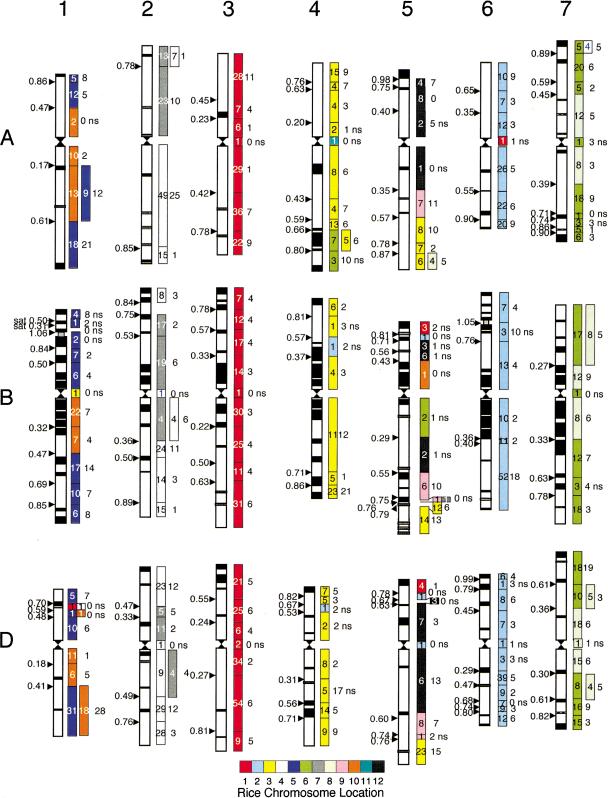
Rice–wheat genome relationships. Rice genome view showing the wheat chromosome arm location for the most similar wheat gene sequences. Each colored box represents a rice–wheat gene sequence match at ≥ 80% identity. When the wheat EST mapped to more than one wheat chromosome, the other color-coded locations are positioned adjacent to the first. Homologous wheat chromosome locations are grouped together. Rice BAC/PAC sequences that did not match any wheat sequence as well as redundant matches are omitted. The rice centromere location is indicated by `C'.
Figure 2.
Wheat–rice genome relationships. (Left) The wheat C-banding karyotype indicating the positions of deletion breakpoints. (Right) Each deletion bin is color-coded according to the rice chromosome matching a significant number (χ2 P < 0.01) of wheat ESTs mapped to that bin. The number of those matches is indicated inside the colored rectangle, and the number of matches to all other rice chromosomes is adjacent. Nonsignificant association with a rice chromosome or insufficient data is indicated by “ns” adjacent to the bin.
Table 1.
Total Number of BAC/PAC Clones Analyzed, BAC/PAC Clones With Mapped Wheat EST Sequences, Percent Sequenced for Each of the Rice Chromosomes, and Percent Short Arm Wheat ESTs Represented
|
Number of BAC/PAC clones
|
||||||
|---|---|---|---|---|---|---|
| Rice chromosome | Percent sequenced | Total | Total ordered | With mapped ESTs | With single-bin ESTs | Percent short arm wheat ESTs |
| 1 | 122 | 420 | 394 | 240 | 170 | 39 |
| 2 | 80 | 284 | 235 | 152 | 120 | 30 |
| 3 | 47 | 162 | 155 | 109 | 90 | 36 |
| 4 | 123 | 229 | 218 | 99 | 88 | 11 |
| 5 | 44 | 126 | 125 | 71 | 53 | 28 |
| 6 | 86 | 231 | 199 | 112 | 82 | 36 |
| 7 | 73 | 227 | 172 | 84 | 73 | 62 |
| 8 | 74 | 204 | 164 | 76 | 66 | 38 |
| 9 | 15 | 27 | 27 | 20 | 13 | 15 |
| 10 | 115 | 189 | 146 | 54 | 53 | 17 |
| 11 | 16 | 29 | 29 | 11 | 11 | 60 |
| 12 | 46 | 82 | 72 | 27 | 27 | 50 |
Deletion Mapping
Deletion mapping was performed by hybridizing the cDNA clone corresponding to each EST to a Southern blot of DNA from a panel of wheat genetic stocks, each missing a different terminal portion of a chromosome arm (Qi et al. 2003). Absence of a particular restriction fragment in the lane for a particular stock indicates that the locus is distal to the corresponding deletion breakpoint (Fig. 2). The regions between adjacent breakpoints are referred to as bins. The deletion mapping of 4485 ESTs representative of unigenes (http://wheat.pw.usda.gov/NSF/progress_mapping.html) in hexaploid wheat utilized 101 wheat genetic stocks with specific regions of chromatin deleted (Fig. 2; Endo and Gill 1996; Qi et al. 2003) obtained from B.S. Gill (Kansas State University) and the nulli-tetrasomic and ditelosomic aneuploids (Sears 1954; Sears and Sears 1978) obtained from the USDA-Sears collection of wheat genetic stocks (University of Missouri). These genetic stocks allowed for the assignment of fragments to specific bins delineated by the deletion breakpoints on individual chromosomes.
Ordering of Rice BAC/PAC Clones
The correct ordering of rice BAC/PACs was critical for analyzing the rice-to-wheat relationship. No single source among the members of the international rice genome sequencing effort provided an order for all of the BAC/PACs and their overlaps. To estimate the correct ordering of the clones, the following sources of information were used along with manual checking of incongruities: (1) chromosome assignment of BAC/PAC clones provided by the International Rice Genome Sequencing Project (IRGSP) consortium (http://www.tigr.org/tdb/e2k1/osa1/sequencing.shtml), (2) fingerprinting data for the BAC/PAC clones, (3) similarity to sequenced cDNA probes from the genetic map from RGP obtained from The Institute for Genomic Research (TIGR), and (4) BAC/PAC sequence overlaps based on similarity searches of the database against itself. The physical map of the rice cultivar `Nipponbare' genome based on clone fingerprinting was downloaded from the Clemson University Genomics Initiative (CUGI; http://www.genome.clemson.edu/projects/rice/fpc/; Chen et al. 2002), who provided in silico digest fingerprints for the sequenced BAC/PACs in their rice fingerprinted contig (FPC) file. This file was parsed, and the information for assignment of BAC/PAC coordinates within the contigs was entered into SQL tables. Both TIGR and RGP provided tables of sequence matches of BAC/PACs to sequences of genetically mapped cDNAs, and based on the genetic map, centimorgan locations were assigned to the BAC/PACs. This gave an approximate location to the BAC/PACs, that when combined with the ordering, within FPC contigs, provided a framework for ordering the clones. On a finer scale, the sequence overlap between BACs provided the ultimate order. NCBI MegaBLAST was used to scan all 2251 BAC/PACs against themselves, and the results were imported into SQL tables. Specific queries were made to filter true end-to-end overlaps from other types of matches among the BAC/PACs. Overlaps between BAC/PACs were used to resolve discrepancies between the other information sources, but were insufficient alone to order all of the clones. Some overlaps provided extra information for linking FPC contigs that the digest-based fingerprinting failed to join. BAC/PAC clones that were completely contained within the overlap of adjacent clones were eliminated. Applying this methodology to the available sequence data enabled the relative ordering of the available BAC/PACs into a partial tiling path for each of the rice chromosomes, though many sequence gaps remained in the unfinished chromosomes.
Sequence Comparisons
A total of 4485 deletion-mapped ESTs were compared against the sequences from 2251 rice BAC/PACs using NCBI BLAST. Wheat ESTs that were not mapped or were only located to chromosome or arm did not provide a wheat chromosome bin location and were not used in the analyses. The procedure used to filter and summarize the BLAST results was as follows: High-scoring pairs (HSPs) with an E-value greater than 1E-15 were rejected. Given that a cDNA probe sequence may match and align to several contiguous but interrupted regions in the genomic sequence, the BLAST algorithm reported individual matching regions between a probe-BAC pair as independent HSPs. The statistics, such as sum of the bit-scores, total alignment length, and percent identity in the total matched region (%ID) of all the HSPs for any given query-subject pair were calculated and summarized. The query-subject pairs with greater than 80%ID over greater than 50% of the length of the query sequence, but not less than 100 bases, were considered significant matches and used for further analysis. Significance of homology between each wheat bin and rice chromosomes was evaluated using a χ2 test where the number of ESTs with homology to a particular rice chromosome versus other chromosomes was compared to 1:11, observed:expected, for a random distribution. Bins with class sizes less than 4 were not analyzed.
Genome Comparisons
A table was constructed to display the rice BAC/PAC sequence most similar to each of the mapped wheat ESTs as well as sequenced and mapped probes. This allowed us to connect the physical and genetic maps of wheat to the genomic sequence of rice. The rice BAC/PACs with significant matches to wheat sequences are shown in order with the wheat chromosome location for the matching sequence color-coded (Fig. 1). The A, B, and D wheat genomes were used as a single consensus wheat genome for constructing Figure 1. Rice BAC/PAC clones that did not match any wheat sequence were omitted from the figure. Figure 1 was trimmed to eliminate redundant information where more than one mapped wheat EST matched the same rice BAC/PAC region without providing additional information regarding wheat chromosome location. Identical matches of any wheat sequence to overlapping regions of two or more BAC/PACs were represented as only one match. A reciprocal view (Fig. 2) was constructed with the three homologous wheat genomes as independent sources of genome location. The rice location of all sequence matches was compiled and displayed according to the deletion bin location of only single-bin (putative single copy) wheat ESTs, where single-bin/copy genes are those that were mapped to no more than one bin in each of the three homologous chromosomes in a group.
Sequence Analysis and Genome Coverage
At the time the rice genome sequence was accessed for this analysis (May 2002), approximately 60% of the rice genome had been sequenced by the public sequencing consortium (Table 1). Some numbers exceeded 100% because the percent completion included overlapping BAC sequences. Except for rice chromosomes 9 and 11, the distribution of sequenced clones along the chromosomes was fairly homogenous. Using all available wheat sequences, best-matches between 63,928 EST/mRNAs and 1828 rice BAC/PACs were compiled, where the average alignment was 386 base pairs (std. dev. 394 bp). Because of EST redundancy, some of the BAC/PACs had many wheat matches (up to 1473). Among the matched BAC/PACs, the overall mean and median were 36 and 21, respectively. Highly expressed wheat sequences included histones, translation factors, chlorophyll binding protein, heat shock proteins, ribosomal proteins, and rubisco. These filtering criteria resulted in an overall percent similarity between the wheat and rice sequences of 89.5%. More than half of the wheat EST sequences (90,619 out of 155,726) did not match any rice sequence within the specified parameters. Reasons for not identifying a matching sequence included (1) those portions of the rice genome that had not been sequenced, (2) wheat sequences may be derived from a less-conserved noncoding region, (3) short sequences may be adjacent to a masked repeat, (4) stringent filtering, and (5) rapid sequence divergence. Similarly, of the 2251 rice BAC/PAC clones, 423 (19%) did not have any significant matching wheat sequence. These BAC/PAC clones were located on all rice chromosomes.
To avoid EST redundancy, further work utilized 4485 mapped ESTs representative of unigenes. These ESTs consisted of 3358 putative single-copy (defined as those assigned to a maximum of one bin per wheat homologous chromosome group) and 1127 multiple-copy genes (25%) that were mapped on all 21 wheat chromosomes. Mapped wheat sequences identifying a rice BAC/PAC sequence at greater than 80% similarity were utilized to construct a comparative sequence map (Fig. 1). For 1247 single-bin mapped wheat ESTs, the corresponding sequences identified in rice ranged from 15 matches to 11 BAC/PACs on rice chromosome (R) R11 (with little genomic sequence available) to 249 matches to 170 BAC/PACs on R1 (Fig. 2). Map locations of wheat cDNAs were based either on linkage analyses of segregating populations or on physical location derived from deletion lines. A total of 217 cDNAs with linkage map location that matched rice genome sequence were sorted with those mapped to deletion bins, thus providing a second framework for comparing colinearity. A total of 2872 deletion-mapped wheat unigenes did not match any rice sequence and were well distributed among the wheat chromosomes in the three genomes, with a higher proportion mapping near the ends of the chromosomes.
RESULTS AND DISCUSSION
Comparative Analysis of Wheat Gene Locations in the Rice Genome
Conservation of gene identity and colinearity between wheat and rice will depend on the rate of genome/gene evolution and rearrangement in both species. Figure 1 provides an overview, from the rice genome perspective, of the genome relationships between rice and wheat at the resolution of the wheat chromosome arm. There are a number of interesting features that become apparent in this rice genome view of the homologous regions of the wheat genome (Fig. 1). The structural relationships between the genomes indicate that for most individual rice chromosomes there is a preponderance of wheat genes from one or two wheat homologous groups. For example, wheat ESTs matching sequences on rice chromosome 1 are largely from wheat chromosome group (W) W3, whereas R2 and R3 are generally related to W6 and W4. For some wheat chromosomes there is homology to two rice chromosomes. R4 and R7 are related to W2, R5 and R10 to W1, and R6 and R8 to W7. Although there are regions of gene content conservation that are apparent in all rice chromosomes, some contain regions related to more than one wheat chromosome. Rice centromere locations are shown; however, for most of the rice chromosomes, the centromere locations did not correspond well with centromere locations in wheat (data not shown). Most of these genome relationships were apparent from earlier RFLP-based comparative maps (Fig. 3; Kurata et al. 1994; Van Deynze et al. 1995a,b,c; Sarma et al. 2000). Rice homology to genes on the long arms of wheat chromosomes predominate, presumably because there are more expressed genes on long versus short arms (Table 1). For R1, R2, and R3, which are mostly related to one wheat chromosome, 39%, 30%, and 36% of the ESTs mapped to the short arms of wheat, respectively. Rice chromosomes 4 and 7 complement their relationship to W2 with 11% and 62% homology to wheat short arms, respectively. Whereas R5 and R10 were related to W1, homology to short arms was only 28% and 17%. Rice chromosomes 6 and 8 were comparable with 36% and 38% homology to wheat short arms. The limited sequence data available for R9, R11, and R12 precluded detailed analyses of their relationship to wheat.
Figure 3.
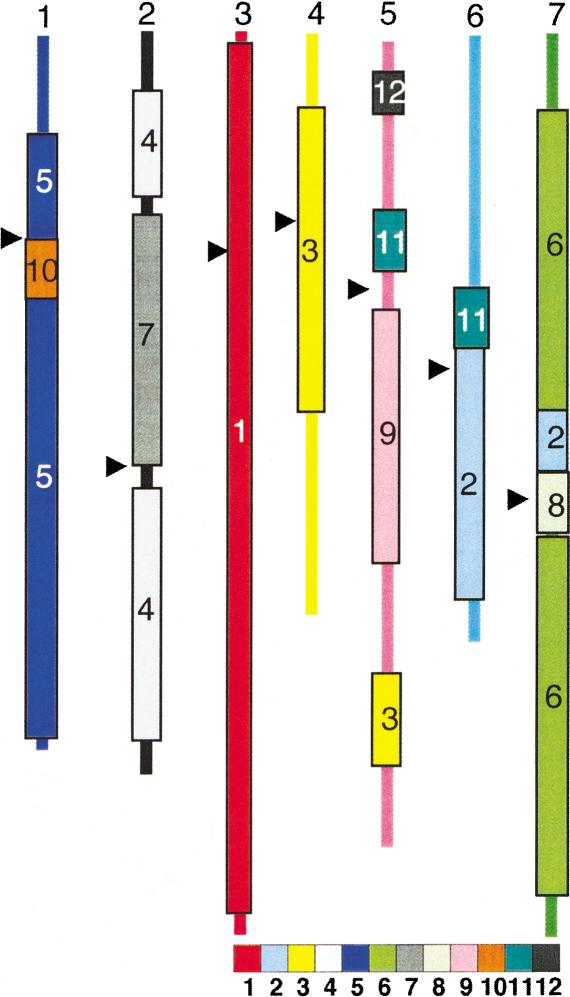
Diagrammatic representation of earlier RFLP-based comparative maps (Kurata et al. 1994; Van Deynze et al. 1995c; Sarma et al. 2000) for the rice and wheat genomes. The seven homologous wheat chromosome groups are color-coded by wheat chromosome for comparison with Fig. 1. The overlaid boxes represent the predicted homologous rice chromosome region delineated by anchor loci and are color-coded by rice chromosome for comparison with Fig. 2.
Features of the rice–wheat genome relationship revealed by this analysis compared to the RFLP-based maps include a high frequency of breakdown in colinearity throughout the genomes, and localized homology between the genomes not previously reported. Prominent features of the rice–wheat genome comparison were grouped into four categories: (A) regions of conserved gene content with one wheat genome location, (B) regions of conserved gene content with multiple wheat genome locations, (C) poorly conserved regions with one wheat genome location, and (D) poorly conserved regions with multiple wheat genome locations. Category A regions are prominent in all the rice chromosomes, whereas category B regions are less common and much more localized. This may be due to a bias in mapping multicopy genes, which could be an artifact of the similarity criteria used or a product of evolution. Category C is more common with notable examples in the centromeric regions of R1, R2, and R3 as well as the long arms of R2 and R8. In category D, wheat ESTs with multiple wheat genome locations are associated with some of the poorly conserved regions of similarity between rice and wheat. These regions appear to be widespread and are especially apparent in the short arms of R3, R6, the long arms of R3, R4, and R10, and the centromeric region of R5. Both arms of R3 and the short arm of R6 have partial homology to genes in the wheat chromosome region involved in the 4AL, 5AL, 7BS ancestral translocation, which artificially increases the number of wheat genome locations. Some regions may be associated with the gradients of recombination rates along chromosome arms that were suggested to promote more rapid rates of transcriptome evolution in distal, high-recombination regions than in proximal, low-recombination regions (Akhunov et al. 2003). The physical location of these nonconserved, multicopy regions in wheat were not consistent across the rice chromosomes; however, a future comparison to physical locations of regions of high recombination or gene density in the rice genome may reveal an association.
These comparisons and interpretations assume the availability of a complete rice genome sequence with correctly ordered BAC/PAC clones. The BAC/PAC orders used for these analyses were derived from the RGP Web site as well as FPC, BAC end sequence, and linkage data. Even with additional rice genome sequence, relative order of (internally consistent) islands of contigous BAC/PACs should not change but in fact, may become connected to each other. Also, because the wheat chromosome bin assignments required polymorphism for homoalleles among the three genomes and among multiple copies within a genome, an estimated 24% of the fragments could not be mapped using these deletion lines and the single restriction enzyme. This combined with technical problems in scoring all bands in all lanes will lead to an underestimation of the number of loci in the wheat genome and an overestimate of gene content conservation between wheat and rice. Bin location estimates gene content within a region but not the degree of colinearity within a chromosome deletion bin. Using the rice genome sequence as a template, one can predict the order of genes within bins in the wheat genome; however, microsynteny studies (Han et al. 1999; Bennetzen and Ramakrishna 2002) suggested that, in most cases, colinearity will need to be verified at the DNA sequence level before committing major resources. The ordering of the mapped wheat ESTs within deletion bins would be a desirable future enhancement for wheat–rice comparative analyses.
The identification and mapping of additional unique wheat ESTs, their relative order, and a complete ordering of the entire rice genome sequence is required in order to provide a more accurate estimate of both gene content and colinearity. Thus, our present ability to identify paralogous genes is limited by the proportion of the rice genome sequenced and ordered. This is because we use the best match from the BLAST analysis, and sequence of the ortholog may be missing. With more of the rice genome sequence, the best rice sequence matches for wheat ESTs may improve the colinearity; however, this analysis indicates that the genome relationships are more complex than previously thought.
To improve the coverage of wheat sequences on the rice genome, wheat ESTs matching rice genome sequences in unpopulated regions will need to be selectively mapped in wheat or associated to an existing EST unigene with a mapped representative. In the present study, ESTs from 37% of the wheat unigenes matched at least one rice sequence at the specified parameters, and 81% of the rice BAC/PAC clones were matched by a wheat EST. The completion of the ordered rice genome sequence will not result in matching sequences for all wheat genes using the specified parameters. Orthologs may have been deleted or evolved more rapidly than a paralog. Rapidly evolving genes, such as disease resistance genes (Leister et al. 1998), will likely be among those not matched. A comparison of genomic regions for average gene similarity may reveal regions showing nonrandom rates of gene evolution associated with features such as heterochromatin or recombination hot spots, all of which should be taken into account for an accurate comparative map.
Comparative Analysis of Rice Gene Locations in the Wheat Genome
A reciprocal view of Figure 1 shows the relationship between the wheat and rice genomes revealing the conservation of gene content and order at the resolution conferred by the chromosome deletions in the wheat genome (Fig. 2). Deletion lines used for mapping in this project provided five to 12 physical locations per chromosome, but within deletion bins, gene order cannot be ascertained directly. Comparisons of gene complement across the three genomes will not be accurate, because the size of the deletion bins varied among the homologs and because such comparisons are sensitive to mapping errors. Only single-bin genes (maximum of three loci mapped in a single homologous chromosome group of wheat) are shown in Figure 2; thus, many more rice chromosomes will contain homologous sequences for genes mapped to these deletion bins when the multiple-copy genes are included. The relative order of genes within bins could be inferred from a comparison to the rice genome sequence but would require verification in wheat using other methods.
In Figure 2, the rice chromosome(s) with the largest number of homologous sequences for ESTs in each wheat bin are color-coded, and the number of ESTs matching sequences on other rice chromosomes is shown adjacent to the bin. Although the previous chromosome relationships (Fig. 1) are apparent, in this figure, the heterogeneity in gene content and homology along the chromosomes reveal the complexity of evolutionary divergence between wheat and rice. It is clear that patterns of conservation differ within and among chromosomes. The long arm of W2 is more closely related to R4, whereas the short arm of W2 is related to R7. Wheat 7 is related to R6 and R8 but the conservation patterns are quite different. Wheat 7 genes from both arms are homologous to sequences on both R6 and R8, but R8 is largely centromeric whereas R6 is distal. A similar pattern was observed for W1 with R10 being proximal and R5 distal. Wheat group 5 appeared to be the least conserved, with genes scattered across all rice chromosomes represented; however, some regions of homology to R3, R9, and R12 are evident across all three homologs. Because little sequence was available from R9 and R11, more wheat ESTs mapping to W5 will probably match those two rice chromosomes with additional sequence data. A similar relationship between W5 and rice based on RFLP was previously reported (Sarma et al. 2000). Genes mapped to wheat group 4 are mostly related to R3, although in this analysis, the ancestral 4AL, 5AL, 7BS translocation in wheat shows up on 4AL as being related to R6 and R3. Wheat groups 3 and 6 showed the best conservation of gene order and content with highly significant associations to R1 and R2, respectively. Over all wheat chromosomes, even in the most conserved regions, deletion bins containing only sequences from one rice chromosome are rare, and they tend to be in the centromeric region, where few genes have been mapped in wheat so far. These data suggest that during cereal evolution there has been an abundance of rearrangements, insertions, deletions, and duplications that will complicate the utilization of many regions of the rice genome for cross-species transfer of information. This disruption in the colinearity of genes will greatly complicate map-based cloning and the selection of linked markers. The concentration of mapped genes in many distal deletion bins of wheat chromosomes concurs with previous investigations showing that gene-rich regions are predominantly near the ends of many wheat chromosomes (for review, see Moore 2000). However, this assumes that genes in all regions are mapped in equal proportions. Previous sequencing studies have reported gene densities of about one gene every 40 kb in wheat or wheat relatives (Keller and Feuillet 2000; San Miguel et al. 2002). However, the observed gene densities in wheat and barley are still greater than the expected average gene density of one gene per 220 kb (Lagudah et al. 2001), as predicted based on cDNA mapping of wheat deletion lines (Gill et al. 1996; Faris et al. 2000). These results suggest that most Triticeae genes might be associated with gene-rich regions. Gene-rich regions are prime candidates for large-scale wheat genomic sequencing projects that will likely be necessary for fully exploiting the information in the wheat genome.
Comparisons to Previous Comparative Maps
Previous comparative maps between wheat and rice (Ahn et al. 1993; Kurata et al. 1994; Van Deynze et al. 1995a,b,c; Devos and Gale 1997) were based largely on linkage analysis and utilized anchor probes (Van Deynze et al. 1998) mapped to multiple species. Such maps have been necessarily low-resolution and imprecise due to the small number of common markers. A diagrammatic representation of a wheat–rice comparative map based on Southern analysis (Fig. 3) illustrates the general chromosome relationships for wheat and rice reported earlier (Kurata et al. 1994; Van Deynze et al. 1995a,b,c; Sarma et al. 2000). There are striking similarities and differences between existing low-resolution comparative maps based on RFLPs and the DNA sequence-based comparative map described above (Figs. 1,2). The primary differences are the number of points for comparison and the precision imparted by the DNA sequence analyses. When comparing wheat–rice relationships in Figure 3 to the deletion bin map in Figure 2, it is important to remember that linkage distances will differ substantially from physical distances. Rice chromosome 1 was the most complete in terms of sequence available and ordering of the BAC/PAC clones. As predicted by earlier maps, this chromosome appeared to be the most conserved relative to wheat and was composed largely of genes that are homologous to W3. Previous maps did not reveal the high level of conservation between W6 and R2, nor the complexity of the relationships between W5 and rice chromosomes.
The large number of wheat genes within bins that had homology to several rice chromosomes is contradictory to RFLP-based comparative maps that indicated a high degree of conservation. In a review of comparative mapping studies, Gaut (2002) re-analyzed published comparative map data and calculated a statistic called “synteny probability” that is based on the odds that moving in either direction along the chromosome, adjacent markers will be colinear. Using this statistic, estimates of the conservation of gene order among grass genomes were much lower than reported earlier (Van Deynze et al. 1995a,b,c; Devos et al. 1998; Wilson et al. 1999). Gaut concluded that grass genomes are evolutionarily labile and less conserved that previously thought. This was supported by recent comparisons of the rice subspecies genomic sequences for indica and japonica (Feng et al. 2002) as well as genomic sequences from two distantly related maize inbred lines (Fu and Dooner 2002). The high frequency of insertions and deletions reported for rice and maize are consistent with the results of the present study, and suggests that these genomes are more fluid at the DNA sequence level than indicated by Southern analyses.
These results can be extended to other members of the Poaceae by comparing cDNA clones that have been mapped and sequenced in those species. One approach has been illustrated using a concentric circle diagram that arranges various grass species by genome size (Gale and Devos 1998). If common gene sequences are mapped in each of the species, the genomes can be cross-referenced at those points for general comparisons. The concentric circles highlight the genome size differences and facilitate multiple species comparisons; however, the utility of this representation is limited for complex genome relationships. Further extension of these results to other species would require ESTs for sequence comparisons with rice and wheat. The enhanced resolution afforded by comparative sequence analyses facilitates cross-referencing of various kinds of information such as quantitative trait loci, mutants, and gene expression. The greater resolution of the comparative DNA sequence-based analysis is most critical for transferring information about the location of specific genes, whereas the greater precision for comparing gene structure and function will greatly benefit evolutionary studies. All species linked in such a way contribute unique and valuable information, because of the wide range of variation in adaptation and evolution as well as the collective intellectual contributions from more scientists bridging many disciplines.
Conclusions
The comparative sequence analysis described herein substantiates much of the gene content and order in earlier comparative maps but at a much finer resolution. However, the increased resolution afforded by sequence analysis of 4485 mapped wheat unigenes revealed numerous discontinuities in gene order between wheat and rice that will complicate any transfer of information and markers between these species. Resolution of sequence similarity among species, genomes, and paralogs is variable among different genes due to evolutionary pressures as well as their respective physical genome location. A completely ordered rice genome sequence and additional analyses are required to resolve orthology/paralogy, rearrangements, and duplications between the wheat and rice genomes. Our results support the view that grass genomes are labile, rapidly evolving entities and that structural and functional relationships are complex. These sequence-based maps will facilitate the use of rice for locating genes of interest in wheat; however, most applications will require extensive cross-species mapping, sequencing, and analysis at the BAC level.
Acknowledgments
This publication is based upon work supported by the National Science Foundation under Cooperative Agreement No. DBI-9975989 and USDA/NRI project No. 2001-35301-10612.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1113003.
Footnotes
[Supplemental material is available online at www.genome.org and also at the GrainGenes Web site: http://wheat.pw.usda.gov/pubs/2003/Sorrells/.]
References
- Ahn, S., Anderson, J.A., Sorrells, M.E., and Tanksley, S.D. 1993. Homoeologous relationships of rice, wheat and maize chromosomes. Mol. Gen. Genet. 241: 483-490. [DOI] [PubMed] [Google Scholar]
- Akhunov, E.D., Goodyear, J.A., Geng, S., Qi, L., Echalier, B., Gill, B.S., Lazo, G., Chao, S., Anderson, O.D., Linkiewicz, A.M., et al. 2003. The organization and rate of evolution of the wheat genomes are correlated with recombination rates along chromosome arms. Genome Res. 5: 753-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J.A., Ogihara, Y., Sorrells, M.E., and Tanksley, S.D. 1992. Development of a chromosomal arm map for wheat based on RFLP markers. Theor. Appl. Genet. 83: 1035-1043. [DOI] [PubMed] [Google Scholar]
- Argumuganathan, K. and Earle, E.D. 1991. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9: 208. [Google Scholar]
- Band, M.R., Larson, J.H., Rebeiz, M., Green, C.A., Heyen, D.W., Donovan, J., Windish, R., Steining, C., Mahyuddin, P., Womack, J.E., et al. 2000. An ordered comparative map of the cattle and human genomes. Genome Res. 10: 1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J.L. and Ramakrishna, W. 2002. Numerous small rearrangements of gene content, order, and orientation differentiate grass genomes. Plant Mol. Biol. 48: 821-827. [DOI] [PubMed] [Google Scholar]
- Chen, M., SanMiguel, P., DeOliveira, A.C., Woo, S.S., Zhang, H., Wing, R.A., and Bennetzen, J.L. 1997. Microcolinearity in sh2-homologous regions of the maize, rice, and sorghum genomes. Proc. Natl. Acad. Sci. 94: 3431-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M., Presting, G., Barbazuk, W.B., Goicoechea, J.L., Blackmon, B., Fang, G., Kim, H., Frisch, D., Yu, Y., Sun, S., et al. 2002. An integrated physical and genetic map of the rice genome. The Plant Cell 14: 521-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos, K.M. and Gale, M.D. 1997. Comparative genetics in the grasses. Plant Mol. Biol. 35: 3-15. [PubMed] [Google Scholar]
- Devos, K.M., Wang, Z.M., Beales, J., Sasaki, Y., and Gale, M.D. 1998. Comparative genetic maps of foxtail millet (Setaria italica) and rice (Oryza sativa). Theor. Appl. Genet. 96: 63-68. [Google Scholar]
- Dickson, D. and Cyranoski, D. 2001. Commercial sector scores success with whole rice genome. Nature 409: 551. [DOI] [PubMed] [Google Scholar]
- Dubcovsky, J. and Dvorak, J. 1995. Ribosomal RNA loci: Nomads in the Triticeae genomes. Genetics 140: 1367-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky, J., Luo, M.-C., Zhong, G.-Y., Bransteiter, R., Desai, A., Kilian, A., Kleinhofs, A., and Dvorak, J. 1996. Genetic map of diploid wheat, Triticum monococcum L., and its comparison with maps of Hordeum vulgare L. Genetics 143: 983-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunford, R.P., Kurata, N., Laurie, D.A,. Money, T.A., Minobe, Y., and Moore, G. 1995. Conservation of fine-scale DNA marker order in the genomes of rice and the Triticeae. Nucleic Acids Res. 23: 2724-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, T.R. and Gill, B.S. 1996. The deletion stocks of common wheat. J. Hered. 87: 295-307. [Google Scholar]
- Faris, J.D., Haen, K.M., and Gill, B.S. 2000. Saturation mapping of a gene-rich recombination hot spot region in wheat. Genetics 154: 823-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Q., Zhang, Y., Hao, P., Wang, S., Fu, G., Huang, Y., Li, Y., Zhu, J., Liu, Y., Hu, X. et al. 2002. Sequence analysis of rice chromosome 4. Nature 420: 316-320. [DOI] [PubMed] [Google Scholar]
- Fu, H. and Dooner, H.K. 2002. Intraspecific violation of colinearity and its implications in maize. Proc. Natl. Acad. Sci. 99: 9573-9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, M.D. and Devos, K.M. 1998. Comparative genetics in the grasses. Proc. Natl. Acad. Sci. 95: 1971-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S. 2002. Evolutionary dynamics of grass genomes. New Phytol. 154: 15-28. [Google Scholar]
- Gill, K.S., Gill, B.S., Endo, T.R., and Boyko, E.V. 1996. Identification and high-density mapping of gene-rich regions in chromosome Group 5 of wheat. Genetics 143: 1001-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff, S.A., Ricke, D., Lan, T.-H., Presting, G., Wang, R., Dunn, M., Glazebrook, J., Sessions, A., Oeller, P., Varma, H., et al. 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92-100. [DOI] [PubMed] [Google Scholar]
- Han, F., Kilian, A., Chao, J.P., Kudrna, D., Steffenson, B., Yamamoto, K., Matsumoto, T., Sasaki, T., and Kleinhofs, A. 1999. Sequence analysis of a rice BAC covering the syntenous barley Rpg1 region. Genome 42: 1071-1076. [DOI] [PubMed] [Google Scholar]
- Hulbert, S.H., Richter, T.E., Axtell, J.D., and Bennetzen, J.L. 1990. Genetic mapping and characterization of sorghum and related crops by means of maize DNA probes. Proc. Natl. Acad. Sci. 87: 4251-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, B. and Feuillet, C. 2000. Colinearity and gene density in grass genomes. Trends Plant Sci. 5: 246-251. [DOI] [PubMed] [Google Scholar]
- Kurata, N., Moore, G., Nagamura, Y., Foote, T., Yano, M., Minobe, Y., and Gale, M. 1994. Conservation of genomic structure between rice and wheat. Bio. Technol. 12: 276-278. [Google Scholar]
- Lagudah, E., Dubcovsky, J., and Powell, W. 2001. Wheat genomics. Plant Physiol. Biochem 39: 335-344. [Google Scholar]
- Leister, D., Kurth, J., Laurie, D.A., Yano, M., Sasaki, T., Devos, K., Graner, A., and Schulze-Lefert, P. 1998. Rapid reorganization of resistance gene homologues in cereal genomes. Proc. Natl. Acad. Sci. 95: 370-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. and Gill, B.S. 2002. The colinearity of the Sh2/A1 orthologous region in rice sorghum and maize is interrupted and accompanied by genome expansion in the Triticeae. Genetics 160: 1153-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, G. 2000. Cereal chromosome structure, evolution, and pairing. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51: 195-222. [DOI] [PubMed] [Google Scholar]
- Moore, G., Devos, K.M., Wang, Z., and Gale, M.D. 1995. Grasses, line up and form a circle. Curr. Biol. 5: 737. [DOI] [PubMed] [Google Scholar]
- Moore, G., Roberts, M., Aragon-Alcaide, L., and Foote, T. 1997. Centromeric sites and cereal chromosome evolution. Chromosoma 35: 17-23. [DOI] [PubMed] [Google Scholar]
- Naranjo, T., Roca, P., Goicoechea, P.G., and Giraldez, R. 1987. Arm homology of wheat and rye chromosomes. Genome 29: 873-882. [Google Scholar]
- Qi, L., Echalier, B., Friebe, B., and Gill, B.S. 2003. Molecular characterization of a set of wheat deletion stocks for use in chromosome bin mapping of ESTs. Funct. Integr. Genomics. 3: 39-55. [DOI] [PubMed] [Google Scholar]
- Rebeiz, M. and Lewin, H.A. 2000. COMPASS of 47,787 cattle ESTs. Anim. Biotechnol. 11: 75-241. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P., Ramakrishna, W., Bennetzen, J.L., Busso, C.S., and Dubcovsky, J. 2002. Transposable elements, genes and recombination in a 215-kb contig from wheat chromosome 5A. Funct. Integr. Genomics 2: 70-80. [DOI] [PubMed] [Google Scholar]
- Sarma, R.N., Fish, L., Gill, B.S., and Snape, J.W. 2000. Physical characterization of the homologous Group 5 chromosomes of wheat in terms of rice linkage blocks, and physical mapping of some important genes. Genome 43: 191-198. [PubMed] [Google Scholar]
- Sears, E.R. 1954. The aneuploids of common wheat. Missouri Agricultural Experiment Station Research Bulletin No. 572.
- Sears, E.R. and Sears, L.M. 1978. The telocentric chromosomes of common wheat. Proc. 5th Int. Wheat Genet. Symp., New Delhi 389-407.
- Sorrells, M.E. 2000. The evolution of comparative plant genetics. In Genomes. Proc. 22nd Stadler Symp., June 6–8, 1998, Columbia, MO (ed. J.P. Gustafson), pp. 183-195. Kluwer Academic Publishers, MA.
- Tanksley, S.D., Ganal, M.W., and Martin, G.B. 1995. Chromosome landing: A paradigm for map-based gene cloning in plants with large genomes. Trends Genet. 11: 63-68. [DOI] [PubMed] [Google Scholar]
- Tarchini, R., Biddle, P., Wineland, R., Tingey, S., and Rafalski, A. 2000. The complete sequence of 340 kb of DNA around the rice Adh1-Adh2 region reveals interrupted colinearity with maize chromosome 4. The Plant Cell 12: 381-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov, A.P., SanMiguel, P.J., Nakajima, Y., Gorenstein, N.M., Bennetzen, J.L., and Avramova, Z. 1999. Colinearity and its exceptions in orthologous adh regions of maize and sorghum. Proc. Natl. Acad. Sci. 96: 7409-7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDeynze, A.E., Dubcovsky, J., Gill, K.S., Nelson, J.C., Sorrells, M.E., Dvorak, J., Gill, B.S., Lagudah, E.S., McCouch, S.R., and Appels, R. 1995a. Molecular-genetic maps for group 1 chromosomes of Triticeae species and their relation to chromosomes in rice and oat. Genome 38: 45-59. [DOI] [PubMed] [Google Scholar]
- VanDeynze, A.E., Nelson, J.C., O'Donoughue, J.S., Ahn, S.N., Siripoonwiwat, W., Harrington, S.E., Ylesias, E.S., Braga, D.P., McCouch, S.R., and Sorrells, M.E. 1995b. Comparative mapping in grasses. Oat relationships. Mol. Gen. Genet. 249: 349-356. [DOI] [PubMed] [Google Scholar]
- VanDeynze, A.E., Nelson, J.C., Yglesias, E.S., Harrington, S.E., Braga, D.P., McCouch, S.R., and Sorrells, M.E. 1995c. Comparative mapping in grasses. Wheat relationships. Mol. Gen. Genet. 248: 744-754. [DOI] [PubMed] [Google Scholar]
- VanDeynze, A.E., Sorrells, M.E., Park, W.D., Ayres, N.M., Fu, H., Cartinhour, S.W., Paul, E., and McCouch, S.R. 1998. Anchor probes for comparative mapping of grass genera. Theor. Appl. Genet. 97: 356-369. [Google Scholar]
- Wilson, W.A., Harrington, S.E., Woodman, W.L., Lee, M., Sorrells, M.E., and McCouch, S.R. 1999. Can we infer the genome structure of progenitor maize through comparative analysis of rice, maize, and the domesticated panicoids? Genetics 153: 453-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, L., Loukoianov, A., Tranquilli, G., Helguera, M., Fahima, T., and Dubcovsky, J. 2003. Positional cloning of wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. 100: 6263-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., Hu, S., Wang, J., Wong, G.K.-S., Li, S., Liu, B., Deng, Y., Dai, L., Zhou, Y., Zhang, X., et al. 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79-92. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://wheat.pw.usda.gov/pubs/2003/Sorrells/; Supplementary materials for this publication.
- http://wheat.pw.usda.gov/NSF/progress_mapping.html; U.S. National Science Foundation Wheat Genome Mapping Progress.
- http://wheat.pw.usda.gov/; GrainGenes home page.
- http://wheat.pw.usda.gov/NSF/curator/assembly.html; NSF Wheat Genome Project—Description of unigene assembly protocol.
- http://www.gramene.org/; Gramene home page.
- http://www.agron.missouri.edu/; MaizeDB home page.
- http://rgp.dna.affrc.go.jp; Japan Rice Genome Project home page.
- http://www.ncbi.nlm.nih.gov; NCBI Entrez home page.
- http://wheat.pw.usda.gov/wEST/; GrainGenes Wheat EST database.
- http://www.tigr.org/tdb/e2k1/osa1/sequencing.shtml; TIGR Web site for the International Rice Genome Sequencing Project Consortium.
- http://www.genome.clemson.edu/projects/rice/fpc/; Clemson University Genomics Initiative Web site for rice BAC fingerprinting data.