Abstract
Developing ordered gene maps from multiple mammalian species coupled with chromosome-painting data provide a powerful resource for resolving the evolutionary history of chromosomes and whole genomes. In this work, we recapitulate the evolutionary history of human chromosome 1 and its homologs in placental mammals, putatively the largest physical unit in the ancestral placental genome. Precise definition of translocation exchange breakpoints in human, carnivore, cetartiodactyl, and rodent-ordered gene maps demonstrate that chromosome breakpoints, previously considered as equivalent, actually represent distinct chromosome positions and exchange events. Multidirectional chromosome painting, using probes from homologs to chromosome 1 in seven mammal species from six orders of placental mammals, confirm the gene-mapping results and indicate that the multiple human chromosome 1 homologs in these species are derived from independent fissions of a single ancestral chromosome. Chromosome painting using human chromosome 1 probes identifies a single human chromosome 1 homolog in phylogenetically distant taxa, the two-toed sloth, cetaceans, and higher primates. The diverse phylogenetic occurrence of a single Hsa1 synteny among the major clades of placental mammals suggests that human chromosome 1 represents an intact ancestral chromosome, which was variously fissioned in the majority of placental species. We find that the number of human chromosome 1 fissions in a specific lineage reflects its general rate of genomic evolution. Further, historic chromosome exchange appears to have been disproportionately clustered in two breakpoint hotspots on the long arm.
Human chromosome 1 (Hsa1) represents the largest physical unit in the human genome, comprising some 285 million base pairs—nearly 9% of the entire genome (Lander et al. 2001, Venter et al. 2001). The genomic synteny of this chromosome is entirely conserved in apes, in some Old World monkeys (O'Brien and Stanyon 1999; Wienberg et al. 2000), and in cetaceans (Bielec et al. 1998). In the majority of other placental mammal species examined at the genomic level, Hsa1 is found as two unordered conserved segments (CSUs) in species with slower rates of genomic evolution, to as many as six CSUs in the more derived karyotypes defined by chromosome painting, such as the spider monkey and dog (Glas et al. 1998; Burt et al. 1999; O'Brien and Stanyon 1999; O'Brien et al. 1999; Breen et al. 1999; Yang et al. 2000). Because previous comparative chromosome painting results showed that Hsa1 was divided into at least two segments in nearly all species tested (>40 placental mammals from 11 orders; Wienberg et al. 2000; Murphy et al. 2001c), it was hypothesized that human chromosome 1 was originally composed of two fragments. One segment corresponds roughly to 1q21.3-pter, whereas the second segment is comprised of the remainder of the long arm (1q21.3-qter) (Chowdhary et al. 1998; Wienberg et al. 2000; Murphy et al. 2001c). Reciprocal painting of nonhuman, chromosome 1 homologs suggested a common evolutionary origin of the apportioned Hsa1 chromosomal segments in different orders. The boundary of the segments mapped repeatedly somewhere in the region 1q21–1q23 (pig: Goureau et al. 1996; cat, primate: Wienberg et al. 1997; rabbit: Korstanje et al. 1999; tree shrew: Müller at al. 1999; Fig. 1). The small differences between reports were discounted as within the margin of error when assigning hybridization signals between distantly related species (Wienberg et al. 2000).
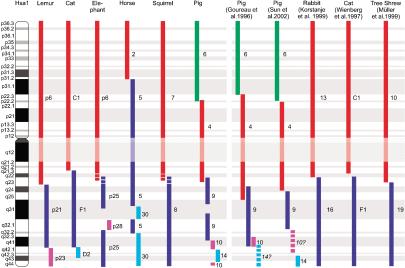
Figure 1.
Ideogram depicting the hybridization signals of chromosome sorts from eight placental mammal species (cat, elephant, horse, squirrel, pig, rabbit, ring tailed lemur, and tree shrew) onto human chromosome 1 (G-banded ideogram labeled at left). Colored blocks below each species name delineate the extent of hybridization observed for the chromosome paint probe on human chromosome metaphase spreads. The six species columns at left display new data, whereas the five species at right are taken from the literature as indicated. The numbers to the right of each colored block refer to the chromosomal origin of each probe from the examined species. For the lemur and elephant probes, the corresponding numbers of the flow karyotype peaks are given. The absence of hybridization signals in the heterochromatic region (Hsa1p11–Hsalq12) is indicated by the shaded bar.
If the two-segment ancestor hypothesis is true, then these would have fused independently in the evolutionary lineages leading to primates and cetaceans, in which a single conserved segment homologous to Hsa1 is present (Bielec et al. 1998). The alternate hypothesis would be that Hsa1 was intact in the ancestral genome of extant placental mammals. If so, then Hsa1 has been fragmented independently in the same chromosomal region in different mammalian lineages, but cannot be distinguished with the existing reciprocal chromosome-painting data. To test these competing hypotheses, we examined homologs to Hsa1 in species from nine orders of placental mammals. First, we refined the cat–human chromosome 1 comparative map and then compared these results with ordered Hsa1 comparative chromosome maps in cow, mouse, and rat species with sufficient gene density to demonstrate close, but nonidentical breakpoints. Reciprocal and multidirectional chromosome painting was performed among index species representing nine orders of placental mammals and interpreted to infer the likely ancestor of chromosome homologs of human chromosome 1. This interpretation takes into account the recent phylogenetic resolution of the 18 orders of placental mammals into four major superordinal clades (Madsen et al. 2001; Murphy et al. 2001a,b). Our results strongly suggest that the ancestral placental mammal karyotype included a single intact chromosome homologous to Hsa1. This chromosome then underwent frequent independent fission events throughout mammalian evolutionary history, but has also remained intact in some species with slower rates of genome evolution. Rates of chromosomal evolution across Hsa1 indicate disparate histories of genomic evolution, with the p arm being relatively conserved in gene order, whereas the q arm has been subject to a higher rate of inter- and intrachromosomal disruption.
RESULTS AND DISCUSSION
Comparative Gene Mapping
Radiation hybrid mapping of additional feline Type 1 gene markers assigned loci to intervals on feline chromosomes C1, D2, or F1, predicted a priori based on previously identified CSOs (Murphy et al. 2000), and further refine the F1-C1 syntenic boundary to the vicinity of 1q21 in the human genome (Fig. 2; see Supplemental Material available online at www.genome.org). RH mapping of LGALS8 and CHRM3, both from Hsa1q42–Hsa1q43, to feline chromosome D2, confirms the presence of a small fragment of Hsa1q in association with Hsa10q on cat chromosome D2 identified previously (Murphy et al. 2000; Yang et al. 2000). This small syntenic segment has also been observed in another carnivore, the dog (Yang et al. 2000; Breen et al. 1999), and in cetartiodactyls, represented by the cow (Band et al. 2000) and the pig (Rettenberger et al. 1995; Frönicke et al. 1996). It is likely that due to the small size of the Hsa1 fragment, this association may have gone undetected in other carnivores and cetartiodactyls examined in previous chromosome-painting studies using human probes.
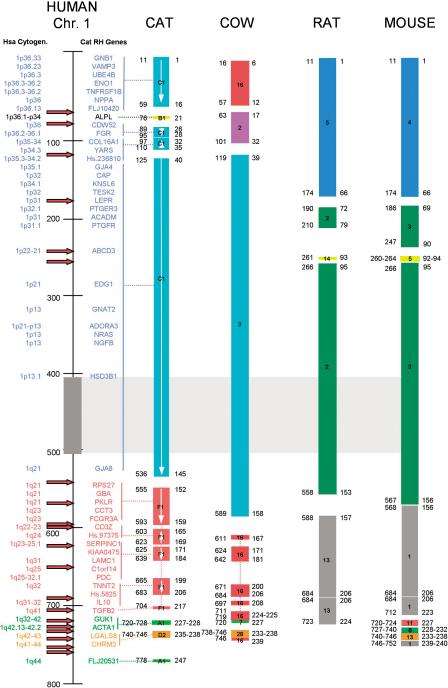
Figure 2.
Comparison of cat-ordered conserved segments with published data from cow, mouse, and rat genomes, shown with respect to human chromosome 1 (Hsa1). See Methods for published sources of cat, rat, and mouse gene order data. The genes mapped in the feline genome are marked for comparison, and groups linked to their respective conserved segment indicated by a line. The human cytogenetic position is shown to the far left. The scale to the right of the human cytogenetic position refers to centiray3000 positions on the human Genbridge-4-based RH map. Each colored block represents an ordered conserved segment (CSO) with the human genome, with the internal number referring to the chromosomal origin of the CSO within the genome of each species. White arrows within each feline CSO indicate orientation (p to q) of the human segment within the cat genome. The gray block and hatched box extending to the right, from roughly 410–510 centirays on the human map, indicates the position of the Hsa1 heterochromatic block. The number on the left of the ends of each block refers to the human centiray3000 position of the most distal gene marker in that conserved segment, whereas the number on the right refers to the physical position (in Mb) in the NCBI assembly of the human genome (Build 30). At left of the human RH map, red arrows indicate unique breakpoints relative to the human genome. Breaks were enumerated such that apparent shared breakpoints across species were counted only once. Note the increased density of rearrangement breakpoints on the distal portion of the long arm of Hsa1.
Alignment of ordered RH gene mapping-based comparative maps representing three orders of mammals (Carnivora, Cetartiodactyla, and Rodentia), revealed 12, 11, 6, and 10 CSOs relative to the human chromosome 1, respectively (Fig. 2). One breakpoint position, 536–611 cR3000 (145–167 Mb in the NCBI sequence Build 30) corresponding to Hsa1q21–Hsa1q22, appeared to be the same by cytogenetic resolution (Chowdhary et al. 1998; Wienberg et al. 2000). However, the gene map alignments show that the breakpoints are not identical, rather, they contain distinct nonoverlapping junction coordinates separated by different gene markers. For example, the cat junction is homologous to 536 cR3000 (GJA8-Fca chromosome C1)-556 cR3000 (RPS27: Fca chromosome F1); the cow junction is 589 cR3000 (TAGLN2: chromosome Bta3)-611 cR3000 (SELE/SELL: chromosome Bta16); the mouse junction is 567 cR3000 (Cd1d1: Mmu3)-568 cR3000 (Spna1: chromosome Mmu1), overlapping the rat junction between 558 cR3000 (Rn.9148: Rno2)-588 cR3000 (Rn.10274: chromosome Rno13). Evidence of different junctions are also consistent when comparisons are based upon the human sequence assembly (Fig. 2). Although the rat gene maps are less dense in this region, it is likely that the rat and mouse break-points in this region are identical, given the strong one-to-one correspondence between mouse–rat CSOs homologous to Hsa1 (Fig. 2) and the relatively close evolutionary relationship between these two species. The distinct physical locations of the evolutionary breakpoints among the three mammalian orders (Carnivora, Cetartiodactyla, and Rodentia) would be consistent with independent origins for the breakpoints. The simplest interpretation of these data place an intact Hsa1 as the ancestral form, rearranged by independent breaks in carnivores, rodents, and cetartiodactyls, but not humans.
Chromosome Painting
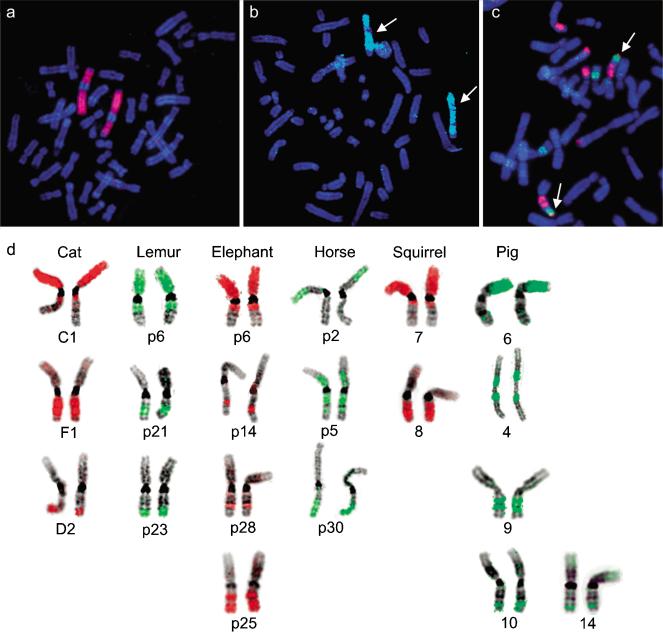
Zoo-FISH experiments hybridizing the Hsa1 paint to Sei whale chromosomes delineated a single homologous chromosome (Fig. 3a), consistent with previous Zoo-FISH characterization of the dolphin genome (Bielec et al. 1998) and strong karyotypic conservation within cetaceans (Arnason 1972). A single Hsa1 homolog is similarly found in the two-toed sloth from the order Xenarthra (Fig. 3b), shown recently to be an early divergent superordinal clade among placental mammals (Murphy et al. 2001b,c), supporting the hypothesis of a primitive intact Hsa1 chromosome in the ancestral placental mammal karyotype.
Figure 3.
Comparative chromosome-painting signals from human and placental mammal painting probes labeled with biotin (green signals detected with Avidin-FITC) and digoxigenin (red signals detected Rhodamine labeled anti-digoxigenin antibodies). FISH signals of the human Hsa1 probe onto metaphase spreads of the Sei whale (a) and the two-toed sloth (b) hybridizing to the euchromatin of one chromosome pair in both species. (c) Ring-tailed lemur probes 3 (red) and 19 (green) delineate neighboring signals on squirrel chromosome 5 (arrows). (d) Hybridization results of flow-sorted chromosome probes from cat, lemur, elephant, horse, squirrel, and pig onto human chromosome 1. The chromosomal origin of each probe is labeled below each hybridization result. In the case of the lemur and elephant, the label refers to the flow-sorted peaks (e.g., p6 = peak six) from which the probes were made.
To provide additional evidence that the boundary between these segments were the same or different, we performed reciprocal painting between humans and six species from different mammalian orders as follows: horse, pig, squirrel, ring-tailed lemur, African elephant, and cow (Fig. 3d). Hybridization signals on human metaphase spreads indicated an evolutionary fusion/fission somewhere in the Hsa1q21–Hsalq23 region (Fig. 1). The precise edges of the homology signals are difficult to resolve with chromosome painting. The FISH signal from the horse chromosome 5 paint clearly extends past the hypothetical fusion/breakpoint around Hsa1q22, into the p arm (Fig. 1). However, equids are well known for their rapid karyotypic evolution (Wichman et al. 1991), and this overlap might be due to a recently derived rearrangement. Therefore, these data alone cannot reject the hypothesis that the CSUs found in various species were conserved from the ancestral eutherian ancestor that had two CSUs (Chowdhary et al. 1998; Wienberg et al. 2000; Murphy et al. 2001c).
We further examined the homology of the two Hsa1 homologs using multidirectional chromosome painting between seven species from different orders (cat, horse, Eastern gray squirrel, African savannah elephant, ring-tailed lemur, pig, and cattle) (Figs. 3c and 4). In multidirectional reciprocal painting experiments, the two Hsa1 homologs that adjoined the 1q21–1q24 region were differentially labeled, hybridized together, and detected with different fluorochromes. If the two CSUs were inherited from a common ancestor and shared equivalent breakpoints, the probes should produce separate signals on two different chromosomes of the target species (i. e., only one color per chromosome). However, if the two CSUs had breakpoints in different chromosomal positions, in the painter versus the painted species, then we would expect to see bicolored FISH signals on a single chromosome in the painted species (e. g., Fig. 3c).
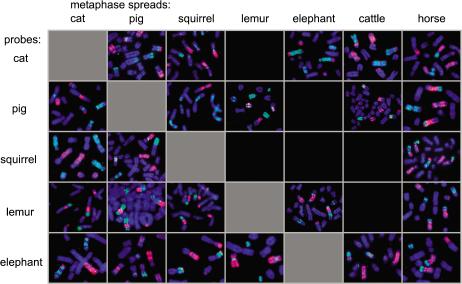
Figure 4.
Presence of adjoining FISH signals in simultaneous hybridizations of Hsa1 homologous chromosomes between mammalian species. Adjoining bicolor signals were obtained on one chromosome of the target genome for the two Hsa1 homolog probes (differentially labeled), indicating a different breakpoint in the karyotypes of the two species examined. In only two experiments (squirrel on cat and pig on squirrel) did the two Hsa1 homologous probes produce only signals on different chromosomes of the target genome, indicating no evidence of a different breakpoint in the two species being compared. The reciprocal experiments (cat on squirrel and squirrel on pig) did show bicolor signals, suggesting evidence of different breakpoints between the two species pairs. Black boxes indicate results that were not reproducible between experiments.
Twenty-four of the thirty multidirectional chromosome-painting experiments provided reproducible FISH signals (Fig. 4). In nearly all species pairs, the results showed chromosomes with adjacent bicolor hybridization signals, supporting the hypothesis that evolutionary breakpoints are different and likely produced by multiple, independent fissions in each evolutionary lineage (Fig. 4). Because of the limited resolution of Zoo-FISH (estimated to be -5 Mbp; Scherthan et al. 1994), and variations in probe quality, the absence of adjoining signals is not evidence of identical break/fusion points, but the presence of two hybridization signals on the same chromosome is evidence of different breakpoints. In summary, the new chromosome-painting data support the hypothesis of a single, ancestral chromosome homologous to human 1. An intact chromosome homologous to human chromosome 1 was found conserved in a third mammalian order (Xenarthra; Fig. 3b), and an additional principal superordinal placental lineage. The chromosome-painting data also reveal that the two major Hsa1 homologous segments present in other species are not identical in genomic composition.
Evolutionary Aspects of Human Chromosome 1 Divergence in Placental Mammals
The alternative to the single ancestral Hsa1 chromosome hypothesis predicts that at least two Hsa1 homologous chromosomes in the placental ancestor were fused independently in the Old World Primates, cetaceans, and sloth. Recent chromosome painting data from the aardvark also identifies an intact Hsa1 chromosome (Yang et al. 2003). Therefore, to accept this alternative “two ancestral chromosomes” hypothesis, however, would require many convergent chromosome fusions between independent Hsa1 homologs near the 1q22 region in each of the four principal placental clades (Madsen et al. 2001; Murphy et al. 2001a,b): Euarchontoglires (apes and Old World monkeys), Laurasiatheria (cetaceans), Xenarthra (two-toed sloth), and Afrotheria (aardvark). In contrast, our gene order data (Fig. 2) and independent bicolor painting results presented in Figures 3 and 4 support independent fragmentation of a single ancestral Hsa1 homolog.
That few modern species still maintain this ancestral single chromosome disposition is not unexpected given the large size of Hsa1, and, hence, the increased probability for breakage per unit evolutionary time. There is a strong correlation in which those species retaining the ancestral single chromosome have slower overall rates of genome evolution (e. g., the dolphin, human, and macaque genomes), whereas species with the higher number of Hsa1 syntenies have the most rearranged genomes (e. g., dog and gibbon; Table 1). Future analyses of other genomes with slow rates of chromosomal evolution may reveal further cases of intact Hsa1 chromosomes.
Table 1.
Rates of Chromosomal Evolution and Hsa1 Fragmentation
| Species | Haploid no. | CSUs relative to the placental ancestora | No. rearrangements relative to ancestral genomeb | Genomic rateb | No. of Hsa1 fragments |
|---|---|---|---|---|---|
| dolphin | 22 | 25 | 3 | 0.034 | 1 |
| cat | 20 | 27 | 6 | 0.068 | 2 |
| human | 23 | 29 | 6 | 0.068 | 1 |
| macaque | 21 | 28 | 7 | 0.080 | 1 |
| mink | 15 | 26 | 10 | 0.114 | 2 |
| lemur | 30 | 35 | 10 | 0.114 | 3 |
| tree shrwe | 31 | 35 | 10 | 0.114 | 2 |
| horse | 32 | 40 | 15 | 0.170 | 3 |
| cow | 30 | 41 | 16 | 0.182 | 3 |
| bat | 16 | 35 | 19 | 0.216 | 3 |
| shrew | 10 | 30 | 20 | 0.227 | 2 |
| pig | 19 | 41 | 22 | 0.250 | 6 |
| gibbon | 22 | 51 | 26 | 0.295 | 4 |
| spider monkey | 17 | 48 | 30 | 0.341 | 6 |
| dog | 39 | 64 | 39 | 0.443 | 7 |
Given the evidence for a single ancestral chromosome, our genomic comparisons suggest a nonuniform breakage history across this chromosome, with strong predisposition for chromosome breakage in two regions of the long arm of Hsa1. The gene-mapping data for the species examined here indicate a clustering of inter- and intrachromosomal rearrangements in the distal part of Hsa1q (Fig. 2). In contrast, regions homologous to Hsa1p exhibit fewer breakpoints and greater conservation of synteny and gene order. The comparative chromosome-painting data also suggest repeated synteny breakage or translocations in the 1q21–1q23 region in members of at least seven mammalian orders. Whether these regions of frequent evolutionary breakage correspond with particular sequence motifs, will depend upon fine mapping of the breakage intervals in each of the species examined and characterization at the sequence level. Sequence analysis of the long arm of human chromosome 1 reveals a higher proportion of segmental duplications, particularly in the vicinity of 1q21–1q22, relative to the short arm (Bailey et al. 2002), which have been shown to be a causative factor in primate genome evolution (Vallente Samonte and Eichler 2002). In addition, studies of interstitial telomere-like sequences in humans and other Old World primates show that this chromosomal region contains such repeats and is susceptible to a high number of breaks after X-irradiation (Azzalin et al. 2001; Ruiz-Herrera et al. 2002). Whether there is some connection between these elements and the increased historic breakage across mammalian orders remains to be seen.
In light of the present results, we now modify our previous 2n = 50 hypothesis for the ancestral placental mammal genome (Murphy et al. 2001c), with two Hsa1-bearing chromosomes, to one of 2n = 48, in which Hsa1 represents the single largest chromosome in the ancestral genome. This study illustrates the advantages of combining chromosome-painting data and ordered gene-mapping data to resolve details of chromosome evolution. Increased application of subregional probes and a broader selection of ordered mammalian genome maps would prove invaluable for identifying regions of historic genomic instability across the mammalian radiation.
METHODS
Primer Design and Radiation Hybrid Mapping
We took a targeted approach to systematically filling in syntenic gaps in the human–cat comparative maps by designing novel conserved PCR primers using multiple mammalian species alignment outputs from BLAST. In most cases, these were based on human–rodent comparison, using genomic sequence information for guidance. The following primer pairs were designed to obtain homologous feline sequence: GNB1-F, GCTCCTGACACCAGACTGTT, GNB1-R, TCGTCTGAGCCAGTGGCAA; ENO1-F, GGTTCTCATGCTGGCAACAA, ENO1-R, CTTCTTTGTTCTCCAGGATGT; GJA4-F, TCATCTTCCGCATCCTCATC, GJA4-R, CGAGATCTTGGCCATCTGA; PTGER3-F, GAGAGCAAGCGCAAGAAGTC, PTGER3-R, GTGCTGATGAAGCACCACGT; GJA8-F, GGGCGACTGGAGTTTCCT, GJA8-R, TAGCCCTTGGCTTTCTGGAT; CD3Z-F, ACAGCCACCAAGGACACCTA, CD3Z-R, GAAGCCAAATTTACAGCAGGA; NCF2-F, TGTCTTGAAGAAGGGCAATG, NCF2-R, CAAGGTAGTTGCAGGGAACA. In addition, sequence-tagged sites (STSs) were also derived from the following feline sequences deposited in GenBank (accession nos. in parentheses): TNFRSF1B (U51429), PTGFR (AF272340), FCGR3 (AB025315), TNNT2 (AF220219), EDG1 (AY011738), and ADORA3 (AY011246). The locus CHRM3 was amplified in the domestic cat using primers from Jiang et al (1998) and sequenced to obtain template for designing a cat-specific STS. DNA sequencing was performed with BigDye Terminator chemistry (Applied Biosystems Inc.), purified using G50 columns (Amersham-Pharmacia), and sequenced using either an ABI 377 or 3700 sequencer. Methods for genotyping and RH map construction have been described in detail elsewhere (Murphy et al. 1999a,b, 2000). Briefly, PCR-based typing was performed on the 93-hybrid 5000-rad feline whole-genome radiation hybrid panel (Murphy et al. 1999a). All feline markers were amplified in 384-well plates using Taq-Gold DNA polymerase (Applied Biosystems Inc.) and scored in duplicate on 2% agarose gels stained with VistaGreen. Two-point linkage analysis and multi-point maximum likelihood ordering was performed with the RHMAP software (Boehnke et al. 1991) as described in Murphy et al. (2000).
Comparative Mapping Data
Human RH locus positions were retrieved from the NCBI UniGene and UniSTS databases, whereas physical positions in the draft human sequence are based upon the NCBI assembly (Build 30). Comparative mapping data from the cattle RH-based gene map is from Band et al. (2000). Rat–human comparative mapping data was derived from Watanabe et al. (1999), and the associated Web site at OtsukaGen: (http://ratmap.ims.utokyo.ac.jp/cgi-bin/comparative_home.pl) and from the Rat Genome Database (http://rgd.mcw.edu/VCMAP). Mouse–human comparative mapping data is derived from the NCBI human–mouse homology maps (http://www.ncbi.nlm.nih.gov/Homology).
Chromosome Painting
Chromosome-specific painting probes to homologs of Hsa1 (listed in Fig. 1) from the horse (Equus caballus,_Coriell Cell Repositories, AG08080), Eastern gray squirrel (Sciurus carolinensi, primary culture), African elephant (Loxodonta africanus, LGD cell bank), Ring-tailed Lemur (Lemur catta,_ Coriell Cell Repositories, AG07099), domestic pig (Sus scrofa, Coriell Cell Repositories, AG08113), domestic cat (Felis catus, primary culture), and cattle (Bos taurus, Coriell Cell Repositories, AG10553) genomes were made by degenerate oligonucleotide-primed PCR (DOP–PCR) from flow sorted chromosomes (Telenius et al. 1992). The same primers (6MW) were used in the primary reaction and to label the chromosome paints with biotin or digoxigenin-dUTP in secondary PCR reactions. Metaphase chromosomes were prepared from the fibroblast cultures listed above and from a cattle cell line (Bos taurus, Coriell Cell Repositories, AG10553) according to classic cytogenetic protocols. The probe concentrations and hybridization conditions were the same as described previously (Frönicke and Wienberg 2001). After hybridization and washing of the slides, biotinylated DNA probes were detected with avidin coupled with Cy-3 (Amersham) or fluorescein isothiocyanate (FITC; Vector). Digoxigenin-labeled probes were detected with antidigoxigenin antibodies conjugated with FITC or Rhodamine (Roche).
Estimating Rates of Chromosomal Evolution
To determine the rate of chromosomal evolution for each lineage, we based all comparisons of Zoo-FISH conserved segments upon a published hypothetical ancestral placental genome (Murphy et al. 2001c). The minimum number of rearrangements was equal to the number of CSUs, minus the lower haploid number of the compared species (O'Brien et al. 1999). To maintain uniformity in the estimate of conserved segments across species, all values are based on chromosome-painting results using human flow-sorted chromosome probes, because different methods and species probes might differ in their ability to discern smaller segments. This number was then divided by the estimated time of divergence from the ancestral Boreoeutherian mammal (-88 million years ago [Mya]; Murphy et al. 2001b) to obtain the rate of chromosomal evolution per lineage.
Acknowledgments
We thank Clarence Smith Jr., Deborah Hirschmann, Jennifer Tabler, and Stan Cevario for technical assistance, and Gary Stone for the flow sorting of various species used in the reciprocal FISH. We also thank Oliver Ryder, San Diego Zoo–CRES, for providing the sloth fibroblast cell line. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1022303.
Footnotes
[Supplemental material is available online at www.genome.org. Feline gene segments have been deposited in GenBank under accession numbers CC596505–CC596511. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: O. Ryder.]
Article published online before print in July 2003.
References
- Arnason, U. 1972. The role of chromosomal rearrangements in mammalian speciation with special reference to cetacea and pinnipedia. Hereditas 70: 113-118. [PubMed] [Google Scholar]
- Azzalin, C.M., Nergadze, S.G., and Giulotto, E. 2001. Human intrachromosomal telomeric-like repeats: Sequence organization and mechanisms of origin. Chromosoma 110: 75-82. [DOI] [PubMed] [Google Scholar]
- Bailey, J.A., Gu, Z., Clark, R.A., Reinert, K., Samonte, R.V., Schwartz, S., Adams, M.D., Myers, E.W., Li, P.W. and Eichler, E.E. 2002. Recent segmental duplications in the human genome. Science 297: 1003-1007. [DOI] [PubMed] [Google Scholar]
- Band, M.R., Larson, J.H., Rebeiz, M., Green, C.A., Heyen, D.W., Donovan, J., Windish, R., Steining, C., Mahyuddin, P., Womack, J.E., et al. 2000. An ordered comparative map of the cattle and human genomes. Genome Res. 10: 1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielec, P.E., Gallagher, D.S., Womack, J.E., and Busbee, D.L. 1998. Homologies between human and dolphin chromosomes detected by heterologous chromosome painting. Cytogenet. Cell. Genet. 81: 18-25. [DOI] [PubMed] [Google Scholar]
- Boehnke, M., Lange, K., and Cox, D.R. 1991. Statistical methods for multipoint radiation hybrid mapping. Am. J. Hum. Genet. 49: 1174-1188. [PMC free article] [PubMed] [Google Scholar]
- Breen, M., Thomas, R., Binns, M.M., Carter, N.P., and Langford, C.F. 1999. Reciprocal chromosome painting reveals detailed regions of conserved synteny between the karyotypes of the domestic dog (Canis familiaris) and human. Genomics 15: 145-155. [DOI] [PubMed] [Google Scholar]
- Burt, D.W., Bruley, C., Dunn, I.C., Jones, C.T., Ramage, A., Law, A.S., Morrice, D.R., Paton, I.R., Smith, J., Windsor, D., et al. 1999. The dynamics of chromosome evolution in birds and mammals. Nature 402: 411-413. [DOI] [PubMed] [Google Scholar]
- Chowdhary, B.P., Raudsepp, T., Fronicke, L., and Scherthan, H. 1998. Emerging patterns of comparative genome organization in some mammalian species as revealed by Zoo-FISH. Genome Res. 8: 577-589. [DOI] [PubMed] [Google Scholar]
- Frönicke L. and Wienberg, J. 2001. Comparative chromosome painting defines the high rate of karyotype changes between pigs and bovids. Mamm. Genome 12: 442-449. [DOI] [PubMed] [Google Scholar]
- Frönicke, L., Chowdhary, B.P., Scherthan, H., and Gustavsson, I. 1996. A comparative map of the porcine and human genomes demonstrates Zoo-FISH and gene mapping-based chromosomal homologies. Mamm. Genome 7: 285-290. [DOI] [PubMed] [Google Scholar]
- Glas, R., Wakefield, M.J., Toder, R., and Graves, J.A.M. 1998. Comparative chromosome painting. ILAR J. 39: wall chart. [PubMed]
- Goureau, A., Yerle, M., Schmitz, A., Riquet, J., Milan, D., Pinton, P., Frelat, G., and Gellin, J. 1996. Human and porcine correspondence of chromosome segments using bidirectional chromosome painting. Genomics 36: 252-262. [DOI] [PubMed] [Google Scholar]
- Jiang, Z., Priat, C., and Galibert, F. 1998. Traced orthologous amplified sequence tags (TOASTs) and mammalian comparative maps. Mamm Genome 9: 577-587. [DOI] [PubMed] [Google Scholar]
- Korstanje, R., O'Brien, P.C., Yang, F., Rens, W., Bosma, A.A., van Lith, H.A., van Zutphen, L.F., and Ferguson-Smith, M.A. 1999. Complete homology maps of the rabbit (Oryctolagus cuniculus) and human by reciprocal chromosome painting. Cytogenet. Cell. Genet. 86: 317-322. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Madsen, O., Scally, M., Douady, C.J., Kao, D.J., DeBry, R., Adkins, R., Amrine, H.M., Stanhope, M.J., deJong, W.W., and Springer, M.S. 2001. Parallel adaptive radiations in two clades of placental mammals. Nature 409: 610-614. [DOI] [PubMed] [Google Scholar]
- Muller, S., Stanyon, R., O'Brien, P.C., Ferguson-Smith, M.A., Plesker, R., and Wienberg, J. 1999. Defining the ancestral karyotype of all primates by multidirectional chromosome painting between tree shrews, lemurs and humans. Chromosoma 108: 393-400. [DOI] [PubMed] [Google Scholar]
- Murphy, W.J., Menotti-Raymond, M., Lyons, L.A., Thompson, M.A., and O'Brien, S.J. 1999a. Development of a feline whole genome radiation hybrid panel and comparative mapping of human chromosome 12 and 22 loci. Genomics 57: 1-8. [DOI] [PubMed] [Google Scholar]
- Murphy, W.J., Sun, S., Chen, Z.Q., Pecon-Slattery, J., and O'Brien, S.J. 1999b. Extensive conservation of sex chromosome organization between cat and human. Genome Res. 9: 1223-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, W.J., Sun, S., Chen, Z.Q., Yuhki, N., Hirschmann, D., Menotti-Raymond, M. and O'Brien, S.J. 2000. A radiation hybrid map of the cat genome: Implications for comparative mapping. Genome Res. 10: 691-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, W.J., Eizirik, E., Johnson, W.E., Zhang, Y.-P., Ryder, O.A., and O'Brien, S.J. 2001a. Molecular phylogenetics and the origins of placental mammals. Nature 409: 614-618. [DOI] [PubMed] [Google Scholar]
- Murphy, W.J., Eizirik, E., O'Brien, S.J., Madsen, O., Scally, M., Douady, C.J., Teeling, E., Ryder, O.A., Stanhope, M.J., de Jong, W.W., et al. 2001b. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science 294: 2348-2351. [DOI] [PubMed] [Google Scholar]
- Murphy, W.J., Stanyon, R., and O'Brien, S.J. 2001c. Evolution of mammalian genome organization revealed by comparative gene mapping. Genome Biol. 2: 0.0005-0.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien, S.J. and Stanyon, R. 1999. Phylogenomics. Ancestral primate viewed. Nature 402: 365-366. [DOI] [PubMed] [Google Scholar]
- O'Brien, S.J., Menotti-Raymond, M., Murphy, W.J., Nash, W.G., Wienberg, J., Stanyon, R., Copeland, N.G., Jenkins, N.A., Womack, J.E., and Marshall Graves, J.A. 1999. The promise of comparative genomics in mammals. Science 286: 458-481. [DOI] [PubMed] [Google Scholar]
- Rettenberger, G., Klett, C., Zechner, U., Kunz, J., Vogel, W., and Hameister, H. 1995. Visualization of the conservation of synteny between humans and pigs by heterologous chromosomal painting. Genomics 26: 372-378. [DOI] [PubMed] [Google Scholar]
- Ruiz-Herrera, A., Garcia, F., Azzalin, C., Giulotto, E., Egozcue, J., Ponsa, M., and Garcia, M. 2002. Distribution of intrachromosomal telomeric sequences (ITS) on Macaca fascicularis (Primates) chromosomes and their implication for chromosome evolution. Hum. Genet. 110: 578-586. [DOI] [PubMed] [Google Scholar]
- Scherthan, H., Cremer, T., Arnason, U., Weier, H.-U., Lima-de-Faria, A., and Frönicke, L. 1994. Comparative chromosome painting discloses homologous segments in distantly related mammals. Nature Genet. 6: 342-347. [DOI] [PubMed] [Google Scholar]
- Sun, H.S., Tuggle, C.K., Goureau, A., Fitzsimmons, C.J., Pinton, P., Chardon, P., and Yerle, M. 2002. Precise mapping of breakpoints in conserved synteny between human chromosome 1 and pig chromosomes 4, 6 and 9. Anim. Genet. 33: 91-96. [DOI] [PubMed] [Google Scholar]
- Telenius, H., Pelmear, A.H., Tunnacliffe, A., Carter, N.P., Behmel, A., Ferguson-Smith, M.A., Nordenskjold, M., Pfragner, R., and Ponder, B.A. 1992. Cytogenetic analysis by chromosome painting using DOP-PCR amplified flow-sorted chromosomes. Genes Chromosomes Cancer 4: 257-263. [DOI] [PubMed] [Google Scholar]
- Vallente Samonte, R. and Eichler, E.E. 2002. Segmental duplications and the evolution of the primate genome. Nat. Rev. Genet. 3: 65-72. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- Watanabe, T.K., Bihoreau, M.T., McCarthy, L.C., Kiguwa, S.L., Hishigaki, H., Tsuji, A., Browne, J., Yamasaki, Y., Mizoguchi-Miyakita, A., Oga, K.et al. 1999. A radiation hybrid map of the rat genome containing 5,255 markers. Nat. Genet. 22: 27-36. [DOI] [PubMed] [Google Scholar]
- Wichman, H.A., Payne, C.T., Ryder, O.A., Hamilton, M.J., Maltbie, M., and Baker, R.J. 1991. Genomic distribution of heterochromatic sequences in equids: Implications to rapid chromosomal evolution. J. Hered. 82: 369-377. [DOI] [PubMed] [Google Scholar]
- Wienberg, J., Stanyon, R., Nash, W.G., O'Brien, P.C., Yang, F., O'Brien, S.J., and Ferguson-Smith, M.A. 1997. Conservation of human vs. feline genome organization revealed by reciprocal chromosome painting. Cytogenet. Cell. Genet. 77: 211-217. [DOI] [PubMed] [Google Scholar]
- Wienberg, J., Froenicke, L., and Stanyon, R. 2000. Insights into mammalian genome organization and evolution by molecular cytogenetics. In Comparative genomics. (ed. M.S., Clark),, pp. 207-244. Kluwer Academic Publishers, Boston, MA.
- Yang, F., Graphodatsky, A.S., O'Brien, P.C.M., Colabella, A., Solanky, N., Squire, M., Sargan, D.R., and Ferguson-Smith, M.A. 2000. Reciprocal chromosome painting illuminates the history of genome evolution of the domestic cat, dog and human. Chromosome Res. 8: 393-404. [DOI] [PubMed] [Google Scholar]
- Yang, F., Alkalaeva, E.Z., Perelman, P.L., Pardini, A.T., Harrison, W.R., O'Brien, P.C.M., Fu, B., Graphodatsky, A.S., and Ferguson-Smith, M.A. 2003. Reciprocal chromosome painting among human, aardvark and elephant (superorder Afrotheria) reveals the likely eutherian ancestral karyotype. Proc. Natl. Acad. Sci. 100: 1062-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://ratmap.ims.utokyo.ac.jp/cgi-bin/comparative_home.pl; Web site at OtsukaGen.
- http://rgd.mcw.edu/VCMAP; Rat Genome Database.
- http://www.ncbi.nlm.nih.gov/Homology; NCBI human–mouse homology maps.