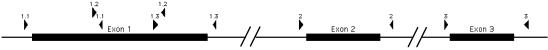Abstract
Peptide mass-signature genotyping (PMSG) is a scanning genotyping method that identifies mutations and polymorphisms by translating the sequence of interest in more than one reading frame and measuring the masses of the resulting peptides by mass spectrometry. PMSG was applied to the RDS/peripherin gene of 16 individuals from a family exhibiting autosomal dominant macular degeneration. The method revealed an A→T transversion in the 5′ splice site of intron 2 that is the likely cause of the disease. It also revealed four different minihaplotypes in exon 3 that represent particular combinations of SNPs at four different locations. This study demonstrates the utility of PMSG for identifying and characterizing point mutations and local minihaplotypes that are not readily analyzed by other approaches.
Peptide mass signature genotyping (PMSG) takes advantage of the diversity of the peptides encoded by a DNA molecule to identify and characterize genetic variants. The DNA region of interest is translated in more than one reading frame to generate a set of peptide analytes whose individual masses are measured by matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS). If the sequence is homogeneous (e.g., from a homozygous individual of known sequence), analytes with mass values predicted by the sequence will be observed in the mass spectrum. These masses, in combination, constitute a “peptide mass signature” characteristic of the sequence. Variant sequences generally yield different peptide mass signatures. Once the peptide mass signature of a mutation or polymorphism has been established, it can serve to identify that variant in subsequent analyses. When two different alleles are present in the same sample (e.g., when the sample is from a heterozygous individual), a combined mass signature will be observed that can be resolved into its two component signatures and thereby reveal the genotype.
PMSG is a sequence scanning method that can detect variation anywhere in the test sequence. In this regard it is comparable to such methods as SSCP, DGGE, or DHPLC. Although these methods report that variation is present, they do not generally distinguish one variant from another; in contrast, PMSG provides a specific signature characteristic of, and often unique to, each particular variant. In addition, PMSG can distinguish particular minihaplotypes that can exist when variation is present at more than one position in the sequence of interest. This sets it apart from dideoxy sequencing, which is incapable of recognizing the cis–trans relationships among multiple sequence variants in a mixed sample.
In the study described here, PMSG was applied to the three-exon human RDS/peripherin gene that encodes a protein found in the rims of the membrane disks in the outer segments of the photoreceptor cells of the retina (Boesze-Battaglia and Goldberg 2002). RDS/peripherin mutations have been implicated in a variety of retinal genetic diseases including retinitis pigmentosa, retinal dystrophy, and macular degeneration (http://www.retina-international.org/sci-news/rdsmut.htm). The family analyzed in this communication was initially identified from a larger set of age-related maculopathy (ARM) families that were subjected to linkage analyses using a genome-wide scan of microsatellite markers (Weeks et al. 2000). Unlike most of the ARM families included in the genome-wide scan, this family was of sufficient size to provide preliminary evidence of linkage to a particular chromosome, in this case chromosome 6. Initial screening of the family by SSCP of amplicons derived from the RDS/peripherin gene, known to be on chromosome 6, failed to identify any potential disease-related variants. However, this was not conclusive evidence because some mutations are hard or impossible to detect using routine SSCP procedures. The possibility of relevant mutation in the RDS/peripherin gene remained; consequently, we decided to analyze the gene again using the PMSG approach.
A PMSG test for variation in each RDS/peripherin exon and its associated splice sites was developed and applied to the 16 members of the ARM family for whom genomic DNA was available. The analysis revealed variation among the family members in exon 2 and in exon 3. For exon 2, three distinct peptide mass signatures were observed, indicating that three different exon 2 alleles were segregating. One signature (subsequently confirmed by dideoxy sequencing) represented an A T transversion at the +3 position of intron 2 and was present in each affected individual and absent in every unaffected individual. Because this mutation is expected to inactivate the 5′ splice site of intron 2, thereby yielding an aberrant gene product, it is the likely cause of the dominant disease phenotype. For exon 3, four distinct peptide mass signatures were observed, indicating that four different exon 3 alleles were segregating in the pedigree. Dideoxy sequencing from the relevant diploid templates showed the presence of SNPs at four different positions within the sequence. Comparison of the observed peptide mass signatures with the signatures predicted for the 16 possible minihaplotypes showed four matches, thereby revealing the molecular nature of the four minihaplotypes and also revealing the diploid genotype of each individual.
RESULTS
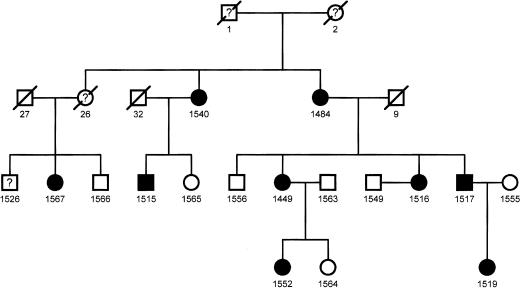
The pedigree of the family of interest is shown in Figure 1. Figure 2 shows the structure of the RDS/peripherin gene and the locations of the nested PCR primers used to amplify the sequences that were expressed as peptides and analyzed by PMSG. Exon 1 is a relatively large exon and was amplified in three overlapping segments; exons 2 and 3 were amplified in a single amplicon each. The amplified portions extended into the flanking intron sequences so that splice site variation could be detected. The amplicons were ligated into an expression vector as described in Methods and transformed into Escherichia coli. The transformed cells were grown on selective media and induced en masse to generate epitope tagged peptides. Two peptides were generated from each region, one encoded in the natural reading frame and the other encoded in a reverse reading frame. The peptides were purified using an affinity tag contributed by the vector sequence and their masses were measured by MALDI-TOF mass spectrometry and compared with the masses of equivalent peptides encoded by the reference sequence.
Figure 1.
This pedigree indicates the affected (black symbols) and unaffected (white symbols) family members (males, boxes; females, circles). Those with four-digit identifiers were included in the initial linkage analysis as well as in the PMSG and dHPLC studies (except 1555). The numbers correspond to the identifiers used in the text, tables, and figures in the present report.
Figure 2.
Locations of primers used for PMSG analysis of the RDS/peripherin gene. The 26-kb gene is represented with the exons to scale but not the introns (intron 1, 17 kb; intron 2, 5.8 kb).
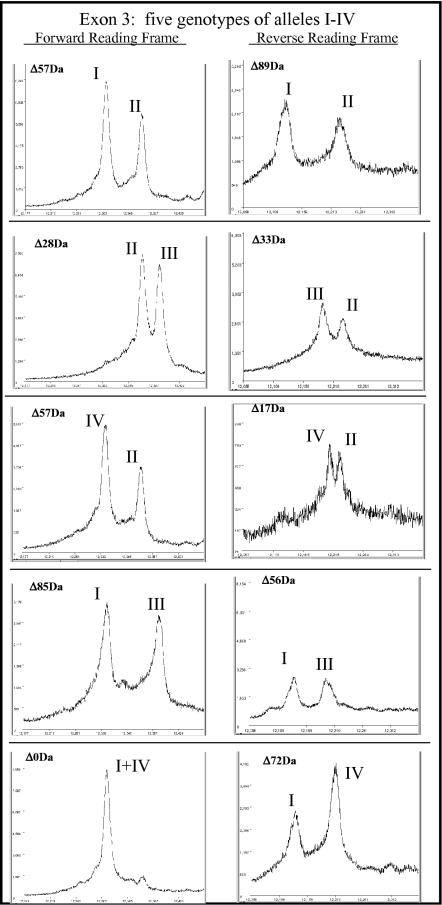
Each of the 16 samples showed a pair of peptide mass signatures for exon 3, indicating that each individual was heterozygous within that region. The individuals fell into five classes with respect to the positions of the peaks in the respective mass spectra, indicating that (at least) five diploid genotypes were represented. Representative mass spectra for the five classes are shown in Figure 3.
Figure 3.
MALDI-TOF mass spectra showing the five genotypes observed in the family. There were four alleles observed in different combinations, as indicated. The allele designations are described in Table 3.
Dideoxy sequencing of all 16 exon 3 amplicons revealed single nucleotide variation (SNPs) at four positions in the sequence. With reference to the mRNA sequence, the four SNPs were C/G at nt 1147, G/A at nt 1166, A/G at nt 1250, and C/T at nt 1291. The pattern of heterozygosity at the four positions in exon 3 for each of the 16 individuals is shown in Table 1. The first three SNPs had been observed previously and correspond to Q/E304, R/K310, and D/G338 in the C-terminal portion of the protein (Jordan et al. 1992). The previously unreported polymorphism at position 1291 is 13 nt downstream of the termination codon and thus does not affect the protein sequence.
Table 1.
Dideoxy Sequencing Results for Variant Positions in Exon 3
| Patient # | 1147 | 1166 | 1250 | 1291 |
|---|---|---|---|---|
| 1540 | C | A/G | A | C |
| 1484 | C | A/G | A | C |
| 1526 | G/C | A | A/G | C/T |
| 1567 | G/C | A | A/G | C |
| 1566 | G/C | A | A/G | C |
| 1515 | G/C | A | A/G | C |
| 1565 | G/C | A/G | A/G | C |
| 1556 | G/C | A/G | A/G | C |
| 1449 | G/C | A | A/G | C/T |
| 1563 | G/C | A | A/G | C |
| 1549 | G | A | G | C/T |
| 1516 | G/C | A | A/G | C/T |
| 1517 | G/C | A | A/G | C/T |
| 1552 | G/C | A | A/G | C |
| 1564 | G | A | G | C/T |
| 1519 | G/C | A | A/G | C/T |
The observed peptide masses were compared with the predicted masses for all 16 possible combinations of the exon 3 SNPs (Table 2). There were four matches, indicating that the four minihaplotypes are G1147A1166G1250C1291, C1147A1166A1250C1291, C1147G1166A1250C1291, and G1147A1166G1250T1291. Each of these minihaplotypes represents a different exon 3 sequence, that is, a different minihaplotype. For convenience, we have designated the minihaplotypes I, II, III, and IV, respectively. The G1147A1166G1250C1291 minihaplotype corresponds to the published mRNA sequence (GenBank NM_000322) and the C1147G1166A1250C1291 minihaplotype corresponds to the RDS/peripherin genomic sequence (GenBank AL049843). Table 3 shows the exon 3 genotype of each of the 16 individuals analyzed.
Table 2.
PMSG Mass Predictions Generated From the DNA Sequencing Results Showing Two Possible Bases at Four Positions in the DNA Being Analyzed
|
Base present at SNP locations
|
Predicted masses
|
|||||
|---|---|---|---|---|---|---|
| 1147 | 1166 | 1250 | 1291 | Forward | Reverse | Allele |
| G | A | G | C | 12311.5 | 12137.3 | I |
| G | A | G | T | 12311.5 | 12209.3 | IV |
| G | A | A | C | 12369.5 | 12127.2 | |
| G | A | A | T | 12369.5 | 12199.3 | |
| G | G | G | C | 12339.5 | 12103.2 | |
| G | G | G | T | 12339.5 | 12175.3 | |
| G | G | A | C | 12397.5 | 12093.2 | |
| G | G | A | T | 12397.5 | 12165.3 | |
| C | A | G | C | 12310.5 | 12236.4 | |
| C | A | G | T | 12310.5 | 12308.5 | |
| C | A | A | C | 12368.5 | 12226.4 | II |
| C | A | A | T | 12368.5 | 12298.4 | |
| C | G | G | C | 12338.5 | 12202.4 | |
| C | G | G | T | 12338.5 | 12274.4 | |
| C | G | A | C | 12396.5 | 12192.3 | III |
| C | G | A | T | 12396.5 | 12264.4 | |
The masses are shown for the natural forward reading frame and the reverse reading frame used for PMSG analysis of exon 3. The final column shows the alleles that were identified in this study
Table 3.
Summary of Exon 3 Patient Genotypes
| Allele | Patient |
|---|---|
| I, II | 1567, 1566, 1552, 1560, 1515 |
| II, III | 1540, 1484 |
| II, IV | 1526, 1449, 1517, 1516, 1519 |
| I, III | 1565, 1556 |
| I, IV | 1549, 1564 |
With each peptide mass signature assigned to a particular minihaplotype, we were able to assign diploid genotypes to all 16 individuals (Table 3). Based on the pedigree data, none of the exon 3 minihaplotypes appears to be responsible for retinal disease but the minihaplotype C1147A1166A1250C1291 was linked to the mutation in exon 2 described below.
PMSG analysis of exon 2 revealed three mass signatures, and thus three different sequence variants, in the exon 2 segment. All of the seven unaffected individuals showed a single signature corresponding to the peptide masses predicted by the published wild-type sequence (natural forward frame 15205.1 daltons and reverse 10647.7 daltons). An additional heterozygous signature was observed in each of the nine affected individuals (natural forward frame 15205.1 daltons and reverse 10598.6 daltons). Dideoxy sequencing showed that each was heterozygous for an A-to-T transversion at position 35793 in the RDS/peripherin gene sequence (GenBank AL049843), a mutation that predicted the observed mass signature. This substitution (IVS2 + 3A T) is in the 5′ splice site of intron 2 and was reported previously in an independent pedigree (Sullivan et al. 1996). DHPLC analysis confirmed that each affected individual was heterozygous in the region of interest, whereas no heterozygosity was detected in the region in the seven unaffected family members and eight unaffected controls. A third mass signature was observed in a single affected individual (1519). This individual also showed the mass signature characteristic of the IVS2 + 3A T mutation, indicating that the variation responsible for the new signature was in trans to the IVS2 + 3A T mutation. Dideoxy sequencing revealed a C-to-T substitution at position 864 in the mRNA sequence. This mutation converts one valine codon to another (V/V209) and is thus silent at the protein level.
DISCUSSION
Advantages of PMSG Over DNA Mass Analysis
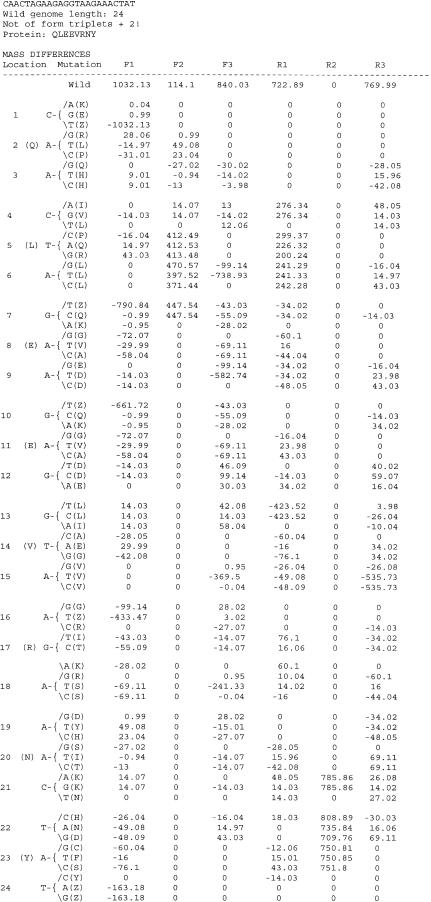
For short DNA amplicons, sequence variation can be detected by MALDI-TOF mass spectrometric analysis of the DNA itself. This approach, however, is not practical for sequences longer than ∼100 nt (nucleotides are >300 daltons each and a 9-dalton resolution is required to distinguish an A from a T), nor is it able to indicate the location of a nucleotide substitution if one is detected. Consider, for example, the two DNA molecules shown in Figure 4. Detection of a DNA mass difference of 9 daltons (7891–7882) would indicate that the upper sequence differs from the lower one by an A-to-T transversion. There are 13 A's in the sequence; however, the mass data carries no information about which A it is. In contrast, the peptide mass difference of 69 daltons (1032-963) indicates that the peptides differ by an arginine-to-serine substitution. Of all possible base substitutions in the parent DNA, only one, an A-to-T transversion at position 18, can give such a result. Thus the analysis reveals the exact nature of the mutation at the DNA and protein levels. Such an exact inference cannot be made in every case, of course, but in most cases the location of the mutation is narrowed down to one or a few possibilities.
Figure 4.
(A) Two DNA sequences that differ by a single A T base substitution and the mass in daltons of the two sequences showing a difference of 9.01 daltons. (B) The peptides that result when the DNAs shown in A are transcribed and translated; the difference in mass is 69.11 daltons.
PMSG in a Single Reading Frame
The utility of detecting mutation by analyzing peptides translated in their natural reading frame from a test sequence was first shown by Garvin et al. (2000), who used the approach to detect mutations 1129insA and C61G of the human BRCA1 gene. As these authors recognized, the approach does more than indicate that variation is present; information about the molecular nature of the variation is present in the sign and magnitude of the mass shift. This can be seen by inspection of Table 4, which shows each of the amino acid substitutions that can result from single nucleotide substitutions in a coding sequence.
Table 4.
Mass Differences for All Amino Acid Substitutions Resulting From Single Nucleotide Transitions or Transversions
| Ile-Leu | 0.00 | Leu-Met | 18.03 | Pro-His | 40.02 |
| Gln-Lys | 0.04 | Ile-Met | 18.03 | Gly-Val | 42.08 |
| Lys-Glu | 0.94 | His-Arg | 19.05 | Ala-Asp | 43.02 |
| Ile-Asn | 0.95 | Asp-His | 22.05 | Ile-Arg | 43.03 |
| Gln-Glu | 0.98 | Asn-His | 23.04 | Leu-Arg | 43.03 |
| Asn-Asp | 0.98 | Leu-His | 23.98 | Cys-Phe | 44.04 |
| Lys-Met | 3.02 | Met-Arg | 25.00 | Gly-Cys | 46.09 |
| Pro-Thr | 3.99 | Pro-Ala | 26.04 | Val-Phe | 48.05 |
| Gln-His | 9.01 | His-Tyr | 26.04 | Asp-Tyr | 48.09 |
| Ser-Pro | 10.04 | Ser-Ile | 26.08 | Asn-Tyr | 49.08 |
| Ile-Thr | 12.05 | Ser-Leu | 26.08 | Cys-Arg | 53.05 |
| Thr-Asn | 12.99 | Ser-Asn | 27.02 | Thr-Arg | 55.08 |
| Asp-Glu | 14.03 | Thr-Lys | 27.06 | Ala-Glu | 58.04 |
| Gly-Ala | 14.03 | Lys-Arg | 28.02 | Gly-Asp | 58.04 |
| Ser-Thr | 14.03 | Ala-Val | 28.05 | Pro-Arg | 59.07 |
| Ile-Val | 14.03 | Gln-Arg | 28.06 | Cys-Tyr | 60.04 |
| Leu-Val | 14.03 | Val-Glu | 29.99 | Ser-Phe | 60.10 |
| Asn-Lys | 14.07 | Trp-Arg | 30.02 | Ser-Arg | 69.11 |
| Leu-Gln | 14.97 | Ala-Thr | 30.03 | Gly-Glu | 72.07 |
| Ile-Lys | 15.01 | Gly-Ser | 30.03 | Leu-Trp | 73.05 |
| Val-Asp | 15.96 | Thr-Met | 30.08 | Ser-Tyr | 76.10 |
| Phe-Tyr | 16.00 | Pro-Gln | 31.01 | Cys-Trp | 83.07 |
| Ala-Ser | 16.00 | Val-Met | 32.06 | Ser-Trp | 99.13 |
| Pro-Leu | 16.04 | Ile-Phe | 34.02 | Gly-Arg | 99.14 |
| Ser-Cys | 16.06 | Leu-Phe | 34.02 | Gly-Trp | 129.16 |
Residue masses from http://www.bmrb.wisc.edu/ref_info/aadata.dat
Most of the mass differences listed in Table 4 are readily detected by MALDI-TOF MS analysis of the 7- to 15-kD peptides that are produced when an exon of typical size (200–300 nt) is expressed as described here. There are, however, some notable and significant exceptions. One substitution (leucine isoleucine) creates no mass shift at all, and another (lysine glutamine) creates a shift so small as to be effectively undetectable. Four additional substitutions change the mass by only about 1 dalton, and four more change it by between 1 and 10 daltons. Reliable detection of these mass shifts cannot be expected with current MALDI technology. The result will be the failure to detect about 15% of all single nucleotide substitutions that result in amino acid substitutions. Mutations that result in synonymous codon substitutions will also go undetected. This is a minor issue for coding sequence DNA, because such mutations are almost always phenotypically silent, but it is of concern with respect to the 5′ and 3′ splice sites, where certain substitutions that inactivate splice site function will also go undetected.
PMSG in Multiple Reading Frames
The limitations of single reading frame mass signature analysis are significantly overcome by collecting data from peptides encoded in more than one reading frame. The choice of which alternative reading frames to use is constrained by the available open reading frames. The number of stop codons in noncoding frames varies from sequence to sequence but it is usually possible to combine an alternate forward or reverse frame to achieve coverage of the entire sequence in additional reading frames. In the future, the addition of stop codon suppression capabilities to the system will maximize the information content of each peptide and make test configurations simpler. Currently, additional reading frames for tests are chosen with the aid of computer programs that predict the detectability of all known and possible single nucleotide substitutions in the sequence. This is illustrated in Figure 5 for a 24-nt sequence CAACTAGAAGAGGTAAGAAACTAT. The figure shows the masses of the peptides encoded in each reading frame of the sequence and the predicted peptide mass differences for each reading frame for all 72 possible single nucleotide substitutions. Note that many substitutions are missed in a single reading frame analysis, but when mass data from multiple reading frames are combined, almost every substitution is revealed. Note also that the Figure 5 data comprise a lookup table whereby a set of observed mass shifts can be assigned to one or a few particular nucleotide substitutions.
Figure 5.
Output of the program used to analyze all possible single base substitutions for a 24mer. F1, F2, and F3 are forward reading frames and R1, R2, and R3 are reverse frames. The change in mass resulting from the indicated substitution is shown for each of the reading frames and the number of substitutions that could not be detected for the stated thresholds are shown.
Table 5 lists the mutations that are not detected for different reading frame combinations at a variety of mass-difference detection thresholds. Note that, for several reading frame pairs, almost every nonsynonymous substitution is detected, and that, for the two reading frame trios shown, all mutations are detected, even at a conservative detection threshold value of 12 daltons.
Table 5.
Summary of the Effect of Multiple Reading Frames and Increased Resolution on Detection of Single Nucleotide Substitutions
|
Detection resolution
|
|||
|---|---|---|---|
| Reading frame | 12 Da | 5 Da | 0.5 Da |
| RF1 | 1. (Q) C-A(K) | 1. (Q) C-A (K) | 1. (Q) C-A (K) |
| 1. (Q) C-G (E) | 1. (Q) C-G (E) | 3. (Q) A-G (Q) | |
| 3. (Q) A-G (Q) | 3. (Q) A-G (Q) | 4. (L) C-A (I) | |
| 3. (Q) A-T (H) | 4. (L) C-A (I) | 4. (L) C-T (L) | |
| 3. (Q) A-C (H) | 4. (L) C-T (L) | 6. (L) A-G (L) | |
| 4. (L) C-A (I) | 6. (L) A-G (L) | 6. (L) A-T (L) | |
| 4. (L) C-T (L) | 6. (L) A-T (L) | 6. (L) A-C (L) | |
| 6. (L) A-G (L) | 6. (L) A-C (L) | 9. (E) A-G (E) | |
| 6. (L) A-T (L) | 7. (E) G-C (Q) | 12. (E) G-A (E) | |
| 6. (L) A-C (L) | 7. (E) G-A (K) | 15. (V) A-G (V) | |
| 7. (E) G-C (Q) | 9. (E) A-G (E) | 15. (V) A-T (V) | |
| 7. (E) G-A (K) | 10. (E) G-C (Q) | 15. (V) A-C (V) | |
| 9. (E) A-G (E) | 10. (E) G-A (K) | 16. (R) A-C (R) | |
| 10. (E) G-C (Q) | 12. (E) G-A (E) | 18. (R) A-G (R) | |
| 10. (E) G-A (K) | 15. (V) A-G (V) | 21. (N) C-T (N) | |
| 12. (E) G-A (E) | 15. (V) A-T (V) | 24. (Y) T-C (Y) | |
| 15. (V) A-G (V) | 15. (V) A-C (V) | ||
| 15. (V) A-T (V) | 16. (R) A-C (R) | ||
| 15. (V) A-C (V) | 18. (R) A-G (R) | ||
| 16. (R) A-C (R) | 19. (N) A-G (D) | ||
| 18. (R) A-G (R) | 20. (N) A-T (I) | ||
| 19. (N) A-G (D) | 21. (N) C-T (N) | ||
| 20. (N) A-T (I) | 24. (Y) T-C (Y) | ||
| 21. (N) C-T (N) | |||
| 24. (Y) T-C (Y) | |||
| RF1 | 1. (Q) C-A (K)* | 1. (Q) C-A (K)* | 1. (Q) C-A (K)* |
| 1. (Q) C-G (E)* | 1. (Q) C-G (E)* | 6. (L) A-C (L) | |
| RF3 | 3. (Q) A-C (H) | 6. (L) A-C (L) | 15. (V) A-C (V) |
| 6. (L) A-C (L) | 15. (V) A-G (V) | 21. (N) C-T (N) | |
| 15. (V) A-G (V) | 15. (V) A-C (V) | 24. (Y) T-C (Y)* | |
| 15. (V) A-C (V) | 18. (R) A-G (R) | ||
| 18. (R) A-G (R) | 21. (N) C-T (N) | ||
| 21. (N) C-T (N) | 24. (Y) T-C (Y)* | ||
| 24. (Y) T-C (Y)* | |||
| RF1 | 1. (Q) C-A (K) | 1. (Q) C-A (K) | 1. (Q) C-A (K) |
| 1. (Q) C-G (E) | 1. (Q) C-G (E) | 3. (Q) A-G (Q) | |
| RF4 | 3. (Q) A-G (Q) | 3. (Q) A-G (Q) | 4. (L) C-T (L) |
| 3. (Q) A-C (H) | 4. (L) C-T (L) | 16. (R) A-C (R) | |
| 3. (Q) A-T (H) | 7. (E) G-A (K) | ||
| 4. (L) C-T (L) | 10. (E) G-C (Q) | ||
| 7. (E) G-A (K) | 10. (E) G-A (K) | ||
| 10. (E) G-C (Q) | 16. (R) A-C (R) | ||
| 10. (E) G-A (K) | 19. (N) A-G (D) | ||
| 16. (R) A-C (R) | |||
| 18. (R) A-G (R) | |||
| 19. (N) A-G (D) | |||
| RF1 | 1. (Q) C-A (K)* | 1. (Q) C-A (K)* | 1. (Q) C-A (K)* |
| 1. (Q) C-G (E)* | 1. (Q) C-G (E)* | 24. (Y) T-C (Y)* | |
| RF6 | 7. (E) G-A (K) | 7. (E) G-A (K) | |
| 24. (Y) T-C (Y)* | 24. (Y) T-C (Y)* | ||
| RF1 | 1. (Q) C-A (K)* | 1. (Q) C-A (K)* | 1. (Q) C-A (K)* |
| 1. (Q) C-G (E) | 1. (Q) C-G (E)* | ||
| RF3 | 3. (Q) A-C (H)* | ||
| RF4 | 18. (R) A-G (R) | ||
| RF1 | 1. (Q) C-A (K)* | 1. (Q) C-A (K)* | 1. (Q) C-A (K)* |
| RF3 | 1. (Q) C-G (E)* | 1. (Q) C-G (E)* | 24. (Y) T-C (Y)* |
| RF6 | 24. (Y) T-C (Y)* | 24. (Y) T-C (Y)* | |
The 24-nt sequence CAACTAGAAGAGGTAAGAAACTAT was analyzed for all possible single nucleotide substitutions. The output shows the nucleotide position, amino acid and nucleotide change, and detection at three different resolution values
Cases where mutation was not detected because the substitution is in a terminal codon and the other reading frame(s) do not penetrate the codon. Analysis of the same mutations when the 24-nt test sequence was internal to a larger polypeptide showed that they were generally detected.
A similar analysis was performed for the RDS/peripherin sequences whose PMSG analysis is described here. Space constraints prevent showing the complete output of the analysis programs, as in Figure 5 and Table 5. The expectations for the exon 2 and 3 sequences are shown in abbreviated form in Table 6. In summary, with three reading frames analyzed, we expect to miss only about 4% and 2.5% of all nonsynonymous mutations in the exon 2 and exon 3 sequences, respectively.
Table 6.
Effect of Using Multiple Reading Frames in PMSG Analysis to Detect All Possible Single Nucleotide Substitutions in Exons 2 and 3 of the RDS/Peripherin Gene
|
12-Da detection threshold
|
5-Da detection threshold
|
||||||
|---|---|---|---|---|---|---|---|
| Total | Nonsynonymous | Total | Nonsynonymous | ||||
| A. Predicted number of substitutions not detected in 252-nt exon 2 sequence (756 possibilities) | |||||||
| RF1 | 231 (30.5%) | 65 (8.6%) | 214 (28.3%) | 52 (6.9%) | |||
| RF1, RF4 | 151 (20.0%) | 54 (7.1%) | 141 (18.6%) | 39 (5.2%) | |||
| RF1, RF2 | 88 (11.6%) | 42 (5.6%) | 80 (10.5%) | 34 (4.5%) | |||
| RF1, RF2, RF4 | 52 (6.9%) | 31 (4.1%) | 44 (5.8%) | 26 (3.4%) | |||
| B. Predicted number of substitutions not detected in 213-nt exon 3 sequence (639 possibilities) | |||||||
| RF1 | 225 (35.2%) | 76 (11.9%) | 213 (33.3%) | 67 (10.5%) | |||
| RF1, RF3 | 152 (23.8%) | 55 (8.6%) | 142 (22.2%) | 44 (6.9%) | |||
| RF1, RF6 | 31 (4.9%) | 21 (3.3%) | 19 (3.0%) | 12 (1.9%) | |||
| RF1, RF3, RF6 | 18 (2.8%) | 16 (2.5%) | 9 (1.4%) | 7 (1.1%) | |||
Not all mutations or sequence variants of interest are single nucleotide substitutions of course, and very nearly all of these (e.g., base deletions, insertions, or two-base substitutions) will create readily detectable mass shifts, and distinctive mass signatures, in the PMSG analysis. Thus we are comfortable in treating the percentages given in Table 6 as upper limits of undetectable mutations—and a measure of the true sensitivity of the PMSG protocols described here.
Minihaplotype Detection and Analysis
In addition to detecting sequence variants, PMSG has the unique ability to detect individual minihaplotypes within the DNA region of interest. Traditional sequencing approaches, in contrast, do not provide minihaplotype information because the sequencing chromatograms from a mixed template that varies at two or more sites do not reveal the cis–trans relationships among the two (or more) SNPs that may be present. To obtain such minihaplotype information by sequencing, it is necessary to clone individual (haploid) molecules and sequence a statistically significant number of them, a laborious and uncertain undertaking. In contrast, minihaplotype information is directly revealed in the peptide mass signature of the diploid because each sequence is translated processively to yield a single peptide species with a mass characteristic of the template nucleic acid. This is amply illustrated by the PMSG analysis of exon 3 described here, in which four distinct minihaplotypes, and five distinct diploid genotypes, were detected in the 16 individuals analyzed.
It is clear that none of the exon 3 minihaplotypes we observed here is causative of dominant retinal disease, al though one of them, C1147A1166A1250C1291, appears to be in linkage disequilibrium with the causative mutation in intron 2. Three of the four exon 3 SNPs (those in the coding region) that vary among the minihaplotypes were observed in an earlier study (Jordan et al. 1992) in which, as in the present analysis, there was no evidence of a deleterious phenotype for any of them. That study also revealed that all three SNPs were very frequent in the human population, each variant appearing in about half of the chromosomes in a set of 160 unaffected individuals. It did not reveal the four minihaplotypes found in the present analysis, however, because the sequencing, SSCP, and ASO methodologies used were unable to distinguish whether the variant nucleotides were in cis or in trans to one another.
The IVS2 + 3A T transversion is a prime candidate to be causative of the macular degeneration phenotype in the family examined here. Adenine is commonly found at the +3 position in 5′ splice sites, and thymine is almost never present at that position (Zhang 1998). The T allele is thus expected to functionally inactivate the splice site and yield a grossly aberrant gene product, an expectation consistent with the dominant phenotype observed in the affected individuals. The conclusion that the T at the IVS2 + 3 position is responsible for the disease phenotype is reinforced by the previous observation of the same mutation in affected individuals in an independent autosomal dominant macular degeneration pedigree (Sullivan et al. 1996). An alternative technique, DHPLC, also showed that variant exon 2 sequences were present in the affected individuals. It could not, however, offer any information about the specific molecular changes that were present, and it did not detect an additional polymorphism in one sample.
The study reported here demonstrates the efficacy of the PMSG process for identifying and scoring variation in genomic DNA, including complex minihaplotypes extending over several hundred nucleotides of sequence. Direct sequencing, in contrast, does not reveal such minihaplotype information because the sequencing chromatograms cannot be deconvolved to reveal the cis–trans relationships among multiple variants in the same sequenced region. PMSG may thus be a method of choice for analyzing DNA regions where local minihaplotype information is of interest.
In the process used here, the DNA sequences of interest were incorporated into specially designed expression vectors and expressed in E. coli in a 96-well format. Cost per test is low, and the analytical steps are very fast, but the entire test process takes at least 24 h because of the cell growth step. Most of these hours could be saved by expressing the peptides in vitro using cell-free transcription and translation (Garvin et al. 2000). Technical problems have prevented us from doing this successfully to date, but once these problems are overcome, PMSG has the promise to become not only accurate and inexpensive, but also very rapid.
METHODS
Samples
DNA samples were provided from the Family Studies Center for Hereditary Eye Diseases in the Department of Ophthalmology at the University of Pittsburgh. All of these individuals were either affected or members of families of individuals with ARM or macular dystrophies. All recruitment methods, data collection, and storage methods were conducted under protocols approved by the University of Pittsburgh Institutional Review Board. Initially, the PMSG method was established with members of a family with a known RDS/peripherin mutation (Gorin et al. 1995). Sixteen additional samples were from members of a single kindred exhibiting a variant of ARM and preliminary evidence of linkage to chromosome 6 from a previous 10-cM microsatellite genome-wide scan; however, the disease-causing gene and gene variant were unknown (Weeks et al. 2000). This family had been previously screened via SSCP of the RDS/peripherin exons, and no disease-causing variants were revealed. All individuals provided informed consent in accordance with protocols approved by the University of Pittsburgh Institutional Review Board (IRB protocol #960127).
DNA Extraction
Genomic DNA was extracted from leukocytes obtained from whole blood and collected into EDTA vacutainer tubes using a simple salting-out procedure, as described previously (Miller et. al. 1988).
PCR
A nested PCR strategy was used to amplify each exon and flanking introns of the RDS/peripherin gene (all primers from Sigma-Genosys). The primers for the first round were as follows: EX1rds5 5′ TCTGGGCTCGTTAAGGTTTG3′, EX1rds3 5′GAGCCTCAGTGTCCCCAATA3′, EX2rds5 5′AGTGGCCCCTGTTGAGAAG3′, EX2rds3 5′GAGGCATGCTCTCCAAGC3′, EX3rds5 5′CCAGCGATTCTCCCAGATT3′, EX3rds3 5′GAGTTGGATGAGGGGGAGAT3′.
Two pairs of nested primers were used, one pair for the natural reading frame and one pair for the reverse reading frame. The primers contain a clamp at the 5′ end followed by an SfiI site. The SfiI recognition sequence, GGCCNNNN/NGGCC, allows for directional cloning using only one enzyme to generate two distinct 3′ overhangs. The primers for the natural reading frame of exon 2 were EX2rdsSfi5b 5′[TATATAGGCCTTTGTGGC C]CCAGCTGTCTGTTTCC3 ′ and EX2rdsSfi3c 5 ′ [TATATAGGCCTCTTTGGCC]AGGCTCTCCTTACCC3′ and for the reverse EX2rdsSfi5D 5′[TATATAGGCCTTTGTGGCC]AGGCTCTCCTTACCC3′ and EX2rdsSfi3D 5′[TATATAGGCCTCTTTGGCC]CCAGCTGTCTGTTTCC3′.
Primers for the natural reading frame of exon 3 were EX3rdsSfi5b 5′ [TATATAGGCCTTTGTGGCC]CTCCTCTCCCACCA3′ and EX3rdsSfi3b 5′ [TATATAGGCCTCTTTGGCC]GGAGTGCACTATTTCTCA 3′ and for the reverse reading frame EX3rdsSfi5F 5′ [TATATAGGCCTTTGTGGCC]GAGTGCACTATTTCTCA 3′ and EX3rdsSfi3F 5′ [TATATAGGCCTCTTTGGCC]TCTCCTCTCCCACCA 3′.
The DNA of interest was PCR amplified in a 20-μL reaction using Taq polymerase (Eppendorf) in a solution containing 10 mM Tris-Cl, 50 mM KCl (pH 8.3), 1.5 mM Mg2+, 200 mM dNTP's (Promega), and 2% DMSO, and each primer was 0.5 μM. For the first amplification, samples were heated to 94°C for 2 min, Taq was added, and then reactions were cycled 35 times for 10 sec at 94°C; 30 sec at 62.6°C; and 20 sec at 72°C before a final extension of 10 min at 72°C and a 4°C hold. The nested reactions used the same solutions and 1 μL of the first round reaction was denatured at 95°C for 2 min., then cycled 25 times through 94°C, 62.6°C, and 72°C for 30 sec each before a final extension of 5 min at 72°C and a 4°C hold.
Cloning, Transformation, and Expression
Amplicons were digested with SfiI (NEB) at 50°C for 2 h. Geneclean (Qbiogene) was used to purify the DNA prior to cloning into DraIII-digested WZ4 expression vector. WZ4 is a modified pET24d+ plasmid (Novagen). The plasmid carries the lacIq gene and the npt1 gene, which confers resistance to kanamycin. Transcription is controlled by T7 polymerase and transcription is terminated at the T7 termination sequence. The coding sequence for the universal epitope (Nelson et al. 1999) was placed immediately downstream of the initial ATG.
Ligation products were transformed into NovaBlue(DE3) competent cells (Novagen) and outgrown in SOC for 1 h at 37°C with shaking at 300 rpm. Selective TB media was then added and the culture continued overnight. The following morning, fresh media was added and cells were incubated for an additional 3 h before IPTG to 1 mM was added and cultures were incubated for one additional hour.
Purification
E. coli were harvested by centrifuging 750 μL of the culture for 1 min at 4000 rpm and media was aspirated. The cell pellet was resuspended in 100 μL 10 mM Tris buffer (pH 7.5), then 100 μL of lysis buffer (2% SDS and 0.6 mM DTT) was added before boiling samples for 5 min at 100°C. After lysis, 100 μL of 1% Triton X-100 was added to the lysate, then a slurry of 50 μL of Ni-NTA beads (Qiagen) suspended in 30% ethanol was added and incubated with shaking for 10 min. The beads were washed with deionized ultrafiltered water and then with 50% acetonitrile before being eluted with the MALDI matrix solution (0.3% TFA/50% acetonitrile saturated with sinapinic acid).
MALDI-TOF Mass Spectrometry
Samples were spotted onto a 384-spot stainless steel plate (Bruker) before being analyzed by MALDI-TOF using a Bruker Autoflex instrument operated in linear mode.
Sequencing
Products from the first PCR reaction were sequenced by the University of Pittsburgh sequencing facility using EX1rds5, EX2rds5, or EX3rds5 as the sequencing primer.
In order to link the SNP's for sequencing, it was necessary to plate the transformation and pick individual bacterial colonies. Amplicons were generated from these clones using T7promoter 5′ GCGAAATTAATACGACTCACTATAGGG 3′ and T7terminator 5′ GCTAGTTATTGCTCAGCGGTGGC 3′ as PCR primers, and then the T7 terminator was used as the sequencing primer.
DHPLC
A 387-bp fragment containing the coding and flanking regions of exon 2 of the RDS gene was amplified using 12.5 pmole of the forward (5′-GAGAAGCCCGGGAAGCCCATC-3′) and 12.5 pmole of the reverse (5′-GAGGCATGCTCTCCAAGCCTG-′) primers, 12.5 μM of each dNTP (Life Technologies), 1.5 mM MgCl2, 1.13U AmpliTaq Gold DNA polymerase and 1X AmpliTaq Gold buffer (Applied Biosystems), and 0.07U Pfu DNA polymerase (Stratagene). An MJ Research PTC-0200 thermocycler (MJ Research) was used to perform the cycling conditions of 95°C for 10 min, then cycled for 34 cycles at 95°C for 20 sec and 64°C for 1 min, followed by 72°C for 1 min.
The PCR products were sized and quantified under nondenaturing DHPLC using the WAVE instrument (Transgenomic) and an appropriate molecular size standard. The optimal melting temperature for heteroduplex formation was determined to be 64°C using WAVEmaker 4.0 software (Transgenomic Inc). A modified buffer gradient was used that went from 47% to 71% buffer B (0.1M TEAA/25% acetonitrile) and simultaneously 53% to 29% buffer A (0.1M TEAA) with a constant flow rate of 0.9 mL/min.
Acknowledgments
We thank Peter Berget, Byron Ballou, Mark Bier, and Randy Nelson for many helpful discussions of the technology and Hans Moravec, Matt Vieta, and Chris Mason for software support of the project. The work was funded by an NIH Phase I SBIR (GM060876–01) awarded to Sequel Genetics Inc. and grants to M.B.G.: NIH RO1-EY 09859, Research to Prevent Blindness, Inc., The Eye & Ear Foundation of Pittsburgh.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.995103.
References
- Boesze-Battaglia, K. and Goldberg, A.F. 2002. Photoreceptor renewal: A role for peripherin/rds. Int. Rev. Cytol. 217: 183-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin, A.M., Parker, K.C., and Haff, L. 2000. MALDI-TOF based mutation detection using tagged in vitro synthesized peptides. Nat. Biotechnol. 18: 95-97. [DOI] [PubMed] [Google Scholar]
- Gorin, M.B., Jackson, K.E., Ferrell, R.E., Sheffield, V.C., Jacobson, S.G., Gass, J.D., Mitchell, E., and Stone, E.M. 1995. A peripherin/retinal degeneration slow mutation (Pro-210-Arg) associated with macular and peripheral retinal degeneration. Ophthalmology 102: 246-255. [DOI] [PubMed] [Google Scholar]
- Jordan, S.A., Farrar, G.J., Kenna, P., and Humphries, P. 1992. Polymorphic variation within “conserved” sequences at the 3′ end of the human RDS gene which results in amino acid substitutions. Hum. Mutat. 1: 240-247. [DOI] [PubMed] [Google Scholar]
- Miller, S., Dykes, D., and Polesky, H. 1988. A simple salting and procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16: 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, R.W., Jarvik, J.W., Taillon, B.E., and Tubbs, K.A. 1999. BIA/MS of epitope-tagged peptides directly from E. coli lysate: Multiplex detection and protein identification at low-femtomole to subfemtomole levels. Anal. Chem. 71: 2858-2865. [DOI] [PubMed] [Google Scholar]
- Sullivan, L.S., Guilford, S.R., Birch, D.G., and Daiger, S.P. 1996. A novel splice site mutation in the gene for peripherin/RDS causing dominant retinal degeneration. Invest. Ophthalmol. Vis. Sci. S1146.
- Weeks, D.E., Conley, Y.P., Mah, T.S., Paul, T.O., Morse, L., Ngo-Chang, J., Dailey, J.P., Ferrell, R.E., and Gorin, M.B. 2000. A full genome scan for age-related maculopathy. Hum. Mol. Genet. 9: 1329-1349. [DOI] [PubMed] [Google Scholar]
- Zhang, M.Q. 1998. Statistical features of human exons and their flanking regions. Hum. Mol. Genet. 5: 919-932. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.retina-international.org/sci-news/rdsmut.htm; Retina International's Mutation Database.