Summary
Factors that influence the orientation of the mitotic spindle are important for the maintenance of stem cell populations and in cancer development. However, screening for these factors requires rapid quantification of alterations of the angle of the mitotic spindle in cultured cell lines. Here we describe a method to image mitotic cells and rapidly score the angle of the mitotic spindle using a simple MATLAB application to analyze a stack of Z-images.
Keywords: mitotic spindle, microtubules, MATLAB, DeltaVision
1. Introduction
Correct orientation of the mitotic spindle in dividing cells is crucial for asymmetric cell division during development as well as for the maintenance of stem cell populations. However, it is often simplest to screen for gene products involved in this process by assaying for quantitative changes in the orientation of mitotic spindles in cultured cell lines. Cultured transformed cell lines are often the system of choice for these measurements because they are easier to grow reproducibly than polarized cell sheets and easier to image than cells in tissues. As spindle orientation may exhibit baseline randomness in cultured cells, enough cells must be assayed to determine whether a particular gene product or treatment is influencing spindle orientation to a significant extent. Cell lines also have the advantage that they can be transfected with proteins to rescue spindle orientation and confirm siRNA depletions. Cultured cells also lack the intrinsic apical and basolateral polarization found in some epithelial model systems for cell polarization which can be an advantage if treatments lead to changes in cell motility or cell adhesion which could complicate the spindle orientation assays. Thus, even though cultured transformed cell lines may not functionally rely on oriented spindles, they still exhibit an orientation relative to the coverslip that is consistent enough to use to screen for the machinery and regulators of spindle orientation.
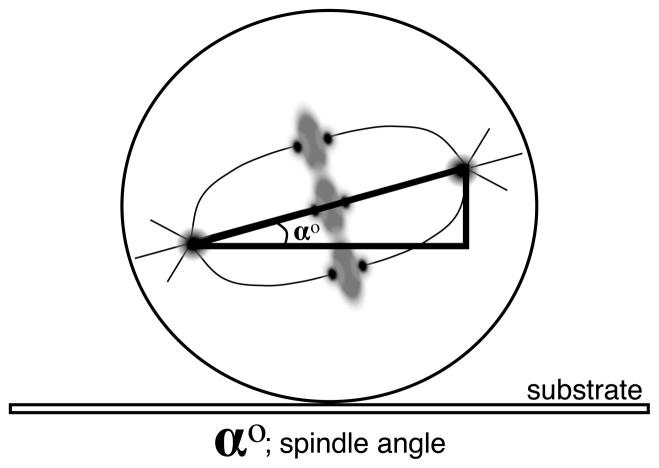
The mitotic spindles of cultured HeLa cells tend to naturally orient to an angle of between 5 and 15 degrees relative to the substrate (Fig. 1). Experimental treatments and loss of the machinery for mitotic spindle orientation can lead to an increase of 30 to 50 degrees in the angle of the mitotic spindle relative to the substrate[1, 2]. Alternate methods for scoring the orientation of the mitotic spindle and cleavage plane have successfully employed patterned substrates[3, 4] or in situ imaging relative to a reference orientation[5] to quantify mitotic spindle orientation. These can be quite effective but are challenging either from the standpoint of cost (for the production of patterned substrates) or with respect to imaging within tissue. Both of these challenges limit the speed and number of cells that can be effectively scored. In contrast, evaluating the orientation of the spindle relative to the cultured substrate requires no special equipment, utilizes standard cell culture methods, and can be applied to common cell lines.
Fig. 1.
Angle (α°) of the long axis of the mitotic spindle generally is 5–15° in cultured HeLa cells. Significant alterations in this angle are often correlated with loss or impairment of the spindle orientation machinery.
2. Materials
2.1 Cell Culture
2.1.1 Cell Culture Reagents
No. 1.5 glass coverslips (12 mm round) (Electron Microscopy Sciences) Acid washed by incubating in 1N HCl solution for 2 hrs, swirling occasionally. Decant acid solution and wash extensively with dH2O 5 times. Wash an additional 5 times with 95% EtOH. Coverslips should be stored in 95% EtOH and flamed prior to use.
10cm2 Falcon polystyrene tissue culture plates.
24-well Falcon polystyrene tissue culture plates.
Fetal bovine serum (FBS) hydroclone, 40 nm filtered. Stored in 50 mL aliquots at −20°C.
Pen-Strep (Gibco) 100X working solution.
L-glutamine.
MEM-alpha (Gibco): Mix 1 10g package (makes 1 L total media) with 850 mLs dH2O, stir to mix. Add 2.2 g NaHCO3 and stir to mix and then adjust to desired pH. Bring volume up to 900 mL with dH2O. Add 100 mL FBS (10% final concentration) and filter sterilize using vacuum-driven filtration system such as 0.22 μm Millipore Durapore bottles. Pen-Strep and L-glutamine can be added prior to filtration if desired.
0.05% Trypsin/EDTA for HeLa cells; 0.25% Trypsin/EDTA for Hct116 cells. (Gibco).
Sterile phosphate buffered saline (PBS): 0.137M NaCl, 2.68 mM KCl, 8 mM Na2HPO4-7H2O, 1.47 mM KH2PO4. PBS is sterilized by autoclaving. 10. Cultured cell lines HeLa, Hct 116, hTERT RPE-1 (see Note 1)
2.1.2 Cell Culture Equipment
Incubator set at 37°C and 5% CO2 for cell maintanence.
Water bath set to 37°C for warming media.
Tissue culture hood.
Microscope.
Hemocytometer.
2.2 Fixation and Labeling
2.2.1 Fixation Reagents
16% paraformaldehyde solution Electron Microscopy Sciences.
Methanol at −20°C.
Donkey serum (Jackson ImmunoResearch): Stored at −20 °C at 60 mg/mL in ddH2O.
Tris-buffered saline (TBS): 50mM Tris, 150mM NaCl. Adjust pH to 7.6. (see Note 3).
Fixative: 4% paraformaldehyde diluted into −20°C methanol (see Note 4).
Blocking Buffer: 20% donkey serum in TBS.
Antibody Dilution Buffer (AbDil): 1X TBS, 1% BSA (Jackson ImmunoResearch), 0.1% Triton-X 100, 0.01% Azide.
Mouse Monoclonal Alpha-tubulin (DM1a) antibodies (Sigma) used at 1:500. Diluted in AbDil.
Mouse Monoclonal Gamma-tubulin antibodies (Sigma): used at a concentration of 1:1000. Diluted in AbDil.
Donkey anti-mouse secondary antibodies with TRITC or FITC labels.
2.2.2 Fixation Equipment
Thomas cover glass staining outfit (Thomas Scientific).
Parafilm.
15 cm2 Petri dish.
Kimwipes.
Beaker (200 mL).
Dumont negative action style tweezers. Style N4 (Electron Microscopy Sciences).
2.3 Imaging
2.3.1 Imaging Reagents
Vectashield mounting medium with DAPI (Vector Labs).
Slides.
2.3.2 Imaging Equipment
DeltaVision deconvolution microscope
2.4 Image Analysis
2.4.1 Image Analysis Software Introduction
To generate meaningful statistics that describe the distribution of spindle angles adopted by cells under a certain set of conditions, large numbers of images (~100) must be analyzed per condition. Doing this manually can be extremely tedious. A better alternative is to write software that automates the identification of spindle poles in batches of image Z-stacks with only occasional direction from the user. Here we describe such software written in MATLAB.
2.4.2 Spindle Pole Location Algorithm
To measure spindle angles, both spindle poles must be located. Labeling gamma tubulin makes this easy since the two poles are often the brightest objects in the image stack, but this is not always true. Our algorithm selects the two brightest objects by default but also provides other candidate poles from which the user can manually select in the event that the simplest assumption is not true.
In the first step, the algorithm creates a maximum intensity projection of the Z-stack. Doing so makes the localization less computationally expensive and makes it much easier to display the results for inspection by the user. The algorithm then integrates the maximum Z intensities over a sliding 5×5 (about 500 nm × 500 nm) pixel window in X and Y. Locations where those integrated intensities are greater than any other integrated intensity within 15 pixels are candidate poles. The two candidate poles with the brightest integrated intensities are selected by the algorithm as the true poles.
% ‘spindle’ is a 3D double array storing the image stack.
% the images are 256x256.
flatspindle = max(spindle,[],3);
intensitysum = zeros(252,252);
for i = 1+2:256-2
for j = 1+2:256-2
roi = flatspindle(i-2:i+2,j-2:j+2);
intensitysum(i,j) = sum(roi(:));
end
end
candidatepoles = zeros(1,4);
m = 0;
for i = 1+15:252-15
for j = 1+15:252-15
roi = intensitysum(i-15:i+15,j-15:j+15)
if intensitysum(i,j) == max(roi(:))
m = m + 1;
% [Y,X,(Z placeholder),Intensity] data is stored.
candidatepoles(m,:) = [i,j,0,intensitysum(i,j);
end
end
end
[scores,I] = sort(candidatepoles(:,4));
finalpoles = I(end-1:end);
Once the final pole objects have been identified in the 2D maximum intensity projection, the objects can be located in Z by finding the maximum integrated intensity within a 5×5×3 vertically sliding cube centered where the poles were found.
pmax = 0;
for l = 1:size(candidatepoles,1)
for k = 2(size(spindle,3)/3)-1
roi = spindle(candidatepoles(l,1)-
2:candidatepoles(l,1)+2,candidatepoles(l,2)-
2:candidatepoles(l,2)+2,k-1:k+1);
sroi = sum(roi(:));
if sroi > pmax
candidatepoles(l,3) = k;
pmax = sroi;
end
end
end
The result is an array, candidatepoles, that stores all of the location information for the pole-like objects (Y,X,Z,Intensity) and a vector, finalpoles, that contains the indices of the two candidate poles that the program will accept for now as the true spindle poles. Once the user has vetted this selection, the position information can be exported in spreadsheet form using a function like xlswrite. The spindle length and angle are easy to calculate at this point.
% [x1,y1,z1] and [x2,y2,z2] are the positions of poles 1 and 2 % pixelsize is the size of a pixel in microns % zscanwidth is the distance between image slices in microns vect = [pixelsize*(x1-x2),pixelsize*(y1-y2),zscanwidth*(z1-z2)]’; L = sqrt(vect(1)^2 + vect(2)^2); spindlelength = sqrt(vect’*vect); spindleangle = (180/pi)*atan(abs(vect(3))/L);
2.4.3 The GUI
In our experience, the algorithm described above correctly identifies the spindle poles about 99% of the time. Nevertheless, most users will want to be able to correct erroneous results in a manner that is not painstaking. This is the purpose of the graphical user interface (GUI). We cannot describe the ~1100 lines of code in great detail here, but we can give an overview of its construction.
2.4.3.1 Creating a GUI
A GUI is simply a customized MATLAB figure with user interface controls. To create one programmatically, you need to write an m-file with instructions for the design of the interface and the functions that execute when buttons are pressed. To combine all of this information into one file, create a master function of the same name as the file that calls the appropriate subfunction. The code below creates a small part of the interface and should be instructive as to how the rest is generated. The MathWorks website has excellent tutorials available for GUI construction.
%% -- SpindleGUI.m --
%% This is the master function carrying the SpindleGUI name.
%% The ‘action’ argument is the name of the sub-function called.
function SpindleGUI(action)
if nargin < 1
%% if no ‘action’ is specified, the initialization function
%% runs
InitializeSpindleGUI
else
feval(action)
end
end
%% This is a partial look at the initialization function
function InitializeSpindleGUI
%% The SpindleGUI figure declaration. The figure handle is stored
%% in ‘SF’.
SF = figure (‘Name’,’SpindleGUI’),…
‘NumberTitle’,’off’,…
‘Position’,
[50,50,1225,800]
,…
‘Resize’,’off’,…
‘MenuBar’,’none’);
%% Axes are placed on the SpindleGUI with this declaration. The
%% axes handle is stored in the ‘ud’ structure.
ud.Axes = axes (‘Parent’, SF,…
‘Units’,’Pixels’,…
‘Position’,
[450,50,700,700]
,…
‘Box’,’on’,…
‘XTick’,[],…
‘YTick’,[]);
%% This button will pull up a file dialog box for the user to
%% select the file to run the algorithm on. Notice the Callback
%% property stores a SpindleGUI command to be executed by the
%% master function. The subfunction will be written below this
%% initialization function.
ud.FileButton = uicontrol(‘Style’,’Push’,…
‘Position’,
[50,500,100,100]
,…
‘String’,’File’,…
‘FontSize’,24…
‘Callback’,’SpindleGUI(“FileButtonCallback”);’);
%% Define a default filepath to retrieve image stack data.
ud.FilePath = ‘My Documents\Data\’;
%% Create a blank image object to store projection images later.
ud.SpindleImage = imagesc(zeros(256,256)); colormap gray;
set(ud.Axes,’XTick’,[],’YTick’,[]);
%% The ‘ud’ structure is written to the Spindle figure userdata
%% property so that it can be recalled by other functions later.
set(SF,’UserData’,ud);
end
%% This is the function that executes when the file button is
%% pressed. The user will first select a file from a dialog box.
%%Then the max Z projection will be calculated and placed on the
%% axes, and the pole locating algorithm will be executed.
function FileButtonCallback
%% Recall the figure’s user data.
ud = get(gcbf,’UserData’);
%% The code for the dialog box. The conditional handles the case
%% that the user clicks ‘cancel’ on the dialog box.
[filename,pathname] = uigetfile([ud.FilePath,’*.tif’]);
if filename == 0
return
end
%% Store the new filepath as the GUI default.
ud.FilePath = pathname;
%% Redefine filename to include the full file path.
filename = [pathname,filename];
%% Query the file for information about its length. Our files are
%% RGB, but gamma tubulin is only in the green channel. We will
%% discard the other two.
info = imfinfo(filename);
L = length(info);
ud.NZScans = L/3;
spindle = zeros(256,256,L/3);
%% Store all of the green channel images in the spindle array.
j = 0;
for i = 2:3:L
j = j + 1;
spindle(:,:,j) = imread(filename,i);
end
%% Convert the spindle array to type double for computation.
spindle = double(spindle);
%% Execute the algorithm reproduced in the earlier sections of
%% this chapter.
3. Methods
3.1 General Cell Culture
Each of the above human cell lines can be routinely cultured in MEM-alpha media plus 10% FBS supplemented with Pen-Strep and l-glutamine.
3.1.1 Splitting and plating cells for analysis
Remove culture medium from cells plated in 10cm2 dish and rinse with 10 mL sterile PBS.
Add 1 mL Trypsin/EDTA and incubate at 37 °C 10 minutes to allow cells to come off the dish.
Add 4 mL MEM-Alpha to inactivate trypsin and triturate cells.
Add a sterile 12 mm2 coverslip to each well of a 24-well plate.
Add 0.5 mL of media to each well.
Remove 9 μL of cell suspension and count cells in a hemocytometer.
Add 25,000 to 35,000 cells per cm2 to each well containing a coverslip (see Note 2).
Culture in a CO2 incubator for a maximum of 48 hours.
3.2 Fixation and labeling for fluorescence microscopy
3.2.1 Fixation
Place Thomas cover glass staining outfit in a 200 mL glass beaker.
Add 100 mLs of fixative to the beaker.
Using negative action tweezers, pull coverslips out of 37°C media and place directly in the fixative, settling them into the rack maintaining a consistent orientation. Incubate in fixative for 10 minutes.
Transfer rack to a 200 mL beaker containing 80 mLs TBS. Wash twice in TBS for 5 minutes each.
3.2.2 Immunofluorescence Labeling
Prepare a chamber for incubating the coverslips by putting a square piece of parafilm on the bottom of a 15 cm2 Petri dish. Tear a Kimwipe in half, twist each half into a strip and place around the inside edge of the dish. Moisten the Kimwipe strips with distilled water.
Using tweezers, place the coverslips cell side up on the parafilm in the 15 cm2 dish prepared in step 1.
Add 20 μL of 20% blocking buffer to each coverslip.
Cover chamber with Petri dish lid and incubate for 45–60 minutes at room temperature or a temperature equivalent to that which you will use for antibodies.
Return coverslips to the Thomas cover glass staining outfit and wash twice more in 80 mLs TBS for 5 minutes each.
Remove coverslips one at a time and return them to the 15 cm2 dish. Wick off excess TBS with a Kimwipe.
Add 6 μL each of gamma-tubulin antibody to the coverslips. Incubate for one hour at room temperature.
Repeat step 5.
Repeat step 6.
Add 6 μL of the secondary antibody with the fluorophore of your choice (we use FITC) to each coverslip. Incubate for one hour at room temperature.
Repeat step 5 with 3 washes instead of 2.
Repeat steps 6–11 with the alpha-tubulin primary antibody and TRITC-labeled secondary antibody.
Place a small drop (~10 μL) of Vectashield on a glass slide (one drop for each coverslip). For imaging with a DeltaVision deconvolution microscope, do not mount more than 3 coverslips per slide.
Acknowledgments
This work was supported by NIH grant GM69429 to LW.
Footnotes
HeLa (ATCC® CCL-2™), Hct 116 (ATCC® CCL-247™), or hTERT RPE-1 (ATCC® CRL-4000™) cell lines are available from the American Type Culture Collection (ATCC). HeLa cells require Biosafety Level 2 cell culture conditions because the cells contain human papillomavirus. Hct 116 human colorectal cancer cells and hTERT RPE-1 telomerase immortalized human retinal pigmented epithelial cells are both cultured at Biosafety Level 1. hTERT RPE-1 cells require a license agreement for commercial customer uses. hTERT RPE-1 cells are distributed for research purposes only. A signed addendum to the ATCC Material Transfer Agreement must be sent to ATCC in advance of shipment.
At this point, cells can be transfected with DNA constructs or siRNA based on individual protocols to screen for effects on the spindle orientation machinery.
If TBS is to be used exclusively with fixed cells, then 0.05% sodium azide can be added as a preservative and the TBS stored at room temperature.
This fixative can be stored at −20°C
References
- 1.Thaiparambil JT, Eggers CM, Marcus AI. AMPK regulates mitotic spindle orientation through phosphorylation of myosin regulatory light chain. Mol Cell Biol. 2012;32(16):3203–17. doi: 10.1128/MCB.00418-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toyoshima F, et al. PtdIns(3,4,5)P3 regulates spindle orientation in adherent cells. Developmental cell. 2007;13(6):796–811. doi: 10.1016/j.devcel.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 3.Fink J, et al. External forces control mitotic spindle positioning. Nature cell biology. 2011;13(7):771–8. doi: 10.1038/ncb2269. [DOI] [PubMed] [Google Scholar]
- 4.Kiyomitsu T, I, Cheeseman M. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nature cell biology. 2012;14(3):311–7. doi: 10.1038/ncb2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Godin JD, et al. Huntingtin is required for mitotic spindle orientation and mammalian neurogenesis. Neuron. 2010;67(3):392–406. doi: 10.1016/j.neuron.2010.06.027. [DOI] [PubMed] [Google Scholar]