Abstract
Background:
Global hypomethylation has been suggested to cause genomic instability and lead to an increased risk of cancer. We examined the association between the global methylation level of peripheral blood leukocyte DNA and breast cancer among Japanese women.
Methods:
We conducted a hospital-based case–control study of 384 patients aged 20–74 years with newly diagnosed, histologically confirmed invasive breast cancer, and 384 matched controls from medical checkup examinees in Nagano, Japan. Global methylation levels in leukocyte DNA were measured by LUminometric Methylation Assay. Odds ratios (ORs) and 95% confidence intervals (CIs) for the associations between global hypomethylation and breast cancer were estimated using a logistic regression model.
Results:
Compared with women in the highest tertile of global methylation level, ORs for the second and lowest tertiles were 1.87 (95% CI=1.20–2.91) and 2.86 (95% CI=1.85–4.44), respectively. Global methylation levels were significantly lower in cases than controls, regardless of the hormone receptor status of the cancer (all P values for trend <0.05).
Interpretation:
These findings suggest that the global methylation level of peripheral blood leukocyte DNA is low in patients with breast cancer and may be a potential biomarker for breast cancer risk.
Keywords: biomarker, breast cancer, global methylation level, peripheral blood leukocyte DNA
Alterations in DNA methylation, both in specific genes and overall in the genome, have been recognised as among the most important molecular alterations in tumour tissue (Widschwendter and Jones, 2002; Esteller, 2008). Global hypomethylation and regional hypermethylation appear to be hallmarks of human tumours: although regional hypermethylation is clearly observed in multiple distinct genomic regions harbouring tumour-suppressor and mutator genes regulating their transcriptional activities (Ushijima, 2005), global hypomethylation is known to cause genomic instability, reactivation of transposable elements, and loss of imprinting, thereby contributing to the development of cancer (Esteller, 2008). As one example, DNA from breast cancer tissues has been shown to be hypomethylated compared with that from normal tissues (Soares et al, 1999; Jackson et al, 2004).
Recently, epidemiologic studies have investigated whether the global methylation level in peripheral blood cells is a useful biomarker for cancers (Terry et al, 2011). Although most of these potential associations have been examined in case–control studies and require careful interpretation of causality, results have suggested that a lower level of global methylation is associated with an increased risk of a number of cancers, including colorectal adenoma (Pufulete et al, 2003; Lim et al, 2008), head and neck squamous cell cancer (Hsiung et al, 2007), bladder cancer (Moore et al, 2008), gastric cancer (Hou et al, 2010) and breast cancer (Choi et al, 2009). Global methylation level may serve as an integrated and intermediate measure for cancer development, reflecting the accumulation of multiple reversible and irreversible factors including age, sex, lifestyle, and environmental exposures, and genetic polymorphisms (Zhang et al, 2011; Zhu et al, 2012).
Folate and related vitamins B are involved in one-carbon metabolism, which has an important role in DNA synthesis, replication and repair and in DNA methylation (Jacob, 2000). Several reports have shown that a lack of folate or related nutrients causes DNA hypomethylation in humans (Jacob et al, 1998; Rampersaud et al, 2000). Moreover, epidemiological studies have shown that folate intake might protect against the development of some cancers (Adzersen et al, 2003; Lajous et al, 2006), albeit that this association is still controversial (Cho et al, 2003; Stolzenberg-Solomon et al, 2006; Kabat et al, 2008; Ma et al, 2009). Although our data showed no significant association of folate, vitamins B2, B6, and B12, and single-nucleotide polymorphisms (SNPs) on methylenetetrahydrofolate reductase (MTHFR), methionine synthase (MTR) or methionine synthase reductase (MTRR) with breast cancer risk (Ma et al, 2009), these factors may affect the association of global methylation with breast cancer risk.
Global methylation has been measured by several methods, such as measurement of the methylation level of the specific repetitive sequences Alu or LINE-1. In the present study, we adopted the LUminometric Methylation Assay (LUMA), which measures DNA methylation in CCGG sequences and provides a robust estimation of overall 5-mC content in dinucleotide CpG sites in the whole genome (Karimi et al, 2006a, 2006b). LUminometric Methylation Assay has been used in studies of the association of global methylation and breast cancer in other ethnic groups, which showed inconsistent results (Delgado-Cruzata et al, 2012; Xu et al, 2012). Here, we examined global methylation of peripheral blood leukocyte DNA by LUMA in patients with breast cancer and matched controls in Japan.
Materials and Methods
Study subjects
This multicenter, hospital-based case–control study of breast cancer was conducted from May 2001 to September 2005 at four hospitals in Nagano Prefecture, Japan. Details of the study have been described previously (Itoh et al, 2009; Ma et al, 2009). Briefly, the case subjects were a consecutive series of women aged 20–74 years with newly diagnosed, histologically confirmed invasive breast cancer who were admitted to one of the four hospitals during the survey period. Of 412 eligible patients, 405 (98%) agreed to participate. Healthy controls were selected from medical checkup examinees in two of the hospitals and confirmed not to have any cancer, with one control matched for each case by age (within three years) and residential area (city or regional area) during the study period. Among the potential control subjects, one declined to participate and two refused to provide blood samples. Consequently, written informed consent was obtained from 405 matched pairs. The study protocol was approved by the Institutional Review Board of the National Cancer Center, Tokyo, Japan.
Data collection
Participants were asked to complete a self-administered questionnaire, which included questions on demographic characteristics, anthropometric factors, smoking habit, family history of cancer, physical activity, medical history, and menstrual and reproductive history. Information for case subjects was obtained on admission to the hospital. Dietary habits were investigated using a 136-item semi-quantitative food-frequency questionnaire (FFQ), which was developed and validated in a Japanese population (Ma et al, 2009). In the FFQ, participants were questioned about how often they consumed the individual food items (frequency of consumption), as well as relative sizes compared with standard portions. Daily food intake was calculated by multiplying frequency by standard portion and relative size for each food item in the FFQ. Daily intakes of nutrients were calculated using the Fifth Revised and Enlarged Edition of the Standard Tables of Food Composition in Japan (The Council for Science and Technology, 2005). We previously confirmed that the questionnaire's assessment of intake of folate and vitamins B2, B6, and B12 was valid (Ma et al, 2009).
Participants provided blood samples at the time they returned their self-administered questionnaire. Whole blood was collected in a 7-ml EDTA-2Na vacutainer, and serum aliquots from the vacutainer tube with clot accelerators were stored at –80 °C until analysis.
Laboratory analysis
Genomic DNA was extracted from the peripheral blood using a Qiagen FlexiGene DNA Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol, followed by further purification by phenol-chloroform extraction and ethanol precipitation.
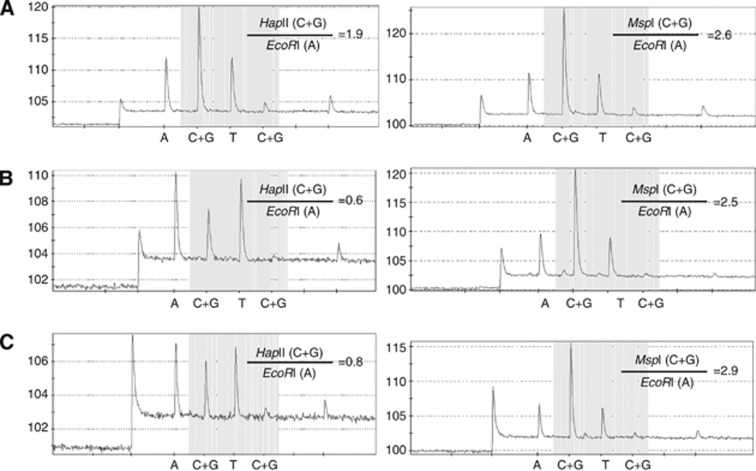
Global DNA methylation was quantified by LUMA. Three hundred nanograms of Genomic DNA was cleaved with HapII+EcoRI or MspI+EcoRI in two separate 200-μl reactions containing 2 μl of 10 × T buffer (330 mM Tris-acetate, 100 mM Mg-acetate, 660 mM K-acetate, 5 mM dithiothreitol), 2 μl of 0.1% BSA, and 5 U of each of the restriction enzymes. The reactions were set up in a PSQ 96 Plate Low (Qiagen) and incubated at 37 °C for 1 h. Then, 20 μl of annealing buffer containing 200 mM Tris-acetate and 50 mM Mg-acetate, pH 7.6, was added to the cleavage reactions, and samples were assayed using the PSQ96MA system (Biotage AB, Uppsala, Sweden). The instrument was programmed to add dNTPs in six steps as follows: step 1, dATPαS; step 2, a mixture of dGTP+dCTP; step 3, dTTP; step 4, a mixture of dGTP+dCTP; step 5, water; and step 6, dATP. Peak heights were calculated using the PSQ96 MA software (Biotage AB). The HpaII/EcoRI and MspI/EcoRI ratios were measured as (dGTP+dCTP)/dATP for each reaction. The HpaII/MspI ratio was then calculated as (HpaII/EcoRI)/(MspI/EcoRI), which corresponds to the proportion of unmethylated CCGG. In this paper, we defined 1-HpaII/MspI as methylation level. Restriction enzymes (HapII, MspI, and EcoRI) were purchased from Takara Bio (1053A, 1150A, and 1040A, respectively; Shiga, Japan). PyroMark Gold Q96 Reagents for pyrosequencing were purchased from Qiagen (972804). DNA quantification was performed using the Quan-iT PicoGreen dsDNA reagent and kit (P7581, Invitrogen, Carlsbad, CA, USA). Figure 1 shows typical Pyrosequencing data by LUMA. The paired samples from cases and matched controls were randomly placed on each plate. The intra-assay coefficients of variation from 20 replications were 6.4% (mean=0.74, standard deviation (s.d.)=0.017).
Figure 1.
Typical pyrograms in LUMA assay of DNA cleaved with HapII+EcoRI (left panels) and MspI+EcoRI (right panels). Methylation levels are derived from 1—{HpaII (C+G)/EcoRI(A)}/{MspI(C+G)/EcoRI(A)}. Genomic DNAs from (A) K-562 cells, (B) Daudi cells, and (C) normal peripheral blood leukocytes.
Five SNPs in MTHFR (rs1801133 and rs1801131), MTR (rs1805087), and MTRR (rs10380 and rs162049) were genotyped by TaqMan SNP Genotyping Assays developed by Applied Biosystems (Foster City, CA, USA). Genotype frequencies were tested for deviation from the Hardy–Weinberg equilibrium with the χ2 test as quality control for genotyping (all P values >0.05).
Statistical analysis
Age-adjusted mean and proportions of baseline characteristics were calculated in case and control subjects. Dietary intakes were adjusted for total energy intake using the residual method (Willett and Stampfer, 1986; Willett, 1998). Characteristics were compared using the Cochran–Mantel–Haenszel test with matched paired strata for categorical variables and the paired t-test for continuous variables.
Global methylation levels were divided into tertiles based on the distribution of methylation levels among controls. Odds ratios (ORs) and 95% confidence intervals (95% CIs) were estimated using a conditional logistic regression model stratified by matched pairs (age and residential area) and adjusted for family history of breast cancer, smoking status, menopausal status, physical activity, number of births, energy-adjusted folate intake, vitamin supplement use, and alcohol drinking. Statistical significance was evaluated with the Wald test. Tertile categories were used as a continuous variable to evaluate linear trends for global methylation levels. Polytomous logistic regression was performed to evaluate the association between methylation level and the hormone receptor status of breast cancer, in which the matching factors are included in the model as covariates. A Wald test for heterogeneity was performed to examine the differences in associations of methylation level and breast cancer risk according to hormone receptor status in breast cancer. Subgroup analysis was performed according to menopausal status, smoking status, intake of folate and vitamins B2, B6, and B12, alcohol drinking, and five SNPs on MTHFR, MTR, or MTRR. A t-test was performed to examine whether global methylation levels differed by these factors in controls. Effect modification was tested using a logistic regression model with the interaction term. All P values were two-sided, with a P value of 0.05 considered statistically significant. We used SAS software (version 9.3, SAS Institute, Cary, NC, USA) for all analyses.
Results
After exclusion of subjects who reported extremely low or high total energy intake (<500 or ⩾4000 kCal) or had no DNA sample, 384 pairs were included in the present analyses.
Table 1 shows the characteristics of the study population. The proportion of women with a family history of breast cancer, ever smokers, and vitamin supplement users was higher in cases than controls, whereas the proportion of those with moderate physical activity in the past 5 years, and high energy-adjusted dietary intake of vitamins B2, B6, B12, and folate was higher in controls.
Table 1. Characteristics of the study population.
| Controls (n=384) | Cases (n=384) | P valuea | |
|---|---|---|---|
| Methylation level by the LUMA method (%), mean (s.d.)b |
70.2 (3.4) |
68.9 (3.5) |
<0.01 |
| Age (years), mean (s.d.) |
54.1 (10.3) |
53.9 (10.7) |
0.07 |
| Postmenopausal women, % |
64 |
56 |
<0.01 |
| Number of births (⩾4 births), % |
3 |
2 |
0.64 |
| Family history of breast cancer, % |
6 |
11 |
0.02 |
| Smoking (ever smoker), % |
7 |
20 |
<0.01 |
| Alcohol drinking (⩾1/week), % |
29 |
26 |
0.28 |
| Moderate physical activity past 5 years (yes), % |
39 |
32 |
0.03 |
| Vitamin supplement user, % |
12 |
18 |
0.02 |
| Age at menarche (years), mean (s.d.)b |
13.2 (1.7) |
13.4 (1.7) |
0.09 |
| History of benign breast disease, % |
8 |
12 |
0.03 |
| Body mass index (kg m−2), mean (s.d.)b |
22.9 (3.2) |
22.7 (3.4) |
0.25 |
|
Dietary intakec | |||
| Vitamin B1 (mg per day), mean (s.d.)b | 1.1 (0.3) | 1.1 (0.3) | 0.13 |
| Vitamin B2 (mg per day), mean (s.d.)b | 1.6 (0.4) | 1.5 (0.4) | 0.01 |
| Vitamin B6 (mg per day), mean (s.d.)b | 1.6 (0.3) | 1.5 (0.3) | 0.04 |
| Vitamin B12 (μg per day), mean (s.d.)b | 8.7 (3.5) | 8.3 (3.1) | 0.03 |
| Folate (μg per day), mean (s.d.)b | 439 (150) | 415 (132) | <0.01 |
| Alcohol (ethanol per week), mean (s.d.)b | 10.3 (24.0) | 8.1 (16.5) | 0.12 |
Abbreviations: LUMA= LUminometric Methylation Assay; s.d.=standard deviation.
P value by the Cochran–Mantel–Haenszel test with matched-pair strata for percent or paired t-test for mean.
Adjusted for age.
Energy-adjusted intake by the residual method.
The mean of global methylation levels was significantly lower in cases (68.9% 95% CI=68.5–69.3%) than controls (70.2% 95% CI=69.8–70.5%). Table 2 shows the association between global methylation level and breast cancer risk, adjusted for the potential confounders. Lower levels of global methylation were associated with a significantly increased risk of breast cancer (trend OR=1.68; 95% CI=1.35–2.09; P value for trend=2.8 × 10−6). We confirmed that the associations did not substantially change using the models, which incorporated the other factors, including body mass index, age at menarche, and history of benign breast disease (trend OR=1.66; 95% CI=1.33–2.07), or vitamin B2, B6, and B12 (trend OR=1.70; 95% CI=1.37–2.13). Compared with women in the highest tertile of global methylation level, ORs for those in the second and lowest tertiles were 1.87 (95% CI=1.20–2.91) and 2.86 (95% CI=1.85–4.44), respectively. There was no significant heterogeneity in the associations of global hypomethylation and breast cancer according to either estrogen receptor status in breast cancer (P value for heterogeneity test=0.155) or progesterone receptor status (P value for heterogeneity test=0.762; Supplementary Table 1). We also evaluated the effect modification of menopausal status and known or probable factors involved in DNA methylation or folate metabolism on the association between global methylation level and breast cancer risk. Of these factors, methylation levels were significantly different according to alcohol drinking (Supplementary Table 2). As shown in Table 3, no significant interaction was observed. In each subgroup, a low level of global methylation tended to be associated with an increased risk of breast cancer.
Table 2. Association between global methylation level and breast cancer risk.
|
Methylation level |
|
||||
|---|---|---|---|---|---|
| Tertile 1 (⩾71.8) | Tertile 2 (69.1–71.8) | Tertile 3 (<69.1) | Trend | P value for trend | |
| Cases/controls |
69/128 |
125/128 |
190/128 |
|
|
| Stratified OR (95% CI)a |
1 (Ref) |
1.70 (1.16–2.50) |
2.67 (1.83–3.88) |
1.63 (1.35–1.96) |
2.9 × 10−7 |
| Multivariate adjusted OR (95% CI)b | 1 (Ref) | 1.87 (1.20–2.91) | 2.86 (1.85–4.44) | 1.68 (1.35–2.09) | 2.8 × 10−6 |
Abbreviations: CI=confidence interval; OR=odds ratio; Ref=reference.
Stratified by matched pairs.
Adjusted for family history of breast cancer (no, yes), smoking (never, past, current), menopausal status (no, yes), physical activity in the last 5 years (no, ⩽2 days per week, ⩽4 days per week, ⩾5 days per week), number of births (0, 1, 2, 3, ⩾4), vitamin use (no, yes), folate intake (continuous), alcohol drinking (non-drinker, occasional drinker, regular drinker of <150 g ethanol per week, regular drinker of ⩾150 g ethanol per week), and stratified by matched pairs.
Table 3. Subgroup analysis of the association between global methylation level and breast cancer risk.
|
Trend |
|||||
|---|---|---|---|---|---|
| Cases/controls | OR | 95% CI | P value for trend | P value for interaction | |
|
Menopausal status | |||||
| Premenopausal | 173/133 | 1.53 | 1.12–2.09 | 7.2 × 10−3 | |
| Postmenopausal |
211/251 |
1.78 |
1.38–2.30 |
8.1 × 10−6 |
0.46 |
|
Smoking | |||||
| Never | 304/354 | 1.66 | 1.35–2.05 | 1.6 × 10−6 | |
| Ever |
76/28 |
1.82 |
0.99–3.36 |
0.054 |
0.78 |
|
Folate (μg per day) | |||||
| ⩽407.4 | 210/192 | 1.55 | 1.18–2.03 | 1.6 × 10−3 | |
| >407.4 |
174/192 |
1.83 |
1.37–2.45 |
4.0 × 10−5 |
0.41 |
|
Vitamin B2 (mg per day) | |||||
| ⩽1.5 | 230/192 | 1.47 | 1.13–1.91 | 4.1 × 10−3 | |
| >1.5 |
154/192 |
1.97 |
1.45–2.66 |
1.1 × 10−5 |
0.15 |
|
Vitamin B6 (mg per day) | |||||
| ⩽1.5 | 208/192 | 1.76 | 1.33–2.33 | 8.0 × 10−5 | |
| >1.5 |
176/192 |
1.61 |
1.22–2.12 |
7.9 × 10−4 |
0.65 |
|
Vitamin B12 (μg per day) | |||||
| ⩽8.0 | 213/192 | 1.64 | 1.25–2.14 | 3.4 × 10−4 | |
| >8.0 |
171/192 |
1.74 |
1.30–2.32 |
1.9 × 10−4 |
0.77 |
|
Alcohol drinking | |||||
| Non-drinker | 240/232 | 1.76 | 1.37–2.26 | 1.0 × 10−5 | |
| Drinker |
142/152 |
1.56 |
1.13–2.14 |
6.3 × 10−3 |
0.56 |
|
MTHFR rs1801131 | |||||
| AA | 252/254 | 1.60 | 1.26–2.02 | 1.2 × 10−4 | |
| AC+CC |
132/130 |
1.87 |
1.32–2.65 |
4.6 × 10−4 |
0.46 |
|
MTHFR rs1801133 | |||||
| CC | 123/112 | 1.89 | 1.31–2.74 | 7.1 × 10−4 | |
| CT+TT |
261/272 |
1.61 |
1.27–2.03 |
6.4 × 10−5 |
0.46 |
|
MTR rs1805087 | |||||
| AA | 235/257 | 1.82 | 1.42–2.34 | 2.2 × 10−6 | |
| AG+GG |
149/126 |
1.45 |
1.05–2.00 |
0.025 |
0.27 |
|
MTRR rs162049 | |||||
| GG | 112/116 | 1.51 | 1.07–2.14 | 0.019 | |
| AG+GG |
272/266 |
1.77 |
1.40–2.25 |
2.6 × 10−6 |
0.46 |
|
MTRR rs10380 | |||||
| CC | 291/302 | 1.78 | 1.42–2.24 | 5.3 × 10−7 | |
| CT+TT | 91/81 | 1.43 | 0.95–2.13 | 0.084 | 0.34 |
Abbreviations: CI=confidence interval; MTHFR=methylenetetrahydrofolate reductase; MTR=methionine synthase; MTRR=methionine synthase reductase; OR=odds ratio.
Adjusted for family history of breast cancer (no, yes), smoking (never, past, current), menopausal status (no, yes), physical activity in the last 5 years (no, ⩽2 days per week, ⩽4 days per week, ⩾5 days per week), number of births (0, 1, 2, 3, 4, >5), vitamin use (no, yes), folate intake (continuous), alcohol drinking (non-drinker, occasional drinker, regular drinker of <150 g ethanol per week, regular drinker of ⩾150 g ethanol per week), age (continuous), residential area (binary).
Discussion
In this case–control study among Japanese women, we found that low levels of global methylation were associated with a significantly increased risk of breast cancer, regardless of the hormone receptor status of the tumour. These associations were not modified by menopausal status, or lifestyle/dietary or genetic factors, which are known to be involved in DNA methylation metabolism, and were analysed in this study. These findings suggest that the global methylation level in leukocyte DNA is a potential biomarker of breast cancer risk.
Our findings are consistent with recent meta-analysis reports (Brennan and Flanagan, 2012; Woo and Kim, 2012) and two retrospective case–control studies for breast cancer (Choi et al, 2009; Cho et al, 2010). Our present and these recent studies also suggest that the mechanisms underlying the associations of global hypomethylation and breast cancer might not be substantially influenced by hormonal factors (Choi et al, 2009; Iwasaki et al, 2012). In addition, findings of associations between the global hypomethylation of leukocyte DNA and the risk of several other cancers might suggest that hypomethylation in leukocyte DNA is a common etiology for cancers of multiple different sites.
In contrast, findings from retrospective case–control studies for breast cancer using LUMA, including our study, have been inconsistent. Xu et al (2012) reported the opposite associations to our findings, namely that LUMA methylation levels were higher in cases than in controls, whereas Delgado-Cruzata et al (2012) reported no difference in LUMA methylation levels between affected and unaffected sisters. Interestingly, the distribution of LUMA methylation levels in the population-based study of Xu et al (2012) was quite different from those in the other two studies, with means (s.d.s) of LUMA global methylation levels of 57.3% (15.7%) in cases and 52.4% (16.7%) in controls. Thus, most of our subjects were in their highest level category. In contrast, the sibling design study in a high-risk population showed a relatively similar range of LUMA methylation levels to ours, with the affected and unaffected sisters having mean (s.d.s) LUMA methylation levels of 67.1% (7.6%) and 67.5% (7.3%), respectively (Delgado-Cruzata et al, 2012). These different results from independent studies may indicate the importance of considering lifestyle and host characteristics in evaluating the associations between global methylation in peripheral blood and cancers. It has been suggested that global methylation in peripheral blood may vary with race/ethnicity and certain lifestyle factors (Zhang et al, 2011; Zhu et al, 2012), suggesting that these factors can affect the associations between global methylation and breast cancer risk. These differences might also have been impacted by differences in the protocols of the LUMA method.
Interpreting possible causal relationships in our findings should be done with caution, as blood samples were collected after the diagnosis of cancer. In particular, associations reported in recent studies, which used pre-diagnostic blood DNA for other cancers, were inconsistent with those from retrospective studies (Nan et al, 2013; Andreotti et al, 2014). In any case, an explanation for the significantly lower global methylation in breast cancer cases remains speculative. Global methylation levels vary with different blood cell types (Wu et al, 2011), although information on the proportion of leukocyte fraction was not available in the present study. Further, the relative proportion of leukocyte fractions may be altered in cancer patients (Terry et al, 2011). It is also theoretically possible that we captured circulating tumour cells or tumour-derived plasma-free DNA (or both; Zhong et al, 2007; Board et al, 2008), which represent global hypomethylation in breast cancer tissues, albeit that these tumour-derived DNAs should be present in only very minute amounts in peripheral blood samples.
Furthermore, a recent prospective study showed LINE-1 hypomethylation was associated with a significantly increased risk of breast cancer (Deroo et al, 2014). In addition, a second study, which included prevalent and incident cancer cases, suggested that global hypomethylation in peripheral blood mononuclear cells may be a useful biomarker for either early detection or cancer risk (Friso et al, 2013). Although the relationship of methylation status between tumour cells and normal cells, including blood cells, remains unclear (Cho et al, 2010), global hypomethylation may represent an integrated exposure signature of multiple known and unknown endogenous and exogenous carcinogenic factors (Zhang et al, 2011; Zhu et al, 2012), which in turn suggests that changes in the global methylation status of leukocytes may be an intermediate step in the causal pathway for breast cancer. It would be interesting to follow time changes in global methylation levels in individuals before and during the course of cancer development in large prospective studies.
In addition to its retrospective case–control design, the present study has several other limitations. We observed significant differences between cases and controls in the several factors, which were reported to be associated with breast cancer or global methylation levels (Ono et al, 2012). However, adjustment for these factors had minimal influence on the association, suggesting little potential for residual confounding. Also, the matched controls were selected from medical checkup examinees; these people may have been health conscious and not representative of the general population, which would have led to selection bias. Because a standard method for assessing global methylation level has not been established, quality control in detecting relatively small differences in global methylation level in peripheral blood leukocytes hinders or prevents comparison among studies with different analytical methods. For instance, previous studies reported that methylation levels in LINE-1 were not correlated with 5-mdC levels or global methylation by LUMA (Choi et al, 2009; Xu et al, 2012); and methylation of the retrotransposon elements (e.g., LINE-1) may not be a reliable surrogate measure of global methylation and may have its own biological functions (Terry et al, 2011). The most appropriate assay for epidemiologic research remains to be determined (Wu et al, 2011; Woo and Kim, 2012). Moreover, our sample size was limited, and the results of subgroup analysis and interaction tests should be interpreted carefully.
In summary, our results suggest that global methylation in leukocyte DNA as analysed by LUMA may be a useful biomarker of breast cancer. The present and previous studies warrant large-scaled prospective studies to elucidate the complex relationship among potential risk factors, changes in global methylation, and breast cancer development.
Acknowledgments
This work was supported by the Ministry of Health, Labor, and Welfare of Japan (Grants-in-Aid for the Third Term Comprehensive Ten-Year Strategy for Cancer Control and for the Research on Applying Health Technology); the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NIBIO); and the Ministry of Education, Culture, Sports, Science, and Technology of Japan and the Japan Society for the Promotion of Science (Grants-in-Aid for Scientific Research on Innovative Areas (221S0001) and for Young Scientists (B) (22700934)). We deeply thank the participants from Nagano Matsushiro General Hospital, Nagano Red Cross Hospital, Nagano Municipal Hospital, and Nagano Hokushin General Hospital. We thank Dr Hiromi Sakamoto for her helpful advice on DNA methylation assay. We also thank Ms Yoko Odaka and Ms Misuzu Okuyama for their technical assistance.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Adzersen KH, Jess P, Freivogel KW, Gerhard I, Bastert G. Raw and cooked vegetables, fruits, selected micronutrients, and breast cancer risk: a case-control study in Germany. Nutr Cancer. 2003;46:131–137. doi: 10.1207/S15327914NC4602_05. [DOI] [PubMed] [Google Scholar]
- Andreotti G, Karami S, Pfeiffer RM, Hurwitz L, Liao LM, Weinstein SJ, Albanes D, Virtamo J, Silverman DT, Rothman N, Moore LE. LINE1 methylation levels associated with increased bladder cancer risk in pre-diagnostic blood DNA among US (PLCO) and European (ATBC) cohort study participants. Epigenetics. 2014;9 (3:404–415. doi: 10.4161/epi.27386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Board RE, Knight L, Greystoke A, Blackhall FH, Hughes A, Dive C, Ranson M. DNA methylation in circulating tumour DNA as a biomarker for cancer. Biomark Insights. 2008;2:307–319. [PMC free article] [PubMed] [Google Scholar]
- Brennan K, Flanagan JM. Is there a link between genome-wide hypomethylation in blood and cancer risk. Cancer Prev Res. 2012;5:1345–1357. doi: 10.1158/1940-6207.CAPR-12-0316. [DOI] [PubMed] [Google Scholar]
- Cho E, Spiegelman D, Hunter DJ, Chen WY, Zhang SM, Colditz GA, Willett WC. Premenopausal intakes of vitamins A, C, and E, folate, and carotenoids, and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2003;12:713–720. [PubMed] [Google Scholar]
- Cho YH, Yazici H, Wu HC, Terry MB, Gonzalez K, Qu M, Dalay N, Santella RM. Aberrant promoter hypermethylation and genomic hypomethylation in tumor, adjacent normal tissues and blood from breast cancer patients. Anticancer Res. 2010;30:2489–2496. [PMC free article] [PubMed] [Google Scholar]
- Choi JY, James SR, Link PA, McCann SE, Hong CC, Davis W, Nesline MK, Ambrosone CB, Karpf AR. Association between global DNA hypomethylation in leukocytes and risk of breast cancer. Carcinogenesis. 2009;30:1889–1897. doi: 10.1093/carcin/bgp143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Cruzata L, Wu HC, Perrin M, Liao Y, Kappil MA, Ferris JS, Flom JD, Yazici H, Santella RM, Terry MB. Global DNA methylation levels in white blood cell DNA from sisters discordant for breast cancer from the New York site of the Breast Cancer Family Registry. Epigenetics. 2012;7:868–874. doi: 10.4161/epi.20830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deroo LA, Bolick SC, Xu Z, Umbach DM, Shore D, Weinberg CR, Sandler DP, Taylor JA. Global DNA methylation and one-carbon metabolism gene polymorphisms and the risk of breast cancer in the Sister Study. Carcinogenesis. 2014;35:333–338. doi: 10.1093/carcin/bgt342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Friso S, Udali S, Guarini P, Pellegrini C, Pattini P, Moruzzi S, Girelli D, Pizzolo F, Martinelli N, Corrocher R, Olivieri O, Choi SW. Global DNA hypomethylation in peripheral blood mononuclear cells as a biomarker of cancer risk. Cancer Epidemiol Biomarkers Prev. 2013;22:348–355. doi: 10.1158/1055-9965.EPI-12-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Wang H, Sartori S, Gawron A, Lissowska J, Bollati V, Tarantini L, Zhang FF, Zatonski W, Chow WH, Baccarelli A. Blood leukocyte DNA hypomethylation and gastric cancer risk in a high-risk Polish population. Int J Cancer. 2010;127:1866–1874. doi: 10.1002/ijc.25190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiung DT, Marsit CJ, Houseman EA, Eddy K, Furniss CS, McClean MD, Kelsey KT. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16:108–114. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- Itoh H, Iwasaki M, Hanaoka T, Kasuga Y, Yokoyama S, Onuma H, Nishimura H, Kusama R, Tsugane S. Serum organochlorines and breast cancer risk in Japanese women: a case-control study. Cancer Causes Control. 2009;20:567–580. doi: 10.1007/s10552-008-9265-z. [DOI] [PubMed] [Google Scholar]
- Iwasaki M, Ono H, Kuchiba A, Kasuga Y, Yokoyama S, Onuma H, Nishimura H, Kusama R, Yoshida T, Tsugane S. Association of postmenopausal endogenous sex hormones with global methylation level of leukocyte DNA among Japanese women. BMC Cancer. 2012;12:323. doi: 10.1186/1471-2407-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson K, Yu MC, Arakawa K, Fiala E, Youn B, Fiegl H, Muller-Holzner E, Widschwendter M, Ehrlich M. DNA hypomethylation is prevalent even in low-grade breast cancers. Cancer Biol Ther. 2004;3:1225–1231. doi: 10.4161/cbt.3.12.1222. [DOI] [PubMed] [Google Scholar]
- Jacob RA. Folate, DNA methylation, and gene expression: factors of nature and nurture. Am J Clin Nutr. 2000;72:903–904. doi: 10.1093/ajcn/72.4.903. [DOI] [PubMed] [Google Scholar]
- Jacob RA, Gretz DM, Taylor PC, James SJ, Pogribny IP, Miller BJ, Henning SM, Swendseid ME. Moderate folate depletion increases plasma homocysteine and decreases lymphocyte DNA methylation in postmenopausal women. J Nutr. 1998;128:1204–1212. doi: 10.1093/jn/128.7.1204. [DOI] [PubMed] [Google Scholar]
- Kabat GC, Miller AB, Jain M, Rohan TE. Dietary intake of selected B vitamins in relation to risk of major cancers in women. Br J Cancer. 2008;99:816–821. doi: 10.1038/sj.bjc.6604540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Johansson S, Ekstrom TJ. Using LUMA: a Luminometric-based assay for global DNA-methylation. Epigenetics. 2006;1:45–48. doi: 10.4161/epi.1.1.2587. [DOI] [PubMed] [Google Scholar]
- Karimi M, Johansson S, Stach D, Corcoran M, Grander D, Schalling M, Bakalkin G, Lyko F, Larsson C, Ekstrom TJ. LUMA (LUminometric Methylation Assay)—a high throughput method to the analysis of genomic DNA methylation. Exp Cell Res. 2006;312:1989–1995. doi: 10.1016/j.yexcr.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Lajous M, Romieu I, Sabia S, Boutron-Ruault MC, Clavel-Chapelon F. Folate, vitamin B12 and postmenopausal breast cancer in a prospective study of French women. Cancer Causes Control. 2006;17:1209–1213. doi: 10.1007/s10552-006-0053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim U, Flood A, Choi SW, Albanes D, Cross AJ, Schatzkin A, Sinha R, Katki HA, Cash B, Schoenfeld P, Stolzenberg-Solomon R. Genomic methylation of leukocyte DNA in relation to colorectal adenoma among asymptomatic women. Gastroenterology. 2008;134:47–55. doi: 10.1053/j.gastro.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma E, Iwasaki M, Kobayashi M, Kasuga Y, Yokoyama S, Onuma H, Nishimura H, Kusama R, Tsugane S. Dietary intake of folate, vitamin B2, vitamin B6, vitamin B12, genetic polymorphism of related enzymes, and risk of breast cancer: a case-control study in Japan. Nutr Cancer. 2009;61:447–456. doi: 10.1080/01635580802610123. [DOI] [PubMed] [Google Scholar]
- Moore LE, Pfeiffer RM, Poscablo C, Real FX, Kogevinas M, Silverman D, Garcia-Closas R, Chanock S, Tardon A, Serra C, Carrato A, Dosemeci M, Garcia-Closas M, Esteller M, Fraga M, Rothman N, Malats N. Genomic DNA hypomethylation as a biomarker for bladder cancer susceptibility in the Spanish Bladder Cancer Study: a case-control study. Lancet Oncol. 2008;9:359–366. doi: 10.1016/S1470-2045(08)70038-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan H, Giovannucci EL, Wu K, Selhub J, Paul L, Rosner B, Fuchs CS, Cho E. Pre-diagnostic leukocyte genomic DNA methylation and the risk of colorectal cancer in women. PLoS One. 2013;8:e59455. doi: 10.1371/journal.pone.0059455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono H, Iwasaki M, Kuchiba A, Kasuga Y, Yokoyama S, Onuma H, Nishimura H, Kusama R, Ohnami S, Sakamoto H, Yoshida T, Tsugane S. Association of dietary and genetic factors related to one-carbon metabolism with global methylation level of leukocyte DNA. Cancer Sci. 2012;103 (12:2159–2164. doi: 10.1111/cas.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pufulete M, Al-Ghnaniem R, Leather AJ, Appleby P, Gout S, Terry C, Emery PW, Sanders TA. Folate status, genomic DNA hypomethylation, and risk of colorectal adenoma and cancer: a case control study. Gastroenterology. 2003;124:1240–1248. doi: 10.1016/s0016-5085(03)00279-8. [DOI] [PubMed] [Google Scholar]
- Rampersaud GC, Kauwell GP, Hutson AD, Cerda JJ, Bailey LB. Genomic DNA methylation decreases in response to moderate folate depletion in elderly women. Am J Clin Nutr. 2000;72:998–1003. doi: 10.1093/ajcn/72.4.998. [DOI] [PubMed] [Google Scholar]
- Soares J, Pinto AE, Cunha CV, Andre S, Barao I, Sousa JM, Cravo M. Global DNA hypomethylation in breast carcinoma: correlation with prognostic factors and tumor progression. Cancer. 1999;85:112–118. [PubMed] [Google Scholar]
- Stolzenberg-Solomon RZ, Chang SC, Leitzmann MF, Johnson KA, Johnson C, Buys SS, Hoover RN, Ziegler RG. Folate intake, alcohol use, and postmenopausal breast cancer risk in the prostate, lung, colorectal, and ovarian cancer screening trial. Am J Clin Nutr. 2006;83:895–904. doi: 10.1093/ajcn/83.4.895. [DOI] [PubMed] [Google Scholar]
- Terry MB, Delgado-Cruzata L, Vin-Raviv N, Wu HC, Santella RM. DNA methylation in white blood cells: association with risk factors in epidemiologic studies. Epigenetics. 2011;6:828–837. doi: 10.4161/epi.6.7.16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Council for Science and Technology: Ministry of Education, Culture, Sports, Science, and Technology, Japan . Standard Tables of Food Composition in Japan, Fifth revised and enlarged edition edn. National Printing Bureau: Tokyo; 2005. [Google Scholar]
- Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat Rev Cancer. 2005;5:223–231. doi: 10.1038/nrc1571. [DOI] [PubMed] [Google Scholar]
- Widschwendter M, Jones PA. DNA methylation and breast carcinogenesis. Oncogene. 2002;21:5462–5482. doi: 10.1038/sj.onc.1205606. [DOI] [PubMed] [Google Scholar]
- Willett W, Stampfer MJ. Total energy intake: implications for epidemiologic analyses. Am J Epidemiol. 1986;124:17–27. doi: 10.1093/oxfordjournals.aje.a114366. [DOI] [PubMed] [Google Scholar]
- Willett WC.1998Nutritional Epidemiology2nd ednOxford University Press: New York [Google Scholar]
- Woo HD, Kim J. Global DNA hypomethylation in peripheral blood leukocytes as a biomarker for cancer risk: a meta-analysis. PLoS One. 2012;7:e34615. doi: 10.1371/journal.pone.0034615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HC, Delgado-Cruzata L, Flom JD, Kappil M, Ferris JS, Liao Y, Santella RM, Terry MB. Global methylation profiles in DNA from different blood cell types. Epigenetics. 2011;6:76–85. doi: 10.4161/epi.6.1.13391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Gammon MD, Hernandez-Vargas H, Herceg Z, Wetmur JG, Teitelbaum SL, Bradshaw PT, Neugut AI, Santella RM, Chen J. DNA methylation in peripheral blood measured by LUMA is associated with breast cancer in a population-based study. FASEB J. 2012;26 (6:2657–2666. doi: 10.1096/fj.11-197251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang FF, Cardarelli R, Carroll J, Fulda KG, Kaur M, Gonzalez K, Vishwanatha JK, Santella RM, Morabia A. Significant differences in global genomic DNA methylation by gender and race/ethnicity in peripheral blood. Epigenetics. 2011;6:623–629. doi: 10.4161/epi.6.5.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong XY, Ladewig A, Schmid S, Wight E, Hahn S, Holzgreve W. Elevated level of cell-free plasma DNA is associated with breast cancer. Arch Gynecol Obstet. 2007;276:327–331. doi: 10.1007/s00404-007-0345-1. [DOI] [PubMed] [Google Scholar]
- Zhu ZZ, Hou L, Bollati V, Tarantini L, Marinelli B, Cantone L, Yang AS, Vokonas P, Lissowska J, Fustinoni S, Pesatori AC, Bonzini M, Apostoli P, Costa G, Bertazzi PA, Chow WH, Schwartz J, Baccarelli A. Predictors of global methylation levels in blood DNA of healthy subjects: a combined analysis. Int J Epidemiol. 2012;41:126–139. doi: 10.1093/ije/dyq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.