Abstract
Osteocytes, positioned within bone's porous structure, are subject to interstitial fluid flow upon whole bone loading. Such fluid flow is widely theorized to be a mechanical signal transduced by osteocytes, initiating a poorly understood cascade of signaling events mediating bone adaptation to mechanical load. The objective of this study was to examine the time course of flow-induced changes in osteocyte gene transcript and protein levels using high-throughput approaches. Osteocyte-like MLO-Y4 cells were subjected to 2 hours of oscillating fluid flow (1 Pa peak shear stress) and analyzed following 0, 2, 8, and 24 hours post-flow incubation. Transcriptomic microarray analysis, followed by gene ontology pathway analysis, demonstrated fluid flow regulation of genes consistent with both known and unknown metabolic and inflammatory responses in bone. Additionally, two of the more highly up-regulated gene products—chemokines Cxcl1 and Cxcl2, supported by qPCR—have not previously been reported as responsive to fluid flow. Proteomic analysis demonstrated greatest up-regulation of the ATP-producing enzyme NDK, calcium-binding Calcyclin, and G protein-coupled receptor kinase 6. Finally, an integrative pathway analysis merging fold changes in transcript and protein levels predicted signaling nodes not directly detected at the sampled time points, including transcription factors c-Myc, c-Jun, and RelA/NF-κB. These results extend our knowledge of the osteocytic response to fluid flow, most notably up-regulation of Cxcl1 and Cxcl2 as possible paracrine agents for osteoblastic and osteoclastic recruitment. Moreover, these results demonstrate the utility of integrative, high-throughput approaches in place of a traditional candidate approach for identifying novel mechano-sensitive signaling molecules.
Keywords: osteocyte, mechanotransduction, fluid flow, shear stress, signaling
1. Introduction
Mounting evidence suggests that osteocytes, positioned within bone mineral's interstitial space, coordinate cellular remodeling leading to functional adaptation in response to mechanically-induced stimuli (Bonewald, 2011; Schaffler et al., 2014). Interstitial fluid flow is one such stimulus (Kufahl and Saha, 1990; Weinbaum et al., 1994) observed upon cyclic whole bone loading (Knothe Tate and Knothe, 2000; Price et al., 2011). Fluid flow exposes osteocytes to enhanced solute transport (Price et al., 2011), streaming potentials (Cowin et al., 1995), and cyclic fluid shear stress transduced via cellular adhesion molecules, the cell membrane, the actin cytoskeleton, and possibly primary cilium (Bonewald and Johnson, 2008).
In vitro, specific responses of osteocytic cells to steady, pulsating, and oscillating fluid flow (OFF) have been widely studied. However, these studies are generally constrained to a limited set of candidate genes or proteins as part of known or suspected signaling pathways. Osteocytic cells elastically deform when subject to physiological peak fluid shear stresses up to 5 Pa (Kwon and Jacobs, 2007; Price et al., 2011), initiating the rapid release of adenosine triphosphate (ATP) and prostaglandin E2 via gap junctions or hemichannels (Batra et al., 2012; Cherian et al., 2005; Genetos et al., 2007; Klein-Nulend et al., 1995). In turn, these factors contribute to cell network propagation of intracellular calcium waves in proportion to flow-induced shear stress in vitro (Huo et al., 2008; Lu et al., 2012) as well as during in situ dynamic bone loading (Jing et al., 2013). Subsequent to OFF, osteocytic cells demonstrate stress amplitude-, frequency-, and duration-dependent shifts in mRNA levels with increased prostaglandin E2-synthesizing Ptgs2 (Cox-2) and a decreased Rankl/Opg mRNA ratio associated with reduced recruitment of bone-resorbing osteoclasts (Kim et al., 2006; Li et al., 2012; Xiong and O’Brien, 2012). Fluid flow also regulates molecules involved with Wnt/β- catenin pathway activation (Kamel et al., 2010; Santos et al., 2009) and protects against osteocyte apoptosis (Cheung et al., 2011; Kitase et al., 2010).
Related studies have broadened our knowledge of skeletal mechano-sensing through transcriptomic (Mantila Roosa et al., 2011; McKenzie et al., 2011; Reijnders et al., 2013; Rolfe et al., 2014; Xing et al., 2005) or proteomic investigations (Li et al., 2011; Zhang and Wang, (2009) of heterogeneous cell populations from whole bones subject to in vivo mechanical loading. Others have analyzed global gene expression specifically in osteocytes isolated from loaded rat trabeculae (Wasserman et al., 2013) and osteocyte-like MLO-Y4 cells subjected to cyclic compressive force stimulation (Chen et al., 2010). However, no single study has taken an integrated transcriptomic and proteomic approach.
In this study we evaluated the utility of two unbiased high-throughput approaches, gene transcript microarrays and protein mass spectrometry, to investigate the response of osteocytes exposed to fluid flow. We mapped a time course of flow-induced fluctuations in both gene transcript levels and protein abundances at corresponding time points. Additionally, in spite of various post-transcriptional modifications, we computationally predicted sequences of critical signaling nodes using an integrative bioinformatics approach. We examined the hypothesis that this broadened inquiry will reveal a mechano-sensitive shift in gene transcript and protein abundances in response to fluid flow, reflecting regulation of known mechano-sensitive signaling pathways as well as novel signaling networks. Our results demonstrate both individual and global shifts in signaling molecules consistent with known regulation of bone metabolism. More importantly, we identified signaling molecules and pathways not previously implicated in mechanotranduction in bone, most notably, up-regulation of Cxcl1 and Cxcl2.
2. Methods
2.1. Cell culture
MLO-Y4 osteocyte-like cells (Kato et al., 1997), courtesy of Dr. Lynda Bonewald (University of Missouri—Kansas City) were maintained in normal growth medium (α-MEM [Invitrogen, Grand Island, NY] with 2.5% CS [Hyclone, Logan, UT], 2.5% FBS [Lonza, Walkersville, MD], 1% Penicillin/Streptomycin) throughout all portions of the experiment. Cells were seeded 48 hours prior to fluid flow on 75 × 38 × 1 mm glass slides coated with 300 μg/ml Type I Collagen (BD Biosciences, Bedford, MA) for 1 hour and washed. Cell seeding density was 1.35 × 104 cells/cm2 so that upon flow exposure, cells were roughly 60% confluent and interconnected by dendritic processes.
2.2. Fluid flow stimulation
Samples were subjected to 2 hours of fully-reversed sinusoidal OFF with a peak shear stress of 1 Pa (10 dynes/cm2) at a frequency of 1 Hz. As previously described (Haut Donahue et al., 2004; Jacobs et al., 1998), glass slides were assembled within parallel plate flow chambers in sterile conditions and placed within an incubator at 37°C and 5% CO2. A rigid-walled inlet tube connected to a Hamilton glass syringe was actuated by a servo pneumatic loading device (EnduraTec, Eden Prairie, MN). Paired controls were maintained in identical, static chambers. After two hours treatment, flowed and static cells were either collected immediately (0 hour time point) or glass slides were transferred to culture dishes and incubated for 2, 8, or 24 hours post-flow in 10 ml fresh medium. When collecting samples, medium was aspirated and cells were lysed directly on glass slides using RLT buffer (Qiagen, Valencia, CA). For each time point, triplicate samples were collected on separate days.
2.3. RNA and protein isolation
Total RNA was isolated using an Rneasy Mini Kit (Qiagen, Valencia, CA) per manufacturer instructions. Protein was collected from the same samples according to Qiagen's supplementary protocol for acetone precipitation using buffer RLT lysate flow-through from RNeasy spin columns. Four volumes of acetone were added to flow-through followed by 30 minutes incubation at −20°C to induce protein precipitation. Samples were centrifuged at 20,000 g for 10 minutes, supernatant discarded, pellet washed with ice-cold ethanol, dried and resuspended in 8 M urea.
2.4. DNA microarray analysis
Analysis of total RNA was carried out as in Waters, et al. (Waters et al., 2011). First, quality was verified using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Biotin-labeled cRNA was synthesized and fragmented using Affymetrix 3' IVT Express reagents for hybridization to Mouse Genome 430A 2.0 GeneChips (Affymetrix, Santa Clara, CA). After hybridization, the arrays were washed and stained with streptavidin-phycoerythrin, and scanned at a resolution of 2.5 microns using an Affymetrix GeneChip Scanner 3000. Quality control parameters were assessed throughout to assure maximum efficiency of transcription, integrity of hybridization, and consistency of qualitative calls. Synthesis and fragmentation of cRNA were assessed using the Agilent 2100 Bioanalyzer. Spike-in control transcripts were monitored to verify hybridization integrity. Raw data files were normalized using the Robust Multi-Array Analysis (Irizarry et al., 2003) and significantly regulated genes identified by one-way ANOVA (unequal variance) with Benjamini Hochberg false discovery rate (FDR) multiple testing correction (Dudoit et al., 2004) and Tukey HSD post-hoc statistics at p<0.05 or p<0.1 using GeneSpring GX 12.5 software. Positive fold-changes were calculated as flow/non-flow quantities while negative fold-changes were calculated as -1/(flow/non-flow). Raw microarray data files have been submitted to the Gene Expression Omnibus under accession number GSE42874.
2.5. Real-time RT-PCR
Complimentary DNA was synthesized from total RNA from original samples using the iScript reverse transcriptase kit (Bio-Rad, Hercules, CA). Quantitative PCR was then carried out in triplicate with the QuantiTect SYBR-Green PCR kit (Qiagen, Valencia, CA) and normalized to β-actin. The following murine-specific primers were used (5’ to 3’ direction): Cxcl1 (forward) GCTTGTTCAGTTTAAAGATGGTAGGC, (reverse) CGTGTTGACCATACAATATGAAAGACG; Cxcl2 (forward) ACAGAAGTCATAGCCACTCTC, (reverse) GCCTTGCCTTTGTTCAGTATC; β-actin (forward) AGATGTGGATCAGCAAGCAG, (reverse) GCGCAAGTTAGGTTTTGTCA. Relative expression levels were calculated as 2^(Ct[βactin]-Ct[Cxcl1,2]) (Zhang and Chen, 2000). Average values (n=3) within each time point were compared with an unpaired Student's t-test using Graphpad Prism 5 (La Jolla, CA).
2.6. Gene ontology (GO) analysis
Gene set enrichment for gene ontology biological process annotation was analyzed using Ingenuity Pathway Analysis (IPA 9.0, Ingenuity Systems, www.ingenuity.com) to identify the most significant cellular processes affected by flow. Canonical pathway analysis identified pathways from the IPA library that were most significant to the data set of gene transcripts demonstrating at least ±1.25 fold-change (p<0.1). The significance of the association between the data set and the canonical pathway was measured in two ways: (1) a ratio of the number of molecules from the data set that map to the pathway divided by the total number of molecules that map to the canonical pathway was determined. (2) Fisher's exact test was used to calculate a p-value determining the probability that the association between the genes in the dataset and the canonical pathway is explained by chance alone. The Functional Analysis identified associated biological functions and/or diseases. A right-tailed Fisher's exact test was used to calculate a p-value determining the probability that each biological function/disease assigned to that data set is due to chance alone. A denominator representing number of molecules associated with each function is not reported as these databases are rapidly changing.
2.7. Proteomic analysis
Total protein was subjected to high mass accuracy liquid chromatography-mass spectrometry (LC-MS) to resolve, detect, and identify individual peptide peaks for protein quantification. Biological and technical replicates (n=3 and 2, respectively) of tryptically digested and isolated peptides from each sample were prepared as previously described (Brown et al., 2012) and subjected to LC-MS analysis using a LTQ-Orbitrap mass spectrometer (MS; Thermo Scientific, Waltham, MA, USA) and an electrospray ionization source manufactured in-house, as previously reported (Brown et al., 2012; Livesay et al., 2008; Xie et al., 2011). The accurate mass and time tag approach (AMT) was utilized to identify and quantify detected peptide peaks by integrating tandem MS data with quantified accurate mass peptide peaks with FDR thresholds as previously described (Angel et al., 2012; Brown et al., 2012). Identified tandem MS spectra were based upon the IPI Mouse database downloaded 10-24-2007, containing 51,489 protein entries. An in-house software program, DAnTE, was used to quantify protein values representing a composite of normalized and correlated peptide values (Polpitiya et al., 2008). Levels of statistical significance were determined from a basic central tendency normalization and one-way ANOVA between flow and non-flow conditions. Proteomics raw data is made available at http://omics.pnl.gov.
2.8. Integrative analysis
From lists of both significantly regulated gene transcripts (p<0.05) and proteins (p<0.1), an integration network for MLO-Y4 cells exposed to fluid flow was generated using MetaCore pathway analysis software (GeneGo, St. Joseph, MI) as similarly conducted by Waters, et al. (Waters et al., 2011). This integration network was developed using the Analyze network algorithm in which root nodes were established from microarray data in conjunction with proteomic data, and additional neighboring nodes and edges established connectivity. These additional nodes were ascertained through MetaCore's proprietary curated database of mouse regulatory interactions. Sub-networks were created by segregation of the full network and prioritized based on significance calculated from the proportion of nodes from the experimental data in the sub-network. To identify significant transcriptional regulators in the integrated dataset, the statistical Interactome tool was used in MetaCore to measure the interconnectedness of genes and proteins in the experimental dataset. Statistical significance of over-connected interactions was calculated using a hypergeometric distribution, where the p value represents the probability of a particular mapping arising by chance for experimental data (Nikolsky et al., 2009).
3. Results
3.1. Transcriptome regulation by fluid flow
We detected approximately 14,000 unique gene products using microarray analysis of whole cell lysates from flowed and non-flowed MLO-Y4 cells. Among these, 171 gene products increased or decreased abundance relative to non-flow controls at one or more of the given time points of 0, 2, 8, or 24 hours post-flow at a significance level of p<0.05 (Supplementary Table 1). Similarly, 423 gene products were regulated at p<0.1 (Supplementary Table 2). As we have done previously (Waters et al., 2011), we then utilized a fold change cut-off to discern gene products of greatest interest. Table 1 identifies all transcripts significantly fluid flow-regulated by at least ±1.5-fold change at any time point, including 13 at p<0.05 and 22 at p<0.1. Among the four time points we examined, the greatest number of fold changes fitting these criteria occurred at 2 hours post-flow incubation; there were four changes at 0 hours, nineteen changes at 2 hours, and one change at 8 hours. The greatest up-regulation occurred in mRNAs encoding C-X-C motif chemokines (Table 1). Cxcl1 increased 2.59-fold immediately after OFF treatment. At 2 hours post-flow, Cxcl1, Cxcl2, and Cxcl5 increased 2.88, 4.33, and 3.48-fold, respectively. By 8 hours post-flow, levels dropped to non-significant fold-changes (1.58, 1.76, and 1.73) and approached non-flow control values by 24 hours (−1.05, 1.05, and 1.06). The greatest fold decrease in all flow-regulated transcripts was in Ccng2 (cyclin G2) with −4.62 at 0 hours. Noteworthy transcripts included in Table 1 despite regulation below the ±1.5-fold cutoff include Nfkb1 and Csf1, both up-regulated 1.2-fold at 2 hours, Creb1, up-regulated 1.2-fold at 8 hours, and Pias1, down-regulated −1.2-fold at 2 hours.
Table 1.
Flow-induced fold changes in gene transcripts revealed by microarray analysis.
| Gene | GO Biological Process | 0 hours | 2 hours | 8 hours | 24 hours |
|---|---|---|---|---|---|
| Cxcl1 | Inflammatory response | 2.59* | 2.88** | 1.58 | −1.05 |
| Cxcl2 | Chemotaxis | 2.54 | 4.33* | 1.76 | 1.05 |
| Cxcl5 | Cytokine production | 2.22 | 3.48** | 1.73 | 1.06 |
| Zc3h12a | Angiogenesis | 1.53 | 2.31** | 1.22 | 1.06 |
| Pdlim5 | Regulation of synaptogenesis | 1.52** | 1.05 | −1.03 | −1.08 |
| Ereg | Angiogenesis | 1.37 | 1.72** | 1.17 | 1.04 |
| Ripk2 | Positive regulation of cytokine-mediated signaling pathway | 1.26 | 1.54* | 1.07 | −1.05 |
| Csf1 | Ossification | 1.19 | 1.19* | 1.05 | −1.01 |
| Nfkb1 | Negative regulation of cytokine production | 1.17 | 1.23* | 1.07 | 1.03 |
| Creb1 | cAMP responsive element binding protein 1 | 1.09 | 1.00 | 1.21* | 1.00 |
| Pik3r1 | Positive regulation of protein phosphorylation | −1.00 | 1.51* | 1.11 | −1.05 |
| Nr2f2 | Negative regulation of transcription | −1.07 | −1.54* | −1.18 | −1.09 |
| Ndrg1 | Mast cell activation | −1.14 | −1.53** | −1.02 | 1.03 |
| Afp | Ovulation from ovarian follicle | −1.14 | 1.45 | 1.97** | 1.01 |
| Osr1 | Metanephros development | −1.16 | −1.51* | −1.21 | −1.04 |
| Pgm2 | Carbohydrate metabolic process | −1.22 | −1.52** | −1.05 | −1.04 |
| Pias1 | Negative regulation of transcription | −1.23 | −1.24* | −1.01 | −1.12 |
| Slc2a1 | Transport | −1.27 | −1.62** | 1.08 | 1.01 |
| Sap30 | Negative regulation of transcription | −1.32 | −1.51* | 1.06 | −1.12 |
| Scd2 | Lipid metabolic process | −1.40 | −1.64* | −1.20 | −1.07 |
| Ero1l | Protein folding | −1.41 | −1.67* | −1.06 | −1.01 |
| Egln1 | Response to hypoxia | −1.48 | −1.90** | −1.11 | −1.03 |
| Selenbp1 | Transport | −1.59 | −2.37** | −1.71 | 1.06 |
| Higd1a | Response to stress | −1.91** | −2.08** | 1.06 | −1.06 |
| Bnip3 | Response to hypoxia | −2.52 | −3.96* | −1.12 | 1.03 |
| Ccng2 | Cell cycle | −4.62** | −2.84 | 1.03 | −1.04 |
Flow vs. non-flow fold increase or decrease for all gene transcripts having at least ±1.5-fold change at one or more time points (exceptions are Nfkb1, Csf1, Creb1, and Pias1) at a significance of p<0.1 (*) or p<0.05 (**). Results are from microarray analysis of MLO-Y4 cells exposed to 2 hours OFF and post-incubated for the given time.
3.2. qPCR confirms up-regulation of Cxcl1 and Cxcl2
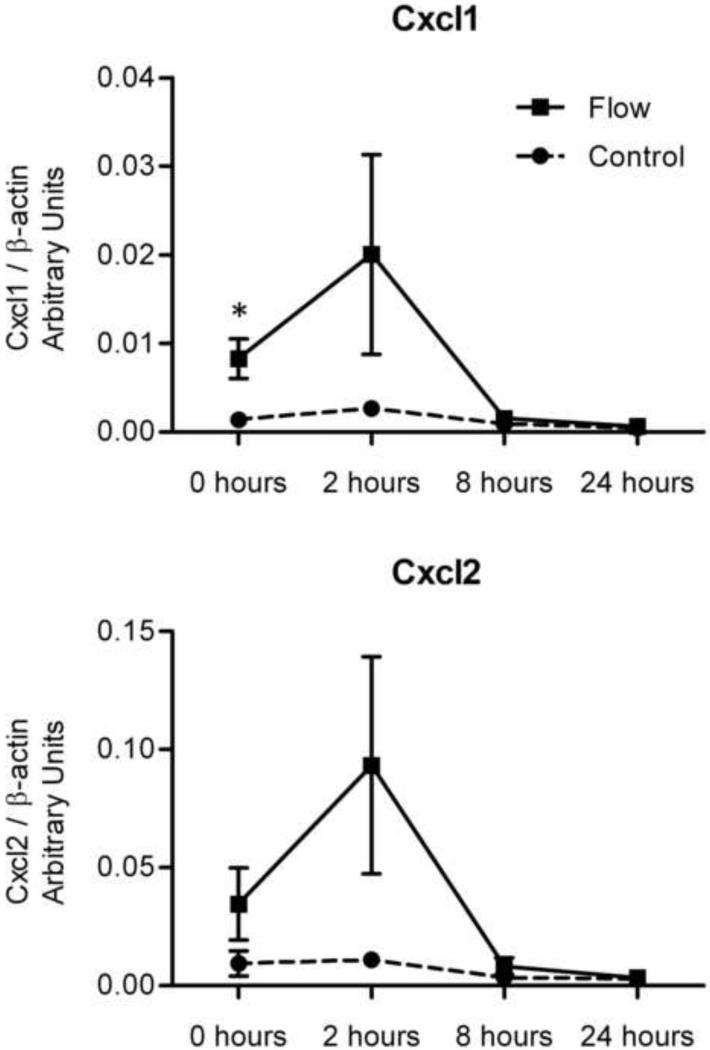
In order to confirm OFF regulation of specific gene transcripts, we performed quantitative RT-PCR on RNA from the same samples. We chose to further examine expression of Cxcl1 and Cxcl2 as these are novel mechano-sensitive genes that also demonstrated the greatest fold-increase of all gene products immediately after flow (0 hours) and 2 hours post-flow. Accordingly, qPCR indicated that Cxcl1 transcript levels exhibited flow-induced fold changes of 6.02 at 0 hours, 7.60 at 2 hours, 1.73 at 8 hours, and 1.52 at 24 hours (Figure 1). Similarly, flow-induced fold changes in Cxcl2 were 3.73, 8.55, 2.46, and 1.21 at these respective time points. Though only Cxcl1 at 0 hours post-flow achieved significance by qPCR (p=0.039), patterns of Cxcl1 and Cxcl2 expression by qPCR are both in agreement with the microarray analysis in Table 1. Also note in Figure 1 that flow-induced increases in Cxcl1 and Cxcl2 transcript levels occurred alongside relatively steady non-flowed transcript levels.
Figure 1. Cxcl1 and Cxcl2 gene transcript levels by real-time RT-PCR.
Quantitative analysis of the same RNA used in microarrays supports observed up-regulation of both Cxcl1 and Cxcl2 in MLO-Y4 cells subject to OFF (blue) relative to non-flow controls (red) over initial 2 hours of post-flow incubation. Graphs indicate mean +/− SEM, n=3 per time point, (*) indicates p=0.039 for 0 hr Cxcl1, p=0.198 for 2 hr Cxcl1, p=0.192 for 0 hr Cxcl2, p=0.149 for 2 hr Cxcl2.
3.3. Gene ontology analysis associates gene changes with cell signaling and inflammatory response
A list of 51 transcripts regulated by at least 1.25 fold-change (p<0.1) was mapped by Ingenuity Pathway Analysis (IPA) software to identify their collective biological associations. These associations are compiled from Ingenuity's curated database of molecular associations in the literature. Table 2 lists select results for consideration in a skeletal context. Top canonical pathway associations suggested signaling via ErbB (p=4.66×10−4) and FAK (p=3.81×10−3); of lower statistical significance were ERK/MAPK (p=1.04×10−2) and NF-κB (p=4.01×10−2). Physiological function associations included hematological and connective tissue development and function, hematopoiesis, and immune cell trafficking. Molecular and cellular functions broadly related to cell morphology, maintenance, proliferation, and movement.
Table 2.
IPA analysis of signaling pathways, physiological functions, and molecular/cellular functions associated with flow-induced changes in gene transcripts.
| Top Canonical Pathways | p-value | Ratio | Associated Molecules |
|---|---|---|---|
| ErbB Signaling | 4.66E-04 | 5/90 | Btc, Ereg, Pak3, Pik3r1, Sos1 |
| Focal Adhesion Kinase Signaling | 3.81E-03 | 4/106 | Pak3, Pik3r1, Sos1, Tln1 |
| Neuregulin Signaling | 5.26E-03 | 4/104 | Btc, Ereg, Pik3r1, Sosl |
| Paxillin Signaling | 5.73E-03 | 4/117 | Pak3, Pik3r1, Sos1, Tlnl |
| Role of IL-17A in Arthritis | 7.27E-03 | 3/64 | Cxcl3, Cxcl6, Pik3r1 |
| ERK/MAPK Signaling | 1.04E-02 | 5/211 | Pak3, Pik3r1, Ppp1r14c, Sos1, Tln1 |
| NF-κB Signaling | 4.01E-02 | 4/181 | Irak4, Peli1, Pik3r1, Tirap |
| Physiological System Development and Function | p-value | # Molecules | |
|---|---|---|---|
| Hematological System Development and Function | <4.89E-02 | 21 | |
| Hematopoiesis | <3.41E-02 | 8 | |
| Immune Cell Trafficking | <4.89E-02 | 15 | |
| Connective Tissue Development and Function | <4.25E-02 | 21 |
| Molecular and Cellular Functions | p-value | # Molecules | |
|---|---|---|---|
| Cell Morphology | <4.89E-02 | 20 | |
| Cellular Function and Maintenance | <4.89E-02 | 21 | |
| Cellular Growth and Proliferation | <4.25E-02 | 45 | |
| Cellular Movement | <4.89E-02 | 31 |
Results from Ingenuity Pathway Analysis software deriving gene ontology associations from transcript levels changing >1.25-fold (p<0.1). The software assigns a p-value based on the probability that the functional assignment is due to chance alone. “Ratio” indicates number of molecules from the data set that map to the pathway divided by the total number of molecules that map to the canonical pathway. Under “# Molecules”, a denominator representing number of molecules associated with each function is not reported.
3.4. Proteomic regulation by fluid flow
Cell lysates from these same flowed and non-flowed MLO-Y4 cells were assessed with high mass accuracy LC-MS, identifying approximately 3,202 unique peptides corresponding to approximately 558 proteins. Among these composite proteins, analysis of variance across all time points revealed 59 proteins significantly regulated at p<0.1 and 24 at p<0.05 for at least one time point (Table 3). Three are underlined as noteworthy signaling changes in osteocytes. The most highly up-regulated was S100-A6—also known as Calcyclin—which increased 2.43-fold (p=0.002) at 8 hours post-flow but decreased by −1.75-fold (p=0.014) at 24 hours. S100-A6 was also the only protein with changes at two time points significant at p<0.05. Nucleoside diphosphate kinase B (NDK) increased 1.97-fold (p=0.039) at 8 hours. G protein-coupled receptor kinase 6 (GRK-6) increased 1.51-fold (p=0.005) at 8 hours. Note that 18 of the 25 fold-changes significant at p<0.05 occurred at the 8 hour time point. This draws a distinction from transcriptomic data, which demonstrated greatest overall flow regulation at 2 hours post-flow.
Table 3.
Flow-induced fold changes in protein levels inferred from liquid chromatography-mass spectrometry.
| Protein | GO Biological Process | 0 hours | 2 hours | 8 hours | 24 hours |
|---|---|---|---|---|---|
| Hnrnpu | mRNA Processing | 1.25 | 1.09 | 1.26* | −1.06 |
| FUSE-binding protein 2 | mRNA Processing | −1.03 | −1.04 | −1.03 | −1.48* |
| Cavin-1 | Transcription | −1.03 | 1.01 | −1.35* | 1.28 |
| HMG-I(y) | Transcription | −1.31 | −1.14 | 1.05 | −1.43* |
| Efemp1 | Transcription | −1.08 | 1.05 | −1.27* | 1.08 |
| Dpy-30L | Transcription | −1.32 | −1.07 | −1.25 | −2.23* |
| Psmc4 | Protein catabolism, folding, and transport | −1.34 | −1.34 | −1.40* | −1.08 |
| Tcp-1 zeta | Protein catabolism, folding, and transport | 1.07 | 1.11 | −1.36* | 1.00 |
| Rab GDI beta | Protein catabolism, folding, and transport | −1.05 | −1.05 | −1.31* | −1.05 |
| Nucleoside diphosphate kinase B | Biosynthetic processes | 1.36 | 1.22 | 1.97* | −1.15 |
| C1-THF synthase | Biosynthetic processes | −1.07 | −1.04 | −1.76* | −1.13 |
| Valyl tRNA synthase | tRNA aminoacylation | −2.07 | 1.14 | −1.39* | −1.17 |
| Nucleolin | Nucleotide binding | −2.48 | −1.24 | −1.69* | 1.27 |
| CCN family member 2 | Regulation of cell growth | 1.59 | 1.33* | 1.43 | 1.10 |
| MCG11809 | Translation | −1.04 | 1.01 | −1.25* | −1.02 |
| Putative uncharacterized protein | DNA replication | −1.20 | 1.27* | −1.18 | 1.18 |
| Hemoglobin alpha adult chain | Heme, oxygen binding | −1.14 | −1.27 | 1.55* | −1.22 |
| S100-A6 (Calcyclin) | Not indicated | −1.11 | −1.14 | 2.43* | −1.75* |
| Deoxyuridine triphosphatase | Not indicated | −1.23 | 1.04 | −1.51* | 1.13 |
| Fibronectin | Not indicated | −1.02 | −1.08 | −1.40* | −1.27 |
| similar to Tubulin, alpha 3C isoform 3 | Not indicated | 1.20 | 1.11 | 1.48* | −1.06 |
| G protein-coupled receptor kinase 6 | Not indicated | 1.19 | −1.18 | 1.51* | −1.04 |
| similar to ribosomal protein L39 | Not indicated | −1.09 | 1.04 | −1.28* | 1.34 |
| ribosomal protein SA | Not indicated | 1.03 | 1.30* | −1.02 | 1.16 |
Flow vs. non-flow fold increase or decrease in MLO-Y4 protein levels inferred from peptides detected by liquid chromatography-mass spectrometry. Proteins listed with associated gene ontology biological process and fold-change at each post-incubation time point. n=3, (*) indicates p<0.05. Proteins of special interest (discussed further in text) are underlined.
3.5. Integrative analysis reveals signaling nodes not detected by microarray or mass spectrometry
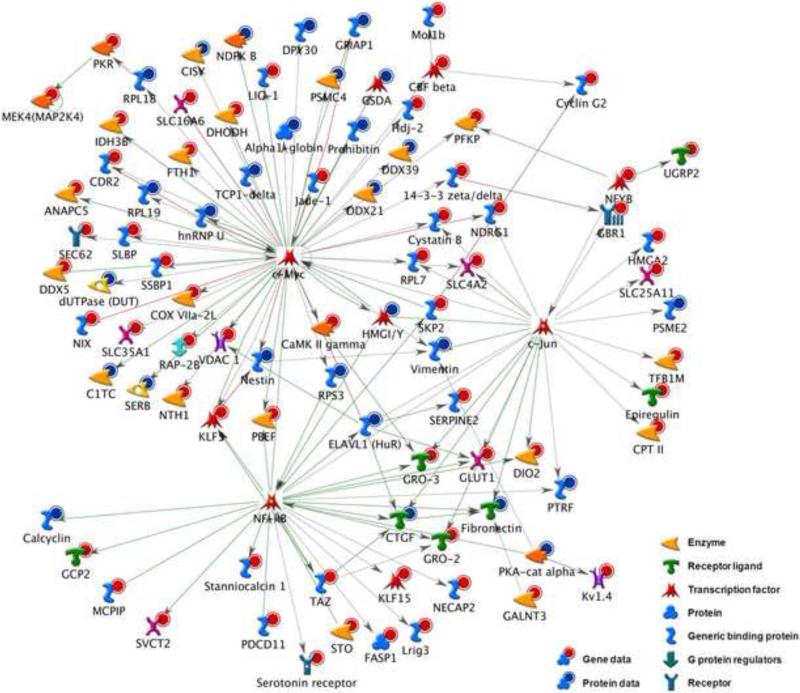
From these microarray and mass spectrometry results, a regulatory network was computed using MetaCore software to integrate both transcriptional data (171 genes regulated at p<0.05) and proteomic data (59 proteins at p<0.1). Figure 2 depicts gene and protein associations (red and blue circles, respectively) for the three most significantly enriched transcription factors in the dataset: c-Myc (p=2.09×10−7), c-Jun (p=1.82×10−6), and RelA/NF-κB (p=8.39×10−6). These three transcription factors are over-connected to seed nodes relative to what would be expected by chance alone.
Figure 2. Integrative network analysis.
MetaCore network analysis integrating both gene data (red circles) and protein data (blue circles) into regulatory sub-networks active in the response of MLO-Y4 cells to OFF. The inferred central transcription factors c-Myc (p=2.09×10−7), c-Jun (p=1.82×10−6), and RelA/NF-κB (p=8.39×10−6) are the most significantly over-connected to the observed dataset.
4. Discussion
The goal of this study was to establish the utility of high-throughput transcriptomic and proteomic analyses to unravel the mechano-sensitive response of MLO-Y4 cells subjected to OFF. We hypothesized that this global analysis would reflect regulation of genes and proteins previously known to be responsive to fluid flow while also identifying novel mechano-sensitive signaling molecules and pathways. Gene transcription and protein levels demonstrated a shift attributable to two variables: fluid flow or non-flow treatment, and time in post-treatment incubation. Non-flowed MLO-Y4 cells displayed widely shifted expression levels throughout 24 hours post-incubation due to changes in environmental stimuli before and after sham flow treatment, including fresh medium, transfer to treatment chambers, and incubator acclimation. As such stimuli are controlled across flow and non-flow samples, we attribute variation from the non-flow temporal pattern to the fluid flow stimulus. Within each time-point, we quantified this variation as fold-change in detected gene transcripts and proteins.
Accumulating evidence indicates that osteocytes are able to regulate both anabolic effector cells (mesenchymal stem cells, pre-osteoblasts, and osteoblasts) and catabolic effector cells (osteoclasts) (Hoey et al., 2011; Raheja et al., 2008; Schaffler et al., 2014). Our proposal that OFF elicits diverse cellular signaling functions is bolstered by gene ontology analysis (Table 2) implicating known anabolic, mechano-sensitive pathways—FAK and ERK/MAPK —alongside inflammatory IL-17A signaling and immune cell trafficking. These latter functions suggest possible osteocytic activation of the catabolic arm of bone remodeling: recruitment of macrophage-derived osteoclasts. Indeed, this is consistent with the dose-dependent contribution of Cxcl2 toward osteoclast recruitment (Ha et al., 2011; Oue et al., 2012), up-regulation of macrophage colony-stimulating factor, Csf1 (Table 1) (Boyle et al., 2003) and widespread up-regulation of cytokine transcripts. The molecule Nfkb1, up-regulated by flow, encodes pro-inflammatory NF-κB and the NF-κB inhibitor Pias1 (Liu et al., 2005) was suppressed by flow. The ligand for NF-κB, commonly known as RANKL, is necessary for osteoclast formation and catabolic remodeling (Xiong and O’Brien, 2012). The RANKL/OPG ratio could not be determined since regulation was not statistically significant.
The transcriptome also provides context to explore novel mechano-sensitive pathways. Most notably, OFF up-regulated the levels of mRNAs encoding inflammatory chemokines Cxcl1 and Cxcl2 more than all other transcripts detected at 0 hours. With supporting qPCR data, this strongly suggests that these genes are up-regulated by OFF. Previous microarray analyses also reported early up-regulation of Cxcl1 in loaded rat ulnas (Xing et al., 2005) and Cxcl2 in MLO-Y4 cells following cyclic compression (Chen et al., 2010). We propose that the products of these genes—pro-inflammatory chemokine ligands—may serve as migratory, proliferation, or differentiation cues for short-lived, translocating osteolineage cells (Park et al., 2012). An influx of inflammatory factors like that accompanying woven bone formation (McKenzie et al., 2011) commonly precedes tissue repair and remodeling. Osteocytes increase mRNA expression of a similar chemokine Cxcl12 (Sdf1) following mouse ulnar loading, as do MLO-Y4 cells exposed to OFF, and antagonism of the CXCL12 receptor CXCR4 attenuates both load-induced bone formation (Leucht et al., 2013) and fracture repair (Toupadakis et al., 2012). Yet, CXCL12 alone is insufficient for increasing proliferation or differentiation of a stem cell line in vitro, suggesting that additional factors mediate mechanical adaptation (Leucht et al., 2013). CXCL1, CXCL2, and CXCL5 each bind to receptor CXCR2, which is expressed by both murine and human bone marrow mesenchymal stem cells (Chamberlain et al., 2008). CXCR2 ligands stimulate migration in a variety of cell types, including endothelial progenitors (Jones et al., 2009; Miyake et al., 2013), hematopoietic stem cells (Fukuda et al., 2007; Yoon et al., 2012), and human bone marrow mesenchymal stem cells (Nedeau et al., 2008; Ringe et al., 2007). Up-regulation of Cxcl1 and Cxcl2 suggests a novel paracrine means by which osteocytic cells exposed to fluid flow might increase local bone formation.
Using LC-MS, we detected peptides corresponding to 558 proteins, including 24 proteins in Table 3 differentially expressed with 95% significance. Three up-regulated proteins are of particular interest: NDK is an enzyme that converts ADP to ATP which is released by mechanically stimulated osteocytes (Genetos et al., 2007). Thus, NDK may play a further critical, as yet unappreciated role in mechanotransduction. S100-A6, also known as Calcyclin, was the most highly up-regulated protein following exposure to OFF. Interestingly, Calcyclin is a calcium-binding protein which, when over-expressed, increases proliferation and expression of alkaline phosphatase mRNA in pre-osteoblastic MC3T3-E1 cells (Hwang et al., 2004). Lastly, GRK-6 is an enzyme critical to G protein-coupled receptor activation of downstream signaling events and has not previously been implicated in mechanotransduction. Together, regulation of these proteins exemplifies the capability of this proteomics approach for identifying novel factors that regulate bone cell activity in response to a mechanical stimulus.
Next, using an integrative informatics analysis to combine both transcript and protein data, we identified central signaling network control nodes implicated by the collective data. Intriguingly, neither mRNA data from microarrays nor peptides from MS-based proteomics alone revealed a flow-responsive role for the two most highly over-connected signaling nodes, transcription factors c-Myc and c-Jun, exemplifying how central control molecules may be revealed computationally from combinations of data platforms. Future studies could directly examine activation (e.g., via ChIP-Seq) of transcription factors c-Myc, c-Jun, and RelA/NF-κB. To our knowledge, this integrative approach has not yet been employed to examine mechanotransduction in any cell type.
Our results demonstrate a modest magnitude of flow regulation of gene transcription and protein expression. Such fold-changes may bring about a cumulative response to a stimulus that alters bone density over days to months in vivo. In the short term, mRNA fold-changes may be potentiated or inhibited by post-transcriptional modification, thus explaining some of the differences between detected gene transcript fluctuations and protein levels. Also, the 0, 2, 8, and 24 hour time-points represent snap-shots of changes so that a continuum of intervening fluctuations and encoded proteins remains undetected. For instance, Ptgs2—which encodes anabolic Cox-2 and has been shown to be up-regulated by pulsatile fluid flow (PFF) and OFF (Kamel et al., 2010; Li et al., 2012; Litzenberger et al., 2010)—was not significantly regulated in our observations, though expression increased 1.37-fold at 2 hours and 1.45 at 8 hours. The greatest and most numerous changes in transcript levels occurred at 2 hours post-flow while protein levels followed with greatest regulation at 8 hours. By 24 hours, most transcript and protein levels returned close to those of non-flow controls, indicating that this 24 hour window is sufficient for capturing the predominant osteocytic OFF response. Progressive examination of intervening time-points, for instance 1, 3, 6, and 12 hours post-flow, would provide further insight into the dynamic response to OFF.
Though our monolayer fluid flow stimulation is highly reproducible, this mechano-stimulus remains a rough approximation of three-dimensional canalicular fluid flow. The MLO-Y4 cell line also does not express all genes and proteins detected in primary osteocytes, which have proven difficult to isolate in cell culture. Specifically, Sost, the gene encoding Sclerostin (Mabilleau et al., 2010; Woo et al., 2011) and strongly implicated in bone mechanotransduction, is not expressed by MLO-Y4 cells. A more recently developed osteocytic cell line, IDG-SW3, may express Sost when terminally differentiated (Woo et al., 2011). We speculate that expression of Sost will adjust levels of numerous associated transcripts and proteins. Indeed, a comparison of our data with the results of a future transcriptomic/proteomic analysis of IDG-SW3 cells exposed to OFF could provide critical insights into the roles of Sclerostin in mechanotransduction by osteocytes.
This complimentary analysis of gene transcript and protein levels has enabled the detection of both conventional and previously unreported signaling molecules at play in osteocytic MLO-Y4 cells subjected to fluid flow. Application of high-throughput quantification allows a progression away from a candidate approach for discovering mechano-sensitive signaling pathways and towards the integration of data into predictive networks. These signaling networks hold future potential for pinpointing opportunities for therapeutic interventions. More important presently, these studies suggest that OFF up-regulates both Cxcl1 and Cxcl2, supporting the concept that OFF regulates both arms of bone remodeling.
Supplementary Material
Acknowledgements
A portion of this research was performed using the Environmental Molecular Sciences Laboratory (EMSL), a national scientific user facility sponsored by the Department of Energy's Office of Biological and Environmental Research and located at the Pacific Northwest National Laboratory. Pacific Northwest National Laboratory is operated by Battelle Memorial Institute for the U.S. Department of Energy under contract DE-AC05-76RLO-1830. This work was supported by AG13087-15 from the National Institute on Aging, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest statement
The authors have no conflicts to disclose.
References
- Angel TE, Jacobs JM, Spudich SS, Gritsenko MA, Fuchs D, Liegler T, Zetterberg H, Camp DG, 2nd, Price RW, Smith RD. The cerebrospinal fluid proteome in HIV infection: change associated with disease severity. Clin Proteomics. 2012;9:3. doi: 10.1186/1559-0275-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra N, Burra S, Siller-Jackson AJ, Gu S, Xia X, Weber GF, DeSimone D, Bonewald LF, Lafer EM, Sprague E, Schwartz MA, Jiang JX. Mechanical Stress-Activated Integrin A5β1 Induces Opening of Connexin 43 Hemichannels. PNAS. 2012;109:3359–3364. doi: 10.1073/pnas.1115967109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonewald LF. The amazing osteocyte. J Bone Miner Res. 2011;26:229–238. doi: 10.1002/jbmr.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonewald LF, Johnson ML. Osteocytes, mechanosensing and Wnt signaling. Bone. 2008;42:606–615. doi: 10.1016/j.bone.2007.12.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- Brown JN, Ortiz GM, Angel TE, Jacobs JM, Gritsenko M, Chan EY, Purdy DE, Murnane RD, Larsen K, Palermo RE, Shukla AK, Clauss TR, Katze MG, McCune JM, Smith RD. Morphine produces immunosuppressive effects in nonhuman primates at the proteomic and cellular levels. Mol. Cell Proteomics. 2012;11:605–618. doi: 10.1074/mcp.M111.016121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain G, Wright K, Rot A, Ashton B, Middleton J. Murine Mesenchymal Stem Cells Exhibit a Restricted Repertoire of Functional Chemokine Receptors: Comparison with Human. PLoS ONE. 2008;3:e2934. doi: 10.1371/journal.pone.0002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Qing H, He Y, Wang J, Zhu Z, Wang H. Gene expression patterns of osteocyte-like MLO-Y4 cells in response to cyclic compressive force stimulation. Cell. Biol. Int. 2010;34:425–432. doi: 10.1042/CBI20090061. [DOI] [PubMed] [Google Scholar]
- Cherian PP, Siller-Jackson AJ, Gu S, Wang X, Bonewald LF, Sprague E, Jiang JX. Mechanical Strain Opens Connexin 43 Hemichannels in Osteocytes: A Novel Mechanism for the Release of Prostaglandin. Mol. Biol. Cell. 2005;16:3100–3106. doi: 10.1091/mbc.E04-10-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung W-Y, Liu C, Tonelli-Zasarsky RML, Simmons CA, You L. Osteocyte apoptosis is mechanically regulated and induces angiogenesis in vitro. J. Orthop. Res. 2011;29:523–530. doi: 10.1002/jor.21283. [DOI] [PubMed] [Google Scholar]
- Cowin SC, Weinbaum S, Zeng Y. A case for bone canaliculi as the anatomical site of strain generated potentials. J Biomech. 1995;28:1281–97. doi: 10.1016/0021-9290(95)00058-p. [DOI] [PubMed] [Google Scholar]
- Dudoit S, van der Laan MJ, Pollard KS. Multiple Testing. Part I. Single-Step Procedures for Control of General Type I Error Rates. Statistical Applications in Genetics and Molecular Biology. 2004;3 doi: 10.2202/1544-6115.1040. [DOI] [PubMed] [Google Scholar]
- Fukuda S, Bian H, King AG, Pelus LM. The chemokine GRObeta mobilizes early hematopoietic stem cells characterized by enhanced homing and engraftment. Blood. 2007;110:860–869. doi: 10.1182/blood-2006-06-031401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genetos DC, Kephart CJ, Zhang Y, Yellowley CE, Donahue HJ. Oscillating fluid flow activation of gap junction hemichannels induces atp release from MLO-Y4 osteocytes. J. Cell. Physiol. 2007;212:207–214. doi: 10.1002/jcp.21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha J, Choi H-S, Lee Y, Kwon H-J, Song YW, Kim H-H. CXC Chemokine Ligand 2 Induced by Receptor Activator of NF-κB Ligand Enhances Osteoclastogenesis. The Journal of Immunology. 2010;184:4717–4724. doi: 10.4049/jimmunol.0902444. [DOI] [PubMed] [Google Scholar]
- Ha J, Lee Y, Kim H-H. CXCL2 mediates lipopolysaccharide-induced osteoclastogenesis in RANKL-primed precursors. Cytokine. 2011;55:48–55. doi: 10.1016/j.cyto.2011.03.026. [DOI] [PubMed] [Google Scholar]
- Haut Donahue TL, Genetos DC, Jacobs CR, Donahue HJ, Yellowley CE. Annexin V disruption impairs mechanically induced calcium signaling in osteoblastic cells. Bone. 2004;35:656–663. doi: 10.1016/j.bone.2004.04.025. [DOI] [PubMed] [Google Scholar]
- Hoey DA, Kelly DJ, Jacobs CR. A role for the primary cilium in paracrine signaling between mechanically stimulated osteocytes and mesenchymal stem cells. Biochemical and Biophysical Research Communications. 2011 doi: 10.1016/j.bbrc.2011.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo B, Lu XL, Hung CT, Costa KD, Xu Q, Whitesides GM, Guo XE. Fluid Flow Induced Calcium Response in Bone Cell Network. Cell Mol Bioeng. 2008;1:58–66. doi: 10.1007/s12195-008-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang R, Lee EJ, Kim MH, Li S-Z, Jin Y-J, Rhee Y, Kim YM, Lim S-K. Calcyclin, a Ca2+ Ion-Binding Protein, Contributes to the Anabolic Effects of Simvastatin on Bone. J. Biol. Chem. 2004;279:21239–21247. doi: 10.1074/jbc.M312771200. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostat. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jacobs CR, Yellowley CE, Davis BR, Zhou Z, Cimbala JM, Donahue HJ. Differential effect of steady versus oscillating flow on bone cells. Journal of Biomechanics. 1998;31:969–976. doi: 10.1016/s0021-9290(98)00114-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing D, Baik AD, Lu XL, Zhou B, Lai X, Wang L, Luo E, Guo XE. In situ intracellular calcium oscillations in osteocytes in intact mouse long bones under dynamic mechanical loading. FASEB J fj. 2013:13–237578. doi: 10.1096/fj.13-237578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CP, Pitchford SC, Lloyd CM, Rankin SM. CXCR2 mediates the recruitment of endothelial progenitor cells during allergic airways remodeling. Stem Cells. 2009;27:3074–3081. doi: 10.1002/stem.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamel MA, Picconi JL, Lara-Castillo N, Johnson ML. Activation of β-catenin signaling in MLO-Y4 osteocytic cells versus 2T3 osteoblastic cells by fluid flow shear stress and PGE2: Implications for the study of mechanosensation in bone. Bone. 2010;47:872–881. doi: 10.1016/j.bone.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Windle JJ, Koop BA, Mundy GR, Bonewald LF. Establishment of an Osteocyte-like Cell Line, MLO-Y4. Journal of Bone and Mineral Research. 1997;12:2014–2023. doi: 10.1359/jbmr.1997.12.12.2014. [DOI] [PubMed] [Google Scholar]
- Kim CH, You L, Yellowley CE, Jacobs CR. Oscillatory fluid flow-induced shear stress decreases osteoclastogenesis through RANKL and OPG signaling. Bone. 2006;39:1043–1047. doi: 10.1016/j.bone.2006.05.017. [DOI] [PubMed] [Google Scholar]
- Kitase Y, Barragan L, Qing H, Kondoh S, Jiang JX, Johnson ML, Bonewald LF. Mechanical induction of PGE2 in osteocytes blocks glucocorticoid-induced apoptosis through both the β-catenin and PKA pathways. J Bone Miner Res. 2010;25:2657–2668. doi: 10.1002/jbmr.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein-Nulend J, Semeins CM, Ajubi NE, Nijweide PJ, Burger EH. Pulsating fluid flow increases nitric oxide (NO) synthesis by osteocytes but not periosteal fibroblasts--correlation with prostaglandin upregulation. Biochem. Biophys. Res. Commun. 1995;217:640–648. doi: 10.1006/bbrc.1995.2822. [DOI] [PubMed] [Google Scholar]
- Knothe Tate ML, Knothe U. An ex vivo model to study transport processes and fluid flow in loaded bone. Journal of Biomechanics. 2000;33:247–254. doi: 10.1016/s0021-9290(99)00143-8. [DOI] [PubMed] [Google Scholar]
- Kufahl RH, Saha S. A theoretical model for stress-generated fluid flow in the canaliculi-lacunae network in bone tissue. J Biomech. 1990;23:171–80. doi: 10.1016/0021-9290(90)90350-c. [DOI] [PubMed] [Google Scholar]
- Kwon RY, Jacobs CR. Time-dependent deformations in bone cells exposed to fluid flow in vitro: investigating the role of cellular deformation in fluid flow-induced signaling. Journal of Biomechanics. 2007;40:3162–3168. doi: 10.1016/j.jbiomech.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucht P, Temiyasathit S, Russell A, Arguello JF, Jacobs CR, Helms JA, Castillo AB. CXCR4 antagonism attenuates load-induced periosteal bone formation in mice. J. Orthop. Res. 2013;31:1828–1838. doi: 10.1002/jor.22440. [DOI] [PubMed] [Google Scholar]
- Li J, Rose E, Frances D, Sun Y, You L. Effect of oscillating fluid flow stimulation on osteocyte mRNA expression. Journal of Biomechanics. 2012;45:247–251. doi: 10.1016/j.jbiomech.2011.10.037. [DOI] [PubMed] [Google Scholar]
- Li J, Zhang F, Chen JY. An integrated proteomics analysis of bone tissues in response to mechanical stimulation. BMC Syst Biol. 2011;5(Suppl 3):S7. doi: 10.1186/1752-0509-5-S3-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litzenberger JB, Kim J-B, Tummala P, Jacobs CR. β1 Integrins Mediate Mechanosensitive Signaling Pathways in Osteocytes. Calcified Tissue International. 2010;86:325–332. doi: 10.1007/s00223-010-9343-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Yang R, Wong KA, Getman C, Stein N, Teitell MA, Cheng G, Wu H, Shuai K. Negative regulation of NF-kappaB signaling by PIAS1. Mol. Cell. Biol. 2005;25:1113–1123. doi: 10.1128/MCB.25.3.1113-1123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesay EA, Tang K, Taylor BK, Buschbach MA, Hopkins DF, LaMarche BL, Zhao R, Shen Y, Orton DJ, Moore RJ, Kelly RT, Udseth HR, Smith RD. Fully automated four-column capillary LC-MS system for maximizing throughput in proteomic analyses. Anal. Chem. 2008;80:294–302. doi: 10.1021/ac701727r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XL, Huo B, Park M, Guo XE. Calcium response in osteocytic networks under steady and oscillatory fluid flow. Bone. 2012;51:466–473. doi: 10.1016/j.bone.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabilleau G, Mieczkowska A, Edmonds ME. Thiazolidinediones induce osteocyte apoptosis and increase sclerostin expression. Diabetic Medicine. 2010;27:925–932. doi: 10.1111/j.1464-5491.2010.03048.x. [DOI] [PubMed] [Google Scholar]
- Mantila Roosa SM, Liu Y, Turner CH. Gene expression patterns in bone following mechanical loading. J Bone Miner Res. 2011;26:100–112. doi: 10.1002/jbmr.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie JA, Bixby EC, Silva MJ. Differential Gene Expression from Microarray Analysis Distinguishes Woven and Lamellar Bone Formation in the Rat Ulna following Mechanical Loading. PLoS ONE. 2011;6:e29328. doi: 10.1371/journal.pone.0029328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake M, Goodison S, Urquidi V, Gomes Giacoia E, Rosser CJ. Expression of CXCL1 in human endothelial cells induces angiogenesis through the CXCR2 receptor and the ERK1/2 and EGF pathways. Lab Invest. 2013;93:768–778. doi: 10.1038/labinvest.2013.71. [DOI] [PubMed] [Google Scholar]
- Nedeau AE, Bauer RJ, Gallagher K, Chen H, Liu Z-J, Velazquez OC. A CXCL5- and bFGF-dependent effect of PDGF-B-activated fibroblasts in promoting trafficking and differentiation of bone marrow-derived mesenchymal stem cells. Experimental Cell Research. 2008;314:2176–2186. doi: 10.1016/j.yexcr.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolsky Y, Kirillov E, Zuev R, Rakhmatulin E, Nikolskaya T. Functional analysis of OMICs data and small molecule compounds in an integrated“knowledge-based” platform. Methods Mol. Biol. 2009;563:177–196. doi: 10.1007/978-1-60761-175-2_10. [DOI] [PubMed] [Google Scholar]
- Oue E, Lee J-W, Sakamoto K, Iimura T, Aoki K, Kayamori K, Michi Y, Yamashiro M, Harada K, Amagasa T, Yamaguchi A. CXCL2 synthesized by oral squamous cell carcinoma is involved in cancer-associated bone destruction. Biochemical and Biophysical Research Communications. 2012;424:456–461. doi: 10.1016/j.bbrc.2012.06.132. [DOI] [PubMed] [Google Scholar]
- Park D, Spencer JA, Koh BI, Kobayashi T, Fujisaki J, Clemens TL, Lin CP, Kronenberg HM, Scadden DT. Endogenous Bone Marrow MSCs Are Dynamic, Fate-Restricted Participants in Bone Maintenance and Regeneration. Cell Stem Cell. 2012;10:259–272. doi: 10.1016/j.stem.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polpitiya AD, Qian W-J, Jaitly N, Petyuk VA, Adkins JN, Camp DG, 2nd, Anderson GA, Smith RD. DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics. 2008;24:1556–1558. doi: 10.1093/bioinformatics/btn217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C, Zhou X, Li W, Wang L. Real-time measurement of solute transport within the lacunar-canalicular system of mechanically loaded bone: Direct evidence for load-induced fluid flow. Journal of Bone and Mineral Research. 2011;26:277–285. doi: 10.1002/jbmr.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raheja LF, Genetos DC, Yellowley CE. Hypoxic osteocytes recruit human MSCs through an OPN/CD44-mediated pathway. Biochemical and Biophysical Research Communications. 2008;366:1061–1066. doi: 10.1016/j.bbrc.2007.12.076. [DOI] [PubMed] [Google Scholar]
- Reijnders CMA, van Essen HW, van Rens BTTM, van Beek JHGM, Ylstra B, Blankenstein MA, Lips P, Bravenboer N. Increased expression of matrix extracellular phosphoglycoprotein (MEPE) in cortical bone of the rat tibia after mechanical loading: identification by oligonucleotide microarray. PLoS ONE. 2013;8:e79672. doi: 10.1371/journal.pone.0079672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringe J, Strassburg S, Neumann K, Endres M, Notter M, Burmester G-R, Kaps C, Sittinger M. Towards in situ tissue repair: Human mesenchymal stem cells express chemokine receptors CXCR1, CXCR2 and CCR2, and migrate upon stimulation with CXCL8 but not CCL2. J. Cell. Biochem. 2007;101:135–146. doi: 10.1002/jcb.21172. [DOI] [PubMed] [Google Scholar]
- Rolfe RA, Nowlan NC, Kenny EM, Cormican P, Morris DW, Prendergast PJ, Kelly D, Murphy P. Identification of mechanosensitive genes during skeletal development: alteration of genes associated with cytoskeletal rearrangement and cell signalling pathways. BMC Genomics. 2014;15:48. doi: 10.1186/1471-2164-15-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos A, Bakker AD, Zandieh-Doulabi B, Semeins CM, Klein-Nulend J. Pulsating fluid flow modulates gene expression of proteins involved in Wnt signaling pathways in osteocytes. J. Orthop. Res. 2009;27:1280–1287. doi: 10.1002/jor.20888. [DOI] [PubMed] [Google Scholar]
- Schaffler MB, Cheung W-Y, Majeska R, Kennedy O. Osteocytes: master orchestrators of bone. Calcif. Tissue Int. 2014;94:5–24. doi: 10.1007/s00223-013-9790-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toupadakis CA, Wong A, Genetos DC, Chung D-J, Murugesh D, Anderson MJ, Loots GG, Christiansen BA, Kapatkin AS, Yellowley CE. Long-term administration of AMD3100, an antagonist of SDF-1/CXCR4 signaling, alters fracture repair. J. Orthop. Res. 2012;30:1853–1859. doi: 10.1002/jor.22145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman E, Webster D, Kuhn G, Attar-Namdar M, Müller R, Bab I. Differential load-regulated global gene expression in mouse trabecular osteocytes. Bone. 2013;53:14–23. doi: 10.1016/j.bone.2012.11.017. [DOI] [PubMed] [Google Scholar]
- Waters KM, Jacobs JM, Gritsenko MA, Karin NJ. Regulation of gene expression and subcellular protein distribution in MLO-Y4 osteocytic cells by lysophosphatidic acid: Relevance to dendrite outgrowth. Bone. 2011;48:1328–1335. doi: 10.1016/j.bone.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinbaum S, Cowin SC, Zeng Y. A model for the excitation of osteocytes by mechanical loading-induced bone fluid shear stresses. J Biomech. 1994;27:339–60. doi: 10.1016/0021-9290(94)90010-8. [DOI] [PubMed] [Google Scholar]
- Woo SM, Rosser J, Dusevich V, Kalajzic I, Bonewald LF. Cell line IDG-SW3 replicates osteoblast-to-late-osteocyte differentiation in vitro and accelerates bone formation in vivo. Journal of Bone and Mineral Research. 2011;26:2634–2646. doi: 10.1002/jbmr.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Liu T, Qian W-J, Petyuk VA, Smith RD. Liquid Chromatography-Mass Spectrometry-based Quantitative Proteomics. J. Biol. Chem. 2011;286:25443–25449. doi: 10.1074/jbc.R110.199703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing W, Baylink D, Kesavan C, Hu Y, Kapoor S, Chadwick RB, Mohan S. Global gene expression analysis in the bones reveals involvement of several novel genes and pathways in mediating an anabolic response of mechanical loading in mice. Journal of Cellular Biochemistry. 2005;96:1049–1060. doi: 10.1002/jcb.20606. [DOI] [PubMed] [Google Scholar]
- Xiong J, O'Brien CA. Osteocyte RANKL: New insights into the control of bone remodeling. Journal of Bone and Mineral Research. 2012;27:499–505. doi: 10.1002/jbmr.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K-A, Cho H-S, Shin H-I, Cho J-Y. Differential regulation of CXCL5 by FGF2 in osteoblastic and endothelial niche cells supports hematopoietic stem cell migration. Stem Cells Dev. 2012;21:3391–3402. doi: 10.1089/scd.2012.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W-B, Wang L. Label-free quantitative proteome analysis of skeletal tissues under mechanical load. Journal of Cellular Biochemistry. 2009;108:600–611. doi: 10.1002/jcb.22291. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen Q. Changes of Matrilin Forms during Endochondral Ossification MOLECULAR BASIS OF OLIGOMERIC ASSEMBLY. J. Biol. Chem. 2000;275:32628–32634. doi: 10.1074/jbc.M002594200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.