Abstract
Transposable elements (TEs) comprise a major component of eukaryotic genomes, and exhibit striking deviations from random distribution across the genomes studied, including humans, flies, nematodes, and plants. Although considerable progress has been made in documenting these patterns, the causes are subject to debate. Here, we use the genome sequence of Arabidopsis thaliana to test for the importance of competing models of natural selection against TE insertions. We show that, despite TE accumulation near the centromeres, recombination does not generally correlate with TE abundance, suggesting that selection against ectopic recombination does not influence TE distribution in A. thaliana. In contrast, a consistent negative correlation between gene density and TE abundance, and a strong under-representation of TE insertions in introns suggest that selection against TE disruption of gene expression is playing a more important role in A. thaliana. High rates of self-fertilization may reduce the importance of recombination rate in genome structuring in inbreeding organisms such as A. thaliana and Caenorhabditis elegans.
Transposable elements (TEs) in many organisms have been shown to accumulate differentially among chromosomal regions, including regions of contrasting recombination rates (Charlesworth and Langley 1989; Duret et al. 2000; Boissinot et al. 2001; Bartolome et al. 2002), gene density (Medstrand et al. 2002), and base composition (Lander et al. 2001). One possible explanation for these patterns is that TEs have insertion preferences for particular regions. Evidence for insertion bias is strong for some TEs (Jakubczak et al. 1991), although for the majority there is little evidence to support insertion preference as an explanation for TE distribution (Nuzhdin et al. 1997). An alternative is the effects of differential selective constraints on TEs in different regions of the genome (Charlesworth and Langley 1989). First, TE's are almost exclusively found outside of coding regions (Duret et al. 2000; Nekrutenko and Li 2001; Bartolome et al. 2002; Pavlicek et al. 2002), suggesting that the majority of insertions into exons are strongly deleterious and rapidly eliminated by selection. Additionally, evidence from patterns of TE insertion polymorphism in natural populations of some species indicate that insertions segregating in noncoding genomic regions are almost always at low frequencies, consistent with the hypothesis that TE abundance is controlled by the action of purifying selection (Charlesworth and Langley 1989; Wright et al. 2001). If TE abundance in noncoding DNA is determined by a balance between the forces of transposition and natural selection, regional genome effects on the strength or efficacy of natural selection will play a significant role in controlling TE distribution.
Several models of selection against TEs in noncoding DNA have been proposed. First, abundance may be controlled by weak selection against the direct effects of insertions into noncoding regions (Charlesworth and Langley 1989; Biemont et al. 1997). In particular, insertions into introns and regulatory regions may have, on average, slightly deleterious consequences on fitness (Long et al. 2000; Lander et al. 2001) by causing disruptions in gene expression. Similarly, element transposition may impose a significant cost on the host due to expression of TE gene products, leading to selection against TE activity (Nuzhdin et al. 1996). Alternatively, the action of natural selection may be only indirectly associated with TE mobility, by the deleterious effects of ectopic recombination between elements located at distinct sites in the genome, which can cause major chromosomal rearrangements and gene deletions (Langley et al. 1988).
Under the ectopic exchange model, lower rates of ectopic exchange are expected in regions of reduced recombination (Virgin and Bailey 1998), allowing TEs to accumulate in these chromosomal locations (Langley et al. 1988). However, under the insertion model, the action of positive and negative selection at linked sites may also weaken the efficacy of selection against deleterious insertions in regions of reduced recombination (Duret et al. 2000; Eickbush and Furano 2002), a process known as the Hill-Robertson effect (Hill and Robertson 1966). Although further modelling is required to assess the action of Hill-Robertson interference on TEs, the effects of linked selection may thus also allow elements to accumulate in regions of reduced recombination. Unlike the ectopic exchange model, however, the insertion model predicts that TE insertions should accumulate in regions of low gene density; even within noncoding DNA, TE insertions are less likely to interfere with gene expression in regions with a low proportion of coding DNA.
Results showing higher copy numbers of TEs in regions of reduced recombination in several species (Boissinot et al. 2001; Bartolome et al. 2002) are consistent with this expectation of both models. Although most of these studies have examined the effects of large-scale heterogeneity in recombination, a recent genetic analysis in maize also provided evidence for a strong reduction in the rate of recombination in TE-rich regions at a very local level (Fu et al. 2002). Furthermore, a recent study of TE frequencies in populations of Drosophila melanogaster has found evidence for an effect of TE size on the action of selection, which provides additional support for the model of ectopic exchange exchange (Petrov et al. 2003). In contrast with these results, however, a recent study showed a positive correlation between recombination rate and DNA transposon abundance in C. elegans, and no effect of recombination on retrotransposon abundance (Duret et al. 2000), suggesting that the predictions of all selection models do not hold. Although the explanation for the differences among species is unclear, C. elegans is highly inbreeding in natural populations (Graustein et al. 2002). In highly inbred species, low effective rates of recombination across the genome may remove effects of recombination rate heterogeneity on genome structure (Charlesworth and Wright 2001; Morgan 2001). First, unless population size or physical recombination rates are very high, the action of Hill-Robertson interference is expected to be present genome-wide (McVean and Charlesworth 2000), and any recombination-based heterogeneity associated with the efficacy of selection may be weak or absent. In addition, high homozygosity in self-fertilizing populations is expected to lead to infrequent ectopic recombination events, due to fewer chances for ectopic pairing, which appears to be promoted by heterozygosity (Charlesworth and Charlesworth 1995; Morgan 2001; Wright et al. 2001). Patterns of heterogeneity in TE distributions observed in the genomes of highly inbred species should thus reflect other effects of natural selection, or the effects of transposition preferences, and variation in recombination rate is less likely to play a role.
To further evaluate this hypothesis, we investigate the effects of recombination rate and gene density on TE distributions in the self-fertilizing plant Arabidopsis thaliana. A. thaliana shows a high diversity of TEs, most of which are widely dispersed across the genome (Surzycki and Belknap 1999; Le et al. 2000). This diversity includes all major superfamilies of retrotransposons (Class I elements), DNA transposons (Class II elements), as well as a recently identified third class of TEs (Class III; Le et al. 2000; Kapitonov and Jurka 2001), with similarities to bacterial rolling-circle transposons (Kapitonov and Jurka 2001). Genome analysis has provided preliminary evidence for an accumulation of many TE families in pericentromeric regions (Copenhaver et al. 1999; Kumekawa et al. 2000), but the relative importance of various characteristics of genome structure in driving this pattern has not been investigated.
RESULTS
TE Accumulation in Low-Recombining Regions Surrounding the Centromere
Central regions of reduced recombination, including the centromeres, pericentromeric regions, and heterochromatic knobs, have been shown to have a strong reduction in rates of recombination (Copenhaver et al. 1999; Haupt et al. 2001), and we first compare TE abundance in these regions with the chromosome arms. All classes of TEs show evidence for accumulation in these regions of reduced recombination surrounding the centromere (Arabidopsis Genome Initiative 2000; Table 1). In total, TE-derived DNA represents -27% of the central regions of reduced recombination, compared with only 3% of DNA in the rest of the genome. Two superfamilies of elements, gypsy- like and CACTA, show a particularly striking accumulation in the centromeric regions, and are almost exclusively found in these regions, and we therefore consider these elements separately. These TEs, particularly the gypsy-like element Athila, have many centromeric copies arranged in tandem, often as truncated or fragmented elements, located near the centromeric core (Pelissier et al. 1995; Kumekawa et al. 2000). Such elements have been hypothesized to be associated with centromere function (Malik and Henikoff 2002), and they may have acquired new mutational mechanisms for their spread in the centromere. However, all other elements, which are not found as tandem arrays, also show significant accumulation in pericentromeric regions of reduced recombination.
Table 1.
TE Copy Number (Per MB) and Genome Characteristics in Regions of Reduced Recombination Compared With the Chromosome Arms
| Reduced recombination | Chromosome arms | |
|---|---|---|
| Class Ia | 16.3*** | 3.5 |
| gypsy | 33.0*** | 0.3 |
| Class II | 30.6*** | 10.2 |
| CACTA | 6.5*** | 0.16 |
| Class III | 16.9*** | 6.7 |
| GC content | 0.368** | 0.361 |
| Exon density (per Mb) | 417** | 1300 |
| Fraction of coding DNA | 0.131*** | 0.326 |
| Intron size (bp)b | 188.9** | 167.2 |
Significance levels are given for the Mann-Whitney U test, *, P <0.05
Class I elements include those insertins that could be definitively classified as copia-like, LINE-like, and SINE-like
Excludes contribution of TE insertions to intron length
, P <0.01
, P <0.001
In addition to low levels of crossing-over, A. thaliana pericentromeric regions also have low exon density, and the fraction of DNA that is coding is much lower compared with the rest of the genome (Table 1; for review, see Arabidopsis Genome Initiative 2000). Similar to recent studies in D. melanogaster (Comeron and Kreitman 2000), average intron length is significantly larger in the regions of reduced recombination, even when the contribution of TE insertions to intron size is factored out (Table 1). Overall differences in base composition are also significant, but the difference in mean base composition between regions is very weak. Surprisingly, GC content is slightly higher in the pericentromeric regions compared with the chromosome arms (Table 1). This contrasts with the pattern at the local level, where base composition surrounding TE insertions has been found to show an elevated frequency of AT (Le et al. 2000).
Because gene density is higher in regions of normal recombination, one simple explanation for the differential accumulation of TEs in low-recombining regions is that it reflects the action of strong purifying selection against insertions into exons. We find an almost complete absence of insertions into exons (Table 2), suggestive of very strong selection. Note that we exclude from this category (1) annotated genes matching solely to TE-derived gene products, (2) fusion proteins encoding predicted genes which fuse coding sequence from TE insertions and adjacent genes, and (3) acquired genes that are host genes internal to TE insertions (Yu et al. 2000). Although both fusion proteins and acquired genes may be expressed and functional, they do not represent the insertion of TEs into pre-existing host coding regions. TE insertions are also strongly under-represented in introns in A. thaliana (Table 2); in both the centromere and the chromosome arms, there is a strong reduction in the frequency of TE insertions in introns. Because most insertions into introns and exons appear to be under strong purifying selection, this might be the sole explanation for their low abundance in the chromosome arms, where the density of coding regions is higher. When the abundance of TEs within intergenic DNA is compared, however, TEs are still significantly in excess in regions of low recombination (Table 2), suggesting that the difference in the abundance of coding DNA alone cannot explain their differential accumulation.
Table 2.
Location of TE Insertions in Relation to Coding Regionsa
|
Reduced recombination
|
Chromosome arms
|
|||||
|---|---|---|---|---|---|---|
| Element class | intergenic | intron | coding | intergenic | intron | coding |
| Class Ib | 19.4*** (0.049) | 2.99* (0.0004) | 1.4* (0.0007) | 5.8 (0.014) | 0.378 (0.0002) | 0.07 (0.0002) |
| Class IIb | 35.5*** (0.046) | 9.4*** (0.023) | 0 | 18.1 (0.018) | 1.5 (0.0001) | 0 |
| Class III | 20.9*** (0.014) | 3.2 (0.002) | 0.56 (0.0005) | 12.5 (0.006) | 1.1 (9.8 × 10-4) | 0 |
Upper values in each cell are the number of elements per megabase of DNA in each category, and values in parentheses are the fraction of DNA in each location occupied by TEs. Significance levels are shown for comparisons between the regions of reduced recombination compared with the chromosome arms by the Mann-Whitney U test
Class I and Class II elements are shown excluding gypsy and CACTA-like elements, respectively. Two-tailed significance
, P <0.05; **, P <0.01
, P <0.001
Recombination Rate Does Not Explain TE Distribution in Intergenic Regions of A. thaliana
The high number of TEs in pericentromeric regions is consistent with either model of selection, as these regions are both gene-poor and recombination-poor. Furthermore, the dense methylation and difference in chromatin structure in heterochromatic regions may make these insertions more effectively silenced (Singer et al. 2001), preventing the potentially deleterious expression of TE-derived gene products in these regions. It is therefore important to examine whether, in addition to this comparison between genomic regions, there are general correlations between TE abundance and recombination rate. Given the underrepresentation of TEs in coding regions, we consider only the TE content in intergenic regions for this analysis. Excluding the centromere-specific TEs (gypsy and CACTA), no correlation is observed between the rate of recombination and TE abundance in intergenic DNA (Table 3). Note that this whole-genome correlation with recombination is not significant, despite accumulation of elements near the centromere; because the pericentromeric regions represent a low proportion of total DNA, even this effect is not detected in a genome-wide correlation. Only gypsy and CACTA, which are almost exclusively present in the central regions of reduced recombination, show a significant negative correlation. If we consider only the chromosome arms, no negative correlation is observed for any class of TE, and all three major classes of element, in fact, show a positive relationship with TE frequency. This positive relationship can be explained at least in part by a negative correlation between coding density and recombination rate along the chromosome arms (Table 3). Using a partial correlation correcting for the effect of coding density, there is no significant effect of recombination rate on Class II or III elements, and we observe a weaker positive relationship between recombination rate and TE abundance for Class I TEs (Table 3). Provided that the rate of ectopic recombination correlates with the rate of crossing over, these results suggest that selection against ectopic exchange does not drive TE distribution in A. thaliana.
Table 3.
Spearman Rank Correlation Coefficients Between Element Frequency, Fraction of Coding DNA and Recombination Ratea
|
Total genome
|
Chromosome arms
|
|||
|---|---|---|---|---|
| Recombination rate | Fraction of coding DNAb | Recombination rate | Fraction of coding DNAb | |
| Class I | -0.005 (0.067*) | -0.375*** (-0.322***) | 0.127** (0.0898*) | -0.187*** (-0.202***) |
| gypsy | -0.378*** (-0.257***) | -0.426*** (-0.469***) | 0.060 (0.124***) | -0.057 (-0.117**) |
| Class II | -0.011 (-0.006) | -0.318*** (-0.232***) | 0.080* (0.021) | -0.100*** (-0.155***) |
| CACTA | -0.247*** (-0.193***) | -0.271*** (-0.276***) | 0.023 (0.0011) | -0.074* (-0.057) |
| Class III | 0.035 (0.09**) | -0.244*** (-0.218***) | 0.076* (0.037) | -0.064 (-0.164***) |
| Fraction of coding DNA | 0.099* | — | -0.139** | — |
Values given in parentheses are partial correlations, factoring out effects of coding fraction and recombination rate, respectively
Direct correlations between coding density and TE abundance are calculated by making bins that exclude TE contributions to DNA length. Partial correlations, factoring out recombination rate effects, use the original bins for accurate estimation of recombination rates over real physical distance. Two-tailed significance
, P <0.05
, P <0.01
, P <0.001
Effect of Gene Density on TE Abundance in Intergenic DNA
Our initial analysis suggested that selection against insertions into exons and introns is playing an important role in TE distribution in the A. thaliana genome, given their strong underrepresentation compared with intergenic DNA. We also wish to test whether regions of high gene density show an underrepresentation of insertions in intergenic regions, due to a higher density of untranslated and cis-regulatory regions. Alternatively, silencing of TE insertions by DNA methylation and associated changes in chromatin structure might interfere with the expression of nearby genes (Gendrel et al. 2002). In a given sample of DNA along the chromosome, increases in TE-derived DNA could force the amount of coding sequence to be smaller, violating the standard assumptions of correlation analysis. To avoid this effect, we excluded the contribution of TEs to sequence length when sampling bins of constant size (100 kb) across the genome to look at effects of coding density on TE frequency. In contrast to recombination rate, all element families show a significant negative correlation between the fraction of coding DNA and TE frequency in intergenic regions (Table 3). If we consider only the chromosome arms, the negative correlation remains significant for Class I and II elements, but not Class III (Table 3). Furthermore, a partial correlation analysis, factoring out the effects of recombination rate, still reveals a residual negative correlation between coding density and TE abundance for both the whole genome and the chromosome arms (Table 3).
Whereas the above analysis suggests that selection against insertions near coding regions is influencing TE distribution, the corresponding correlation coefficients are low, and it is unclear how strong the action of purifying selection would be to explain the data. To assess the insertion model more directly, we use a maximum likelihood framework to test whether a simple model that includes an effect of coding density on the amount of selective constraint in intergenic regions provides a significant improvement to the likelihood of the data compared with a model that assumes elements are randomly distributed in intergenic regions (see Methods). Because the high fraction of TE-derived DNA and the high frequency of nested insertions in the centromere will violate the assumptions of independence among insertions, we consider only the chromosome arms for this analysis, and once again exclude TE contributions to sequence length.
For all three TE classes, a model that includes increases in selective constraint in intergenic regions with an increasing amount of coding DNA provides a significant improvement to the likelihood compared with a model of random distribution in intergenic regions, consistent with the action of purifying selection against insertions near genes (Table 4). We can use this model to estimate α, a measure of the amount to which increases in coding DNA increase the selective constraint in intergenic regions. Estimates of α range from 0.27 for Class II elements to 0.64 for Class I. Additional factors, such as variation across individual element families and genomic regions in α, the presence of nested insertions, and the influence of historical selection pressures predating the evolution of self-fertilization may all play a role in causing residual departures from expectation. Nevertheless, we can use our analysis to roughly estimate the total fraction of intergenic DNA that is under purifying selection against TE insertions along the chromosome arms (see Methods). Using the total amounts of intergenic and coding DNA in our sample from the chromosome arms, our estimates of α would imply that between 18% and 44% of intergenic DNA is under selective constraint against TE insertions. Similarly, given an estimated average of 1308 bp of coding DNA per gene along the chromosome arms, this implies that each gene has on average 235–837 bp of intergenic DNA that are under selective constraint against TE insertions. Given the short distance between genes in the compact A. thaliana genome (average length of intergenic DNA, 2013 bp), this conclusion appears reasonable. Computer simulations of insertion into unconstrained regions using the observed values of the abundance of coding and intergenic DNA and the maximum likelihood estimates of parameters confirm that the observed correlation coefficients between TE frequency and coding density are expected with this level of selective constraint (Table 4). This illustrates that even fairly strong selection against insertions in intergenic DNA will generate similar correlations between coding density and TE frequency along the chromosome arms to those observed.
Table 4.
Likelihood Ratio Test for Selection on Insertions in Intergenic DNA
| Element class | α̂ (95% C.I.)a | 2 ΔL | rs, model (95% C.I.)b |
|---|---|---|---|
| Class I | 0.64 (0.52, 0.70) | 31.1*** | -0.178 (-0.124, -0.232) |
| Class II | 0.27 (0.04, 0.42) | 4.9* | -0.07 (-0.01, -0.126) |
| Class III | 0.34 (0.1, 0.49) | 6.7** | -0.085 (-0.027, -0.144) |
Significance of likelihood ratio test
Maximum likelihood estimate and approximate 95% credibility interval for the estimate of α, using the χ2 approximation
Mean and 95% confidence interval of the Spearman rank correlation coefficient between TE frequency and coding density from 10,000 computer simulations of random insertion using the maximum likelihood estimates of α and ν
, P <0.05
, P <0.01
, P <0.001
Multivariate Analysis of TE Distribution
It is important to assess whether our analyses on the basis of pairwise correlations and the three major TE Class categories are robust to a more global, multivariate analysis. To investigate this, we used principal components analysis to summarize multivariate patterns of TE abundance in intergenic DNA for all individual TE element families, excluding CACTA and gypsy-like elements. The first axis in this data reduction, which explains 23% of the variance, shows a general increase with TE abundance; all TE families increase in abundance with an increasing value for factor 1 (see Supplemental Data, available online at www.genome.org). We performed a multiple linear regression, using factor 1 as the dependent variable, with coding density, recombination rate, and GC content as independent variables. The model, which explains 20% of the overall variance in global TE abundance, provides confirmation of our primary conclusions. As expected, this analysis shows no significant effect of recombination rate on this multivariate measure of TE abundance (P ≫ 0.05), whereas there is a highly significant negative effect of coding density (regression coefficient = -0.42, P ≪ 0.05). The model also shows a significant positive effect of base composition (regression coefficient = 0.40, P ≪ 0.05), presumably as a result of the increased GC content close to the centromeres (Table 1).
DISCUSSION
In the genome of the self-fertilizing A. thaliana, there are strong effects of chromosome position on TE abundance, with a large accumulation of TEs close to the centromere. However, although there is a global difference between regions of contrasting recombination, our results suggest that recombination rate is relatively unimportant in causing this pattern, and the effects of selection against TE disruption of gene expression may be the primary mode of selection affecting TE distribution in this self-fertilizing species. Our results suggesting a low rate of ectopic recombination in A. thaliana are in accordance with a recent analysis of long terminal repeat (LTR) retrotransposons in this species (Devos et al. 2002), which indicated that small deletions within elements occurred at a much higher rate than ectopic recombination events between LTRs.
Along the chromosome arms, most TE families show slight positive correlations with recombination rate. This is similar to observations for DNA transposons in the holocentric genome of the self-fertilizing nematode C. elegans, in which a general positive correlation between recombination rate and TE abundance is observed (Duret et al. 2000), whereas no effect of recombination is seen for retrotransposons. This correlation may result from the effects of biased transposition (Duret et al. 2000). However, a negative correlation between recombination and gene density in the euchromatic portions of A. thaliana, and a generally higher gene density in regions of low recombination in C. elegans (Barnes et al. 1995; Wilson 1999) suggest that these effects may at least in part also be driven by selection against insertions near genes.
Under the deleterious insertion model, higher gene density is expected to reduce the abundance of TEs in intergenic regions, due to a higher fraction of regulatory regions. However, our results (Table 2) and those in C. elegans (Duret et al. 2000) suggest that there is also an increase in TE abundance in introns in regions of low gene density. Nevertheless, genome analysis in both A. thaliana and C. elegans has provided evidence for a higher proportion of pseudogenes in regions of low gene density (Copenhaver et al. 1999; Harrison et al. 2001) and a lower proportion of annotated genes in these regions are known to be expressed (C. elegans Sequencing Consortium 1998; Copenhaver et al. 1999), suggesting that this accumulation may also reflect a relaxation of natural selection against the disruption of gene function. Together, the results from both species are consistent with the hypothesis that the mating system plays an important role in the evolution of genome structure.
It is important to consider whether the results could be consistent with alternative selection models to the deleterious effects of gene disruption. First, the results might also be interpreted as consistent with a model of selection against the deleterious expression of TE-derived gene products. In particular, TE insertions in regions of high gene density may show higher levels of TE-derived gene expression due to differences in chromatin structure, and may consequently impose stronger deleterious effects on host fitness. However, a large fraction of TEs in the Arabidopsis genome lack coding capacity, and many appear capable of non-autonomous transposition (Le et al. 2000; Yu et al. 2000). Given that a low proportion of TE insertions with coding capacity are contributing to distribution patterns, the action of selection against TE-derived gene products seems unlikely to provide a primary explanation for our results.
Secondly, an important assumption of our analyses is that current estimates of recombination rates in A. thaliana are reflective of those in which TE accumulation occurred. If recombination rates have changed substantially recently, our ability to detect an effect of recombination on genome evolution may be compromised. Although this possibility cannot be completely excluded, recent comparative mapping in the outcrossing Arabidopsis lyrata (O. Savolainen, pers. comm.), which diverged from A. thaliana -5 Mya (Koch et al. 2000), suggests that map distances are very similar in these two species, indicating that recombination rates have remained relatively stable over this time period.
Thirdly, studies of recombination hotspots in yeast and plants have provided some evidence that recombination preferentially initiates near genes (for review, see Lichten and Goldman 1995; Schnable et al. 1998). If ectopic exchange events are thus more likely to occur close to coding regions, TEs may show a pattern of underrepresentation in regions of high gene density under the ectopic recombination model. Because our recombination rate estimates are estimated over a large scale, we might not observe this local effect of recombination on TE abundance in our analysis. Furthermore, ectopic exchange events between elements in regions of high gene density may be more deleterious to the organism, allowing greater accumulation in regions with fewer genes. Although more understanding of both the fitness effects of ectopic exchange and the relationship between ectopic exchange and recombination initiation are obviously needed, the lack of a negative correlation between recombination rate and TE abundance once gene density is factored out (from partial correlation analysis) makes these interpretations unlikely.
Testing whether ectopic exchange is of general importance, however, will require comparisons of these results with TE distribution patterns in related outcrossing species (Wright et al. 2001), in which ectopic exchange events are potentially more frequent. Recent evidence for local suppression of recombination in TE-rich intergenic regions in maize (Fu et al. 2002) is consistent with models of ectopic exchange, and it might suggest that low levels of ectopic exchange in the intergenic DNA have allowed for the rapid accumulation of retrotransposons (SanMiguel et al. 1998) in this species. Alternatively, however, this local reduction in recombination might imply that TE silencing by DNA methylation and changes in chromatin structure may themselves be a cause of recombination suppression (Gendrel et al. 2002). If so, ectopic recombination may be generally unimportant in driving TE distributions in organisms that suppress TE activity by methylation, as recombination is effectively suppressed within TE-rich regions. Variation among such organisms in the proportion of neutral insertion sites could then be more important than recombination in driving striking differences in the abundance of transposable elements. For example, recent polyploid evolution or high rates of gene duplication could create a large number of functionally redundant targets for TE insertion, leading to relaxed selection against insertions near duplicate genes (Matzke et al. 1999). However, it is unclear whether the action of selection against deleterious insertions can by itself lead to a stable equilibrium in element abundance (Charlesworth and Langley 1989), and other forces may also be important in controlling TE copy number. Possible additional factors include the presence of self-regulated transposition, or stochastic loss of active elements, both of which may be more frequent in highly inbred and asexual populations (Charlesworth and Langley 1986; Wright and Schoen 1999; Morgan 2001; Wright and Finnegan 2001). Nevertheless, the data indicate that selection against the deleterious effects of the disruption of gene expression may be a general force driving patterns of TE distribution, and they suggest the importance of analyses to uncouple recombination rate and gene density effects in other eukaryotic genomes. Finally, although our results suggest that TE insertions near genes are generally under purifying selection, this does not rule out an important role of some TE insertions in the evolution of gene expression (e.g., Daborn et al. 2002), and unusual TE insertions in promoter and coding regions represent potential candidates for adaptive evolution.
METHODS
Genome Analysis
The genome sequence and positions of predicted genes for all five chromosomes of A. thaliana were extracted from The Arabidopsis Information Resource (TAIR—www.arabidopsis.org). The genome sequence is in the form of pseudomolecules, the concatenated sequence of overlapping sequenced clones for each chromosome. These pseudomolecules comprise the entire genome sequence, with the exception of telomeres, the rDNA islands on chromosomes 2 and 4, and several small gaps denoted as N's (Arabidopsis Genome Initiative 2000). Sequences were divided into 100-kb fragments across each chromosome, and information on base composition and annotated (predicted) gene position was collected, excluding a small number of fragments with estimated gaps or terminal sequence leading to a bin size of <50 kb. Each pseudomolecule was submitted to a BLAST search (NCBI standalone BLAST released for Unix on April 3,2001) against our Arabidopsis TE database (www.tebureau.mcgill.ca/), to determine the positions of each TE superfamily along the entire set of chromosomes. TE positions were then verified manually, to eliminate all redundancies and record the presence of nested insertions. For analysis of gene density and the fraction of coding DNA, all annotated coding regions that showed matches solely to TE-encoded gene products were removed. All putative insertions into annotated exons and introns were verified by individual BLAST searches of annotated coding regions. Matches to small internal fragments of TEs, annotated proteins that represent fusions between TE mobility genes and adjacent genes, and matches to host sequences internal to TE insertions were eliminated.
Recombination Rate Estimation
The physical positions were also recorded for those genetic markers that have both been mapped to the Recombinant Inbred (RI) recombination map (see http://nasc.nott.ac.uk/new_ri_map.html), and have been precisely physically mapped on the basis of flanking sequence, using the marker position information from TAIR. To estimate rates of crossing over, the relationship between genetic and physical distance was examined for each chromosome. Because marker density was relatively low close to the centromere, each chromosome was divided into two fragments, with the boundary representing the centromere. Each fragment had an average of 40 markers, with a total of 400 markers used genome wide. Third-order polynomials were fitted to the relationship between genetic and physical distance (in cM/Mb) in each region, similar to the approach of Kliman and Hey (1993; see Supplemental Data). Rates of crossing over could then be estimated by taking the first derivative of this polynomial at any given physical position. With two exceptions, the polynomial fits explained >97% of the variance in genetic distance; the right arm of chromosome 5 had an R2 of 0.96, whereas the left arm of chromosome 2, represented by 19 markers, had an R2 of 0.87. Boundary regions with low marker density between the central regions of low recombination and the chromosome arms were excluded from correlations with recombination rate.
Central regions of reduced recombination, which include the centromeres and heterochromatic knobs, were also delimited using the genetic marker data in order to compare these regions with the rest of the genome, hereafter called the chromosome arms. The central regions have been shown to have a strong reduction in rates of recombination, by recombination rate estimation that is independent of the genome project data used here (Copenhaver et al. 1999; Haupt et al. 2001). Because of low-marker density, comparisons were restricted to central regions of clearly reduced recombination versus the chromosome arms, with boundary regions of low-marker density excluded from the analysis. In the case of chromosome 1, fine-scale estimates of recombination rates in the centromeric region by Haupt et al (2001) allowed the recombination boundary on the short arm to be precisely delimited to BAC clone T22A15. Estimates of average recombination rate along the arms (4.9 cM/MB) using the polynomial fits were very similar to estimates of the average genome recombination rate, 5.0 cM/MB (Haupt et al. 2001). In the central regions of reduced recombination, average recombination rate using polynomial fits was estimated to be 1.5 cM/MB, although this includes regions with much greater recombination rate reduction, up to perhaps complete suppression in the centromere core (Haupt et al. 2001).
Testing Effects of Coding Density on Purifying Selection Against Insertions
We used a maximum likelihood method to test for the presence of selective constraint in intergenic regions. If TE insertions are inserted randomly within intergenic DNA along the chromosome arms, they should follow a Poisson distribution. Specifically, the likelihood of observing n insertions is the product of the likelihoods in each bin i:
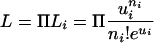 |
Assuming no selective constraint in intergenic regions, the parametric mean μi = (vIi), in which Ii is the number of base pairs of intergenic DNA in bin i estimated directly from the data, and v is the per base pair probability of an insertion. This model assumes complete independence among bins, that is, no local insertion preference, and by constraining v across bins, we assume that TE distribution is random across intergenic DNA. We can calculate the maximum likelihood estimate of v, using the observed values of n and I for each bin. As an alternative model, we allow ui to vary as a function of the observed amount of coding DNA (Ci) in bin i: ui = v(Ii - αCi), where αCi < Ii. Here,
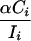 |
is a measure of the proportion of intergenic DNA in bin i that is under selection against TE insertion, and the total proportion of intergenic DNA under selective constraint against TE insertions can be estimated by summing across all bins. We assume that a given insertion site in intergenic DNA is either unconditionally deleterious, with probability
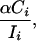 |
or neutral, with probability
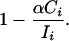 |
By calculating the joint maximum likelihood of v and α, and comparing it with a model in which α = 0, we can test for an effect of coding density on TE distributions. Significant improvement to the likelihood is assessed using the statistic
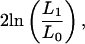 |
in which L0 assumes α = 0, and L1 allows α > 0. This statistic is asymptotically χ2 distributed with 1 degree of freedom. To examine the expected correlation between coding density and TE frequency under a model with these parameter estimates, we also run 10,000 computer simulations of random TE insertion into unconstrained intergenic DNA using the observed sizes of intergenic and coding DNA in each bin. In these simulations, we simply assume a Poisson distributed number of insertions into each bin, with the parametric mean ui in bin i given by the selection equation above. These simulations assume that the only factor influencing TE distribution is random insertion into unconstrained sites.
Acknowledgments
We thank T. Johnson, P. Andolfatto, D. Bachtrog, B. Charlesworth, D. Charlesworth, and D. Schoen for discussion and comments on an earlier version of the manuscript. We also thank three anonymous reviewers for their comments on the manuscript. This work was supported by a National Science and Engineering Research Council (NSERC) of Canada operating and Genomics Project grants to T.E.B., and by a Commonwealth fellowship and an NSERC postgraduate scholarship to S.I.W.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1281503.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- The Arabidopsis Genome Initiative 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815. [DOI] [PubMed] [Google Scholar]
- Barnes, T.M., Kohara, Y., Coulson, A., and Hekimi, S. 1995. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics 141: 159-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolome, C., Maside, X., and Charlesworth, B. 2002. On the abundance and distribution of transposable elements in the genome of Drosophila melanogaster. Mol. Biol. Evol. 19: 926-937. [DOI] [PubMed] [Google Scholar]
- Biemont, C., Tsitrone, A., Vieira, C., and Hoogland, C. 1997. Transposable element distribution in Drosophila. Genetics 147: 1997-1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot, S., Entezam, A., and Furano, A.V. 2001. Selection against deleterious LINE-1-containing loci in the human lineage. Mol. Biol. Evol. 18: 926-935. [DOI] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 282: 2012-2018. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B. and Langley, C.H. 1986. The evolution of self-regulated transposition of transposable elements. Genetics 112: 359-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B. and Langley, C.H. 1989. The population genetics of Drosophila transposable elements. Annu. Rev. Genet. 23: 251-287. [DOI] [PubMed] [Google Scholar]
- Charlesworth, D. and Charlesworth, B. 1995. Transposable elements in inbreeding and outbreeding populations. Genetics 140: 415-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, D. and Wright, S.I. 2001. Breeding systems and genome evolution. Curr. Opin. Genet. Dev. 11: 685-690. [DOI] [PubMed] [Google Scholar]
- Comeron, J.M., and Kreitman, M. 2000. The correlation between intron length and recombination in Drosophila. Dynamic equilibrium between mutational and selective forces. Genetics 156: 1175-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver, G.P., Nickel, K., Kuromori, T., Benito, M.I., Kaul, S., Lin, X., Bevan, M., Murphy, G., Harris, B., Parnell, L.D., et al. 1999. Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286: 2468-2474. [DOI] [PubMed] [Google Scholar]
- Daborn, P.J., Yen, J.L., Bogwitz, M.R., Le Goff, G., Feil, E., Jeffers, S., Tijet, N., Perry, T., Heckel, D., Batterham, P., et al. 2002. A single p450 allele associated with insecticide resistance in Drosophila. Science 297: 2253-2256. [DOI] [PubMed] [Google Scholar]
- Devos, K.M., Brown, J.K., and Bennetzen, J.L. 2002. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12: 1075-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret, L., Marais, G., and Biemont, C. 2000. Transposons but not retrotransposons are located preferentially in regions of high recombination rate in Caenorhabditis elegans. Genetics 156: 1661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush, T.H. and Furano, A.V. 2002. Fruit flies and humans respond differently to retrotransposons. Curr. Opin. Genet. Dev. 12: 669-674. [DOI] [PubMed] [Google Scholar]
- Fu, H., Zheng, Z., and Dooner, H.K. 2002. Recombination rates between adjacent genic and retrotransposon regions in maize vary by 2 orders of magnitude. Proc. Natl. Acad. Sci. 99: 1082-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel, A.V., Lippman, Z., Yordan, C., Colot, V., and Martienssen, R.A. 2002. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297: 1871-1873. [DOI] [PubMed] [Google Scholar]
- Graustein, A., Gaspar, J.M., Walters, J.R., and Palopoli, M.F. 2002. Levels of DNA polymorphism vary with mating system in the nematode genus Caenorhabditis. Genetics 161: 99-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, P.M., Echols, N., and Gerstein, M.B. 2001. Digging for dead genes: An analysis of the characteristics of the pseudogene population in the Caenorhabditis elegans genome. Nucleic Acids Res. 29: 818-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt, W., Fischer, T.C., Winderl, S., Fransz, P., and Torres-Ruiz, R.A. 2001. The CENTROMERE1 (CEN1) region of Arabidopsis thaliana: Architecture and functional impact of chromatin. Plant J. 27: 285-296. [DOI] [PubMed] [Google Scholar]
- Hill, W.G. and Robertson, A. 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8: 269-294. [PubMed] [Google Scholar]
- Jakubczak, J.L., Burke, W.D., and Eickbush, T.H. 1991. Retrotransposable elements R1 and R2 interrupt the rRNA genes of most insects. Proc. Natl. Acad. Sci. 88: 3295-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov, V.V. and Jurka, J. 2001. Rolling-circle transposons in eukaryotes. Proc. Natl. Acad. Sci. 98: 8714-8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliman, R.M. and Hey, J. 1993. Reduced natural selection associated with low recombination in Drosophila melanogaster. Mol. Biol. Evol. 10: 1239-1258. [DOI] [PubMed] [Google Scholar]
- Koch, M., Haubold, M., and Mitchell-Olds, T. 2000. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17: 1483-1498. [DOI] [PubMed] [Google Scholar]
- Kumekawa, N., Hosouchi, T., Tsuruoka, H., and Kotani, H. 2000. The size and sequence organization of the centromeric region of Arabidopsis thaliana chromosome 5. DNA Res. 7: 315-321. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., Fitzhugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Langley, C.H., Montgomery, E., Hudson, R., Kaplan, N., and Charlesworth, B. 1988. On the role of unequal exchange in the containment of transposable element copy number. Genet. Res. 52: 223-235. [DOI] [PubMed] [Google Scholar]
- Le, Q.H., Wright, S., Yu, Z., and Bureau, T. 2000. Transposon diversity in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 97: 7376-7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten, M. and Goldman, A.S. 1995. Meiotic recombination hotspots. Annu. Rev. Genet. 29: 423-444. [DOI] [PubMed] [Google Scholar]
- Long, A.D., Lyman, R.F., Morgan, A.H., Langley, C.H., and Mackay, T.F. 2000. Both naturally occurring insertions of transposable elements and intermediate frequency polymorphisms at the achaete-scute complex are associated with variation in bristle number in Drosophila melanogaster. Genetics 154: 1255-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, H.S. and Henikoff, S. 2002. Conflict begets complexity: The evolution of centromeres. Curr. Opin. Gene. Dev. 12: 711-718. [DOI] [PubMed] [Google Scholar]
- Matzke, M.A., Scheid, O.M., and Matzke, A.J. 1999. Rapid structural and epigenetic changes in polyploid and aneuploid genomes. BioEssays 21: 761-767. [DOI] [PubMed] [Google Scholar]
- McVean, G.A. and Charlesworth, B. 2000. The effects of Hill-Robertson interference between weakly selected mutations on patterns of molecular evolution and variation. Genetics 155: 929-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medstrand, P., Van De Lagemaat, L.N., and Mager, D.L. 2002. Retroelement distributions in the human genome: Variations associated with age and proximity to genes. Genome Res. 12: 1483-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, M.T. 2001. Transposable element number in mixed mating populations. Genet. Res. 77: 261-275. [DOI] [PubMed] [Google Scholar]
- Nekrutenko, A. and Li, W.H. 2001. Transposable elements are found in a large number of human protein-coding genes. Trends Genet. 17: 619-621. [DOI] [PubMed] [Google Scholar]
- Nuzhdin, S.V., Pasyukova, E.G., and Mackay, T.F. 1996. Positive association between copia transposition rate and copy number in Drosophila melanogaster. Proc. R. Soc. Lond. B. Biol. Sci. 263: 823-831. [DOI] [PubMed] [Google Scholar]
- Nuzhdin, S.V., Pasyukova, E.G., and Mackay, T.F. 1997. Accumulation of transposable elements in laboratory lines of Drosophila melanogaster. Genetica 100: 167-175. [PubMed] [Google Scholar]
- Pavlicek, A., Clay, O., and Bernardi, G. 2002. Transposable elements encoding functional proteins: Pitfalls in unprocessed genomic data? FEBS Lett. 523: 252-253. [DOI] [PubMed] [Google Scholar]
- Pelissier, T., Tutois, S., Deragon, J.M., Tourmente, S., Genestier, S., and Picard, G. 1995. Athila, a new retroelement from Arabidopsis thaliana. Plant Mol. Biol. 29: 441-452. [DOI] [PubMed] [Google Scholar]
- Petrov, D.A., Aminetzach, Y.T., Davis, J.C., Bensasson, D., and Hirsh, A.E. 2003. Size matters: Non-LTR retrotransposable elements and ectopic recombination in Drosophila. Mol. Biol. Evol. 20: 880-892. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P., Gaut, B.S., Tikhonov, A., Nakajima, Y., and Bennetzen, J.L. 1998. The paleontology of intergene retrotransposons of maize. Nat. Genet. 20: 43-45. [DOI] [PubMed] [Google Scholar]
- Schnable, P.S., Hsia, A.P., and Nikolau, B.J. 1998. Genetic recombination in plants. Curr. Opin. Plant. Biol. 1: 123-129. [DOI] [PubMed] [Google Scholar]
- Singer, T., Yordan, C., and Martienssen, R.A. 2001. Robertson's Mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene Decrease in DNA Methylation (DDM1). Genes & Dev. 15: 591-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surzycki, S.A. and Belknap, W.R. 1999. Characterization of repetitive DNA elements in Arabidopsis. J. Mol. Evol. 48: 684-691. [DOI] [PubMed] [Google Scholar]
- Virgin, J. B. and Bailey, J.P. 1998. The M26 hotspot of Schizosaccharomyces pombe stimulates meiotic ectopic recombination and chromosomal rearrangements. Genetics 149: 1191-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, R.K. 1999. How the worm was won. The C. elegans genome sequencing project. Trends Genet. 15: 51-58. [DOI] [PubMed] [Google Scholar]
- Wright, S.I. and Finnegan, D. 2001. Genome evolution: Sex and the transposable element. Curr. Biol. 11: R296-R299. [DOI] [PubMed] [Google Scholar]
- Wright, S.I. and Schoen, D.J. 1999. Transposon dynamics and the breeding system. Genetica 107: 139-148. [PubMed] [Google Scholar]
- Wright, S.I., Le, Q.H., Schoen, D.J., and Bureau, T.E. 2001. Population dynamics of an ac-like transposable element in self- and cross-pollinating Arabidopsis. Genetics 158: 1279-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Z., Wright, S.I., and Bureau, T.E. 2000. Mutator-like elements in Arabidopsis thaliana. Structure, diversity and evolution. Genetics 156: 2019-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://nasc.nott.ac.uk/new_ri_map.html; Recombinant Inbred recombination map.
- www.arabidopsis.org; The arabidopsis information resource.
- www.tebureau.mcgill.ca/; Arabidopsis TE database.


